Abstract
The search for developmental mechanisms driving vertebrate organogenesis has paved the way toward a deeper understanding of birth defects. During embryogenesis, parts of the heart and craniofacial muscles arise from pharyngeal mesoderm (PM) progenitors. Here, we reveal a hierarchical regulatory network of a set of transcription factors expressed in the PM that initiates heart and craniofacial organogenesis. Genetic perturbation of this network in mice resulted in heart and craniofacial muscle defects, revealing robust cross-regulation between its members. We identified Lhx2 as a previously undescribed player during cardiac and pharyngeal muscle development. Lhx2 and Tcf21 genetically interact with Tbx1, the major determinant in the etiology of DiGeorge/velo-cardio-facial/22q11.2 deletion syndrome. Furthermore, knockout of these genes in the mouse recapitulates specific cardiac features of this syndrome. We suggest that PM-derived cardiogenesis and myogenesis are network properties rather than properties specific to individual PM members. These findings shed new light on the developmental underpinnings of congenital defects.
Embryonic development encompasses an orchestrated series of cellular events; even subtle alterations in this process can lead to serious disorders. Gene regulatory networks are thought to play key roles during organogenesis. Such developmental networks have been identified in Echinoidea (sea urchin), Drosophila, Ciona intestinalis, and Caenorhabditis elegans (1); the characterization of gene regulatory networks during vertebrate organogenesis lags behind.
Pharyngeal mesoderm (PM) cells are a subset of the head mesoderm, contributing to broad regions of the heart and head musculature. The PM contains initially both paraxial and splanchnic mesoderm cells surrounding the pharynx. Later, these cells migrate to fill the core of the pharyngeal arches, also known as branchial arches (2). Before their differentiation, PM cells express both skeletal muscle and second-heart field markers. Thus, the genetic program controlling early pharyngeal muscle development overlaps with that of the heart; the major molecular players include the transcription factors Tbx1, Pitx2, Tcf21 (capsulin/Pod1), Islet1, and Msc (MyoR) (2–5).
In addition to pharyngeal muscles, PM cells also contribute to the arterial pole of the heart, following the formation of the linear heart tube. Perturbations in the recruitment of PM-derived cells to the heart tube can lead to a wide range of congenital heart defects. Such defects occur in nearly 1% of live births, reflecting the complex cellular processes underlying heart development (6–8). Cardiac and craniofacial birth defects are often linked, because of their anatomical proximity during early embryogenesis and overlapping progenitor populations (2–4). One such congenital defect is DiGeorge syndrome (DGS), the most frequent microdeletion syndrome in humans, with an estimated incidence of 1 in 4,000 live births (9, 10). The clinical features of DGS vary, and may include cardiac defects, craniofacial and aortic arch anomalies, and thymus and parathyroid gland hypoplasia.
The T-box transcription factor 1 (TBX1) is located in the 22q11.2 deleted region, and mutations in TBX1 have been found in some patients with DGS-like phenotype; therefore, TBX1 haploinsufficiency is probably a major contributor to human del22q11 phenotypes and to murine models of the syndrome (11–14).
How does a set of PM transcription factors execute myogenesis and cardiogenesis? What are the relationships between these factors? Could we identify new PM regulators? In this study we have addressed these questions in mice by revealing a hierarchical regulatory network, composed of a set of transcription factors expressed in PM progenitors. Our comprehensive genetic study uncovered molecular evidence for the involvement of the PM regulatory network in myogenesis and cardiogenesis, as well as in the etiology of DGS.
Results
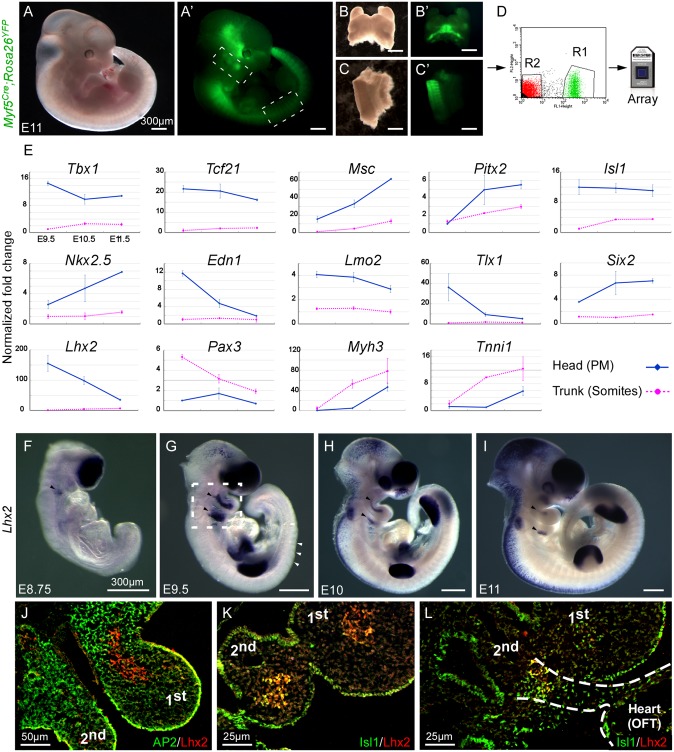
To identify unique regulators of PM myogenic progenitors, we compared gene-expression patterns of PM-derived progenitors, to those derived from the trunk (somites) at early stages of embryonic (E) development in the mouse (E9.5–E11.5). Myf5 is the earliest marker of myogenic commitment (15). In Myf5Cre;Rosa26YFP double-heterozygous embryos the entire skeletal muscle lineage is YFP+ (Fig. 1A′). We FACS-purified PM and trunk myogenic progenitors separately, and evaluated their gene expression profiles using an Affimetrix array (Fig. 1 A–D). Our results confirmed the differential expression of previously described PM-specific transcription factors, such as Tcf21, Isl1, Tbx1, Msc, Pitx2, and Nkx2.5 (Fig. 1E and Fig. S1). Other markers, such as Tlx1 (16), Six2 (17), the endothelial marker Lmo2 (18), the endothelin signaling component Edn1 (19), and retinoic acid-related genes were identified in our screen, and were enriched in PM, compared with the trunk progenitors (Fig. 1E and Fig. S1). As expected Pax3, the key myogenic regulator of trunk skeletal muscles, was not expressed in PM progenitors. Consistent with the fact that myogenic differentiation in head muscle progenitors lags behind that of the trunk, we observed delayed activation of muscle contractile genes, such as myosins (e.g., Myh3) and troponins (e.g., Tnni1) in the PM, relative to trunk muscle progenitors (Fig. 1E). In addition, we identified Lhx2, a LIM domain-containing transcription factor, as a unique PM-specific gene. In situ hybridization revealed that Lhx2 is expressed in the mesodermal core of the pharyngeal arches (Fig. 1 F–I), but is completely absent from the somites. In mice, Lhx2 is a prerequisite for the development of several organs, including the eye, telencephalon, and blood system (20–22), which fits its expression in these tissues (Fig. 1 F–I).
Fig. 1.
Lhx2, a unique PM regulator. (A–D) Experimental design: a Myf5Cre;Rosa26YFP E11.5 embryo is shown under bright light and a fluorescence microscope (A and A′). Dotted lines indicate the dissected regions of the pharyngeal arches (B and B′) and interlimb somites (C and C′). YFP+ cells from these two tissues were isolated by FACS (indicated as R1 in D), and used for RNA transcriptome analysis (E). A comparison of gene expression profiles from head (PM, blue) and trunk (somites, magenta) muscle progenitors. The fold-change corresponds to the difference in signal intensities (E). In situ hybridization for Lhx2 at E8.75 (F), E9.5 (G), E10 (H), and E11 (I) embryonic stages matches the microarray data. (J–L) Transverse sections of control E9.5 embryos, representing the area depicted in G, costained with Lhx2 and either AP2 (J) or Isl1 (K and L). Dotted lines in L indicate the continuum of PM between the pharyngeal arches and the heart. Black arrowheads indicate Lhx2 expression in the PM (F–I), whereas white arrowheads indicate lack of expression in the interlimb somites (G). first/second/third: first/second/third pharyngeal arches. (Scale bars, 300 μm.) Error bars indicate SE.
To obtain detailed expression relationships of Lhx2 relative to other lineages within the pharyngeal arches, we immunostained control E9.5 embryos with antibodies to Lhx2, Isl1, AP2, and Pecam1 (Fig. 1 J–L and Fig. S2). Most cells in the core of the arch express both Lhx2 and Isl1. In contrast, Lhx2 is not expressed in neural crest (AP2) or endothelial (Pecam1) cells (Fig. 1 J–L and Fig. S2). Isl1 is expressed in, and required for, a broad subset of cardiac progenitors in the mouse (23, 24). Isl1 is expressed in the distal part of the PM and these cells contribute to both pharyngeal muscles (and their satellite cells) as well as to the heart (2). A stream of Lhx2+ Isl1+ PM cells can be seen connecting the second pharyngeal arch and the outflow tract (OFT). Taken together, the expression pattern of Lhx2 in Isl1+ PM progenitors suggests that this gene might play a role in both myogenesis and cardiogenesis.
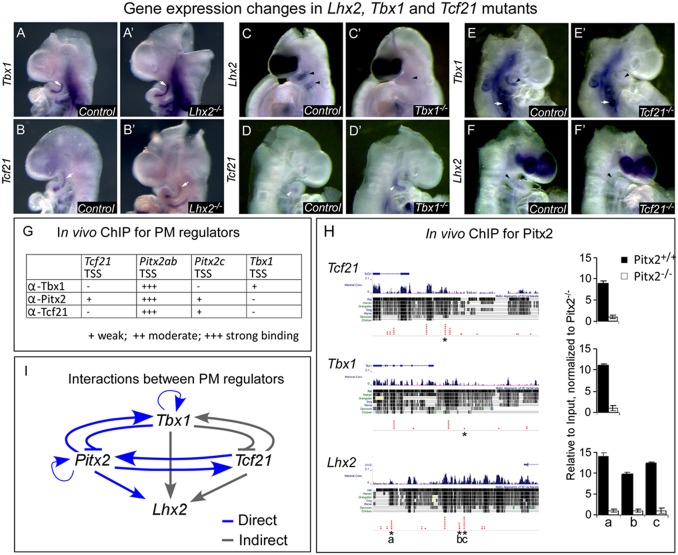
Next, we determined the genetic interactions between the major PM factors at E9.5 (Fig. 2). To systematically examine the epistatic relationships between the major PM regulators, we used several mouse knockout models (Fig. 2 and Fig. S3). Although Tbx1 and Tcf21 expression patterns remained unchanged in Lhx2 mutant embryos (Fig. 2 A–B′), Lhx2 expression was reduced in the PM of Tbx1 mutant embryos at the same developmental stage (Fig. 2 C and C′). These results suggest that Lhx2 acts downstream of Tbx1. The expression levels of Tcf21, Msc, and Pitx2 were slightly increased in the PM of Tbx1 mutant embryos, consistent with findings from a recent screen for Tbx1 target genes (25) (Fig. 2 D and D′, and Fig. S3), suggesting that these factors are regulated by Tbx1.
Fig. 2.
Genetic interactions between members of the PM network. (A–F′) Whole-mount in situ hybridization for the indicated genes (Left) in E9.5 embryos with the indicated genotypes (black rectangles). Arrows/Arrowheads mark the PM; white arrows, unchanged expression; black arrowheads, down-regulated genes; white arrowheads, up-regulated genes. (G) A ChIP experiment using pharyngeal arch tissues at E9.5 with Tbx1, Pitx2, and Tcf21 antibodies. (H) A ChIP-seq experiment using Pitx2 antibody on Pitx2+/+ and Pitx−/− pharyngeal arches, reveals specific interactions with Tcf21, Tbx1, and Lhx2 regulatory regions. (I) A model summarizing direct (blue) and indirect (gray) genetic interactions in the PM regulatory network. TSS, transcription start site (or proximal promoter). Asterisks point to the indicated transcription factor binding site. Number of embryos in each experiment was ≥ 3.
Next we examined how the bHLH factor Tcf21 affects the PM regulators. It was previously shown that a subgroup of pharyngeal muscles was absent in Tcf21/Msc double-knockout embryos (26). Furthermore, these two genes have been shown to regulate the expression of MyoD and Myf5 in craniofacial muscle progenitors (27). The expression of Lhx2, Tbx1, and Pitx2 was reduced in the PM of Tcf21 mutant embryos (Fig. 2 E–F′ and Fig. S3). These findings place Tcf21 in the upper tier of the PM genetic network.
Finally, we have characterized the bicoid-related homeodomain transcription factor Pitx2. Both pharyngeal muscles (derived from the first arch) and extraocular muscles (EOM) were affected in Pitx2 knockout embryos (17, 28). Pitx2 and Tbx1 were shown to be genetically linked in many developmental processes, including cardiac and craniofacial muscle development (3). In Pitx2 knockout embryos, Tbx1 was hardly detected in the PM and Lhx2 was diminished specifically in the mesoderm of the first pharyngeal arch (Fig. S3). We confirmed the observed changes in gene expression using quantitative RT-PCR (qRT-PCR) on isolated pharyngeal arches (first-third) of various mutant embryos (Fig. S4). The results are consistent with the gene-expression patterns observed by in situ hybridization (Fig. 2 and Fig. S3). Notably, some of the analyzed genes (e.g., Pitx2 and Tbx1) are also expressed in the ectoderm and endoderm of the pharyngeal arches; accordingly, their total levels were moderately changed compared with the in situ hybridization results (Fig. S4). Notably, despite some loss of PM cells in Pitx2 mutants at E9.5, which underscores the importance of Pitx2 in PM cell survival (17, 28), the observed changes in gene expression patterns could not be attributed to loss of PM cells (Fig. S4B). Our findings reveal cross-regulation between members of the PM network: Tcf21 and Pitx2 are linked to Tbx1, and Lhx2 lies downstream to these genes.
A key question regarding our findings is whether the observed changes in PM gene expression (Fig. 2 A–F′ and Fig. S3) are a result of direct interactions between the PM transcription factors. ChIP was performed on isolated E9.5 pharyngeal arch tissues using Tbx1, Pitx2, and Tcf21 antibodies to evaluate a potential cross-regulation between PM members. Our results suggest several interactions, the strongest of which are Tbx1, Pitx2, and Tcf21 with the Pitx2 proximal promoter (Fig. 2G). Because of the extensive interactions of Pitx2 with other PM members, we decided to further characterize its specific binding sites using ChIP-seq. Pitx2 binding to Tbx1, Tcf21, and Lhx2 regulatory regions was enriched in isolated E9.5 pharyngeal arch tissues (Fig. 2H). As a control, we compared the binding of Pitx2 to these elements in Pitx2−/−-derived tissues, binding of Pitx2 to nonspecific genomic sites, as well as binding of nonspecific antibody (Fig. 2H and Fig. S5). Although Pitx2 did not bind to the Tbx1 proximal promoter, we could identify its binding to specific sites upstream to the promoter using a ChIP-seq approach (Fig. 2H and Fig. S5). Taken together, our findings suggest that the PM regulatory network involves extensive genetic interactions between its members (Fig. 2I).
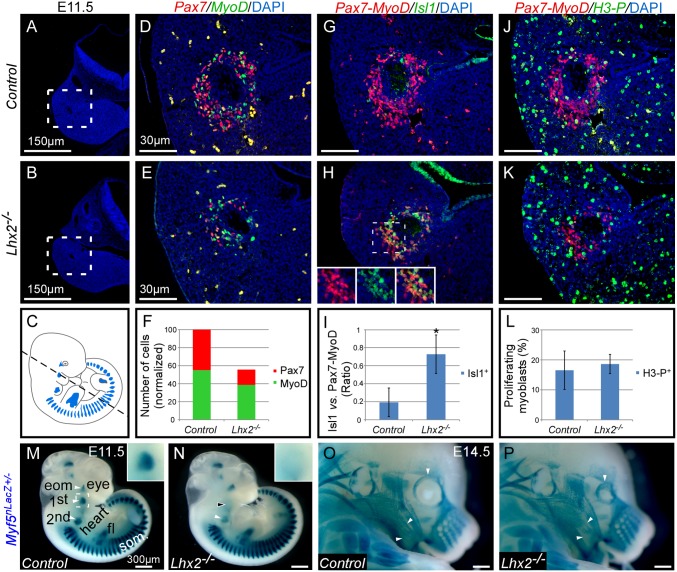
The involvement of Lhx2 in cardiac and craniofacial development was not previously examined, partly because of the fact that Lhx2 knockout mouse embryos die at E15.5 (20). Therefore, we first sought to address its role during head muscle development (Fig. 3). Pax7 marks muscle progenitors, whereas MyoD defines a more committed myogenic state. At E11.5, the total number of myogenic cells (Pax7+ or MyoD+) in the PM of Lhx2 mutant embryos decreased by ∼50% (Fig. 3 A–F). Comparing the ratio between MyoD- and Pax7-expressing cells in control vs. Lhx2 mutants revealed that the Pax7+ population was more affected, suggesting that Lhx2 is required in PM-derived muscle progenitors.
Fig. 3.
Lhx2 is required for specification of pharyngeal muscle progenitors. (A–C) Transverse sections of control (A) and Lhx2 mutant (B) E11.5 embryos, showing the core of the first pharyngeal arch, as indicated in C. Dotted lines in A and B represent the magnified areas in D–K. (D–F) Coimmunofluorescence of Pax7 and MyoD in controls (D) and Lhx2 mutants (E), and quantification of the results (F). (G–I) Coimmunofluorescence of myogenic (Pax7 and MyoD) vs. premyogenic (Isl1) in controls (G) and Lhx2 mutants (H) and quantification of the results (I). (J–L) Coimmunofluorescence of myogenic (Pax7-MyoD) and phosphorylated histone H3 (P-H3), which labels mitotic cells in controls (J) and Lhx2 mutants (K). The percentage of proliferating myoblasts is quantified (L). Quantifications were performed on greater than or equal to six sections from at least two different embryos, as shown in A–L. Error bars indicate SD. Antibodies used and DAPI are written in individual panels, in the color corresponding to the fluorescent staining. (M and N) Myf5 expression (X-Gal staining) in Lhx2 control (M) and mutants (N) E11.5 embryos, which are also heterozygous for the Myf5nLacZ reporter. (M and N, Insets) The area depicted by the dotted line in M. (O and P) Myf5 expression (X-Gal) in Lhx2 control (O) and mutant (P) E14.5 embryos, which are also heterozygous for the Myf5nLacZ reporter. Arrowheads indicate change in muscle patterning. first/second, first/second pharyngeal arch muscle progenitors; fl, forelimb; som, somites.
A decrease in the number of muscle progenitors could be because of either a delay in the specification of PM cells toward the myogenic lineage, a decrease in their proliferation, or elevated apoptosis. To resolve this issue, we compared myogenic (Pax7+-MyoD+) vs. premyogenic PM progenitors expressing Isl1+. Isl1 expression is down-regulated rapidly as head myogenesis ensues, and Isl1 overexpression in chicken embryos delayed myogenic differentiation (29, 30). In Lhx2 mutants, Isl1 expression failed to be down-regulated in the core of the first pharyngeal arch compared with controls (Fig. 3 G–I), but cell proliferation and apoptosis remained comparable (Fig. 3 J–L and Fig. S6). The observed increase in premyogenic Isl1+ cells in Lhx2 mutants was inversely correlated with the number of Pax7-expressing cells, suggesting that Lhx2 is involved in pharyngeal muscle specification.
Myf5 is highly regulated, both spatially and temporally by various factors (31). To further examine the role of Lhx2 during head muscle specification, we used the Myf5nLacZ reporter (32). Myf5 expression in the pharyngeal arches was reduced in Lhx2 mutant compared with control E11.5 embryos, but the trunk and EOM remained unaffected (Fig. 3 M and N) (n ≥ 12). These findings demonstrate that Lhx2 is required for the early activation of Myf5 in the myogenic specification program within the pharyngeal arches. The expression of Myf5 (LacZ staining) was largely restored at E14.5, albeit with some patterning defects (Fig. 3 O and P). Hence, the PM regulatory network acts to provide robustness by allowing the activation of the myogenic program in the absence of a single PM member, consistent with previous studies (16, 33).
Next, we examined whether Myf5 is directly regulated by the members of the PM network by in vivo ChIP. Tbx1, Pitx2, and Tcf21 were associated to the Myf5 evolutionary conserved region (ECR-84), which is part of the mandibular arch enhancer (MAE) (Fig. S5) (31). To further explore the connection between Lhx2 and Myf5, we identified three putative Lhx2 binding sites within the Myf5 MAE. Next, C2C12 cells, transfected with Lhx2-HA construct were used for a ChIP experiment using anti-HA antibody. We found that Lhx2 can bind to one of these sites in C2C12 cells (Fig. S5).
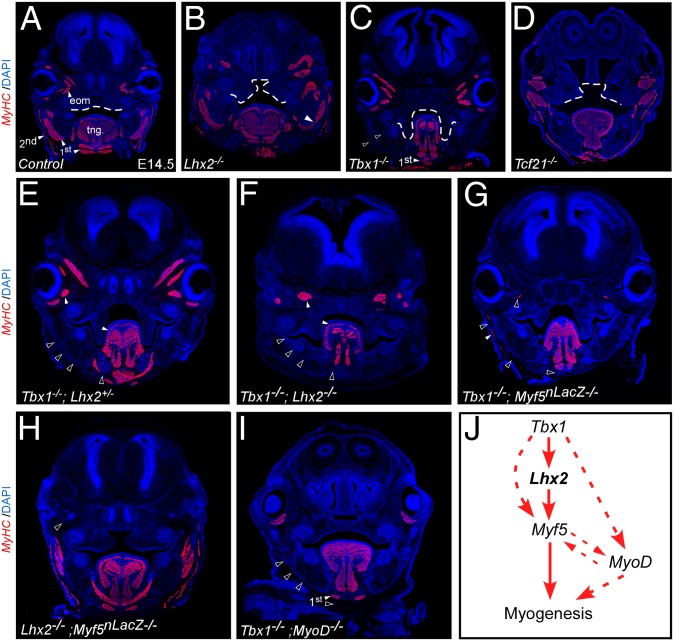
To further validate the robust nature of the myogenic program in the head we compared single knockouts of PM factors. Knockout of Lhx2, Tbx1, and Tcf21 separately revealed muscle patterning defects in all three mutants (Fig. 4 A–D). In agreement with an earlier report (16), pharyngeal muscles are severely perturbed, although not completely eliminated, in Tbx1−/− mutants (Fig. 4C and refs. 16 and 33). To investigate the genetic wiring of the PM regulators, we analyzed the muscle phenotype in double-knockout embryos (Fig. 4 E–I). The muscle phenotype in Tbx1−/− mutants was comparable to that of Tbx1−/−;Lhx2−/+ mutants (Fig. 4 C and E). Remarkably, pharyngeal arch muscles were completely missing in Tbx1−/−;Lhx2−/− double-mutants (Fig. 4F) (n = 2/4). Similarly, pharyngeal muscles were eliminated in most Tbx1−/−;Myf5−/− mutants (Fig. 4G) (n = 3/4), in agreement with ref. 33.
Fig. 4.
Epistatic genetic relationships regulating pharyngeal muscle development. Transverse craniofacial sections of E14.5 mouse embryos stained with MyHC for single- (B–D) and double- (E–J) mutants for the indicated genotypes (n ≥ 4). Dotted line outlines the cleft palate seen in all three single mutants (B–D). (J) A model summarizing the genetic interactions described above. Dotted arrows indicate parallel regulatory interactions affecting head myogenesis, empty arrowheads indicate novel interactions. Skeletal muscle groups are marked in white arrowheads, and their absence in black arrowheads. first/second, first/second pharyngeal arch-derived muscles; tng, tongue.
Taken together, our findings reveal that the Tbx1, Lhx2, and Myf5 genetic circuit is required for pharyngeal muscle specification. Our findings suggest that in the absence of both Myf5 and Lhx2, Tbx1 could initiate myogenesis by activating MyoD via a parallel genetic pathway, as suggested by Sambasivan et al. (33). Accordingly, pharyngeal muscles of Tbx1−/−;MyoD−/− double-mutants were completely missing (with the exception of the digastric muscles in the lower jaw) (Fig. 4I) (n = 2/2). Hence, in the absence of Tbx1 and another factor (e.g., Lhx2, Myf5, or MyoD), pharyngeal muscles are severely perturbed. Consistent with the key role of Tbx1 in this genetic network, Myf5−/−;Lhx2−/− and MyoD−/−;Lhx2−/− double-knockout embryos did not show an enhanced muscle phenotype, compared with each knockout alone (Fig. 4H and Fig. S4). Our findings suggest that the PM network acts to ensure proper myogenesis in the absence of single PM members (Fig. 4J).
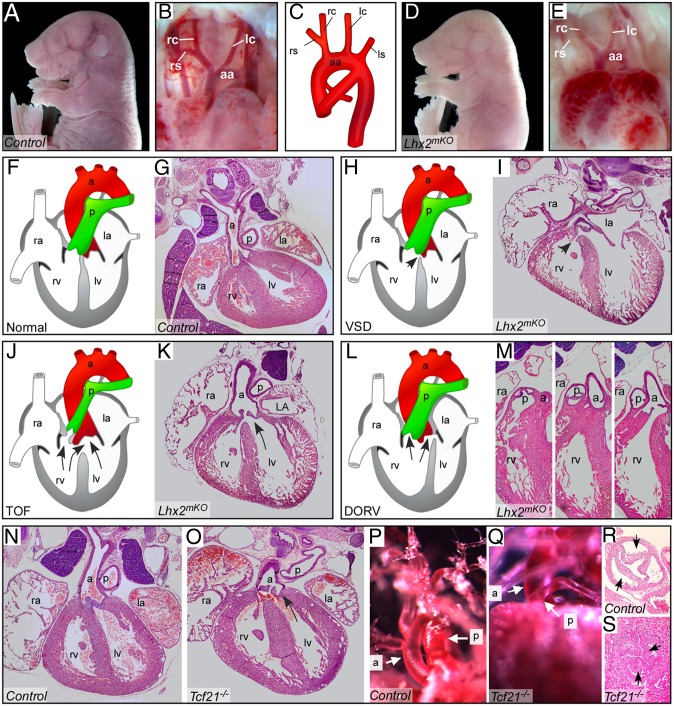
DGS is a common congenital disease involving cardiac and craniofacial defects. The major genetic determinant in its etiology is TBX1, although other genes in the 22q11 region have been shown to be involved. Because Lhx2 lies downstream of Tbx1, we hypothesized that Lhx2 mutant embryos might display DGS phenotypes. Lhx2 mutants die at E14.5–E15.5 from severe anemia and developmental defects (20). The development of the ventricular septum is completed at E15; thus, we analyzed both standard and conditional Lhx2-null embryos around this stage. Lhx2 was ablated in the cardio-craniofacial mesoderm using the MesP1Cre mouse line (34), which prolongs their viability up to birth. Indeed, at E17.5 about 50% of MesP1Cre+/−;Lhx2−/floxed (Lhx2mKO) mutants exhibited DGS-like cardiac defects, including various OFT anomalies, such as ventricular septal defect (VSD), tetralogy of Fallot, and double-outlet right ventricle (Fig. 5 and Table S1) (n = 7/13). Interestingly, aortic arch patterning, one of the most common features of DGS, was normal in all Lhx2mKO mutants (n = 13/13; and in E14.5 Lhx2−/− embryos n = 10/10) (Fig. 5 A–E and Table S1).
Fig. 5.
Lhx2 and Tcf21 mutant embryos display specific DGS-like cardiac defects. (A–E) Whole-mount E17.5 controls (A and B) and Lhx2mKO mutants (D and E), both displaying normally shaped aortic arches (B and D, respectively). Note severe anemia in the mutant (D) embryo, compared with control (A). A scheme illustrating the normal configuration of the aortic arch (C). (F and G) H&E staining of heart paraffin sections in control hearts. (F–M) Lhx2mKO mutants display a simple VSD (H and I, arrow), schematically illustrated (H); tetralogy of Fallot (TOF), characterized by both VSD and overriding aorta (J and K, arrows); double-outlet right ventricle (DORV) (L and M). (N–S) E17.5 Tcf21 mutant embryos display TOF, VSD, and overriding aorta (O) compared with a control heart (N). In addition E17.5 Tcf21 mutant embryos have pulmonic stenosis, shown by vascular casting (Q) and H&E staining (S) compared with controls (P and R, respectively). a, aorta; aa, aortic arch; ls, left subclavian artery; lc, left common carotid artery; la, left atrium; lv, left ventricle; p, pulmonary artery; ra, right atrium rc, right common carotid artery; rs, right subclavian artery; rv, right ventricle. The left side of the mouse is displayed on the right side of the picture in all panels.
We next investigated the genetic interaction between Tbx1 and Lhx2, by measuring the frequency of VSD in compound mutants. Although Tbx1+/− heterozygous embryos had no detectable VSD (n = 11), 20% of Tbx1+/−Lhx2+/− double-heterozygous (compound) embryos had VSD (n = 10) (Table S1). This functional interaction strongly suggests that Tbx1 and Lhx2 are in the same genetic pathway and synergistically regulate heart morphogenesis.
To identify genes lying downstream of Lhx2, we examined the expression of several possible candidates. The expression levels of both Fgf8, which is genetically linked to Tbx1 in the context of DGS (35), and Bmp4, which was shown to act downstream of Lhx2 during eye development (36), were comparable in Lhx2 mutant and control embryos (Fig. S7 A–D). Several recent studies have shown that both cardiac neural crest (affecting caudal PM progenitors) and cranial neural crest cells (affecting rostral/cranial PM progenitors) influence the migration of PM cells into the looping heart, and their subsequent differentiation (2, 37). We therefore examined the expression pattern of several neural crest markers, Dlx5, Twist, and Sox10, as well as the PM marker, Isl1. Although Isl1, Dlx5, and Twist expression seemed to be comparable in Lhx2 mutants and controls, Sox10 expression pattern was slightly perturbed in some mutants, suggesting that neural crest cell migration might play some role in the observed phenotype (Fig. S7 E–J). These findings suggest that perturbation of the PM regulatory network affects cardiac formation both cell-autonomously and noncell-autonomously, via cross-talk with neural crest cells.
Given the regulatory interactions between various network members, we hypothesized that elimination of each of the core factors, one-by-one, might elicit a DGS-like phenotype either directly or by affecting Tbx1 levels. Consistent with this view, Pitx2 is known to affect cardiac development (38, 39). Because Tbx1 levels were reduced in Tcf21 mutants (Fig. 2), we sought to better analyze the heart phenotype of these mutants. Tcf21 mutants display tetralogy of Fallot, including VSD, overriding aorta, pulmonic stenosis (Fig. 5 N–R, Fig. S8, and Table S1), as well as cleft palates (Fig. 4D). Similar to Lhx2mKO mutant embryos, the morphology of the aortic arch remained normal in Tcf21 mutants. Furthermore, hearts of Tcf21−/− mutants displayed regions of epicardial detachments (Fig. S8), in agreement with a recent report (40). Taken together, insights from the PM network composition led us to predict that both Tcf21 and Lhx2, which are genetically linked to Tbx1, might cause cardiac defects. We demonstrate such cardiac anomalies in both Tcf21 and Lhx2 mutant embryos, some of which are shared by DGS patients.
Discussion
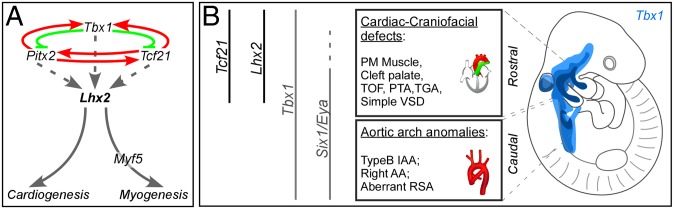
Our results demonstrate that a set of transcription factors expressed in PM progenitors form a regulatory network that coordinates normal heart and craniofacial development (Fig. 6A). The expression of PM members (Tbx1, Pitx2, Tcf21, and Lhx2) is regulated by other members of the network, and involves direct genetic interactions. Lhx2 is a unique player within the PM network; knockout of this gene resulted in a pharyngeal muscle specification defect, as well as DGS-like phenotypes (Fig. 6A). We revealed epistatic relationships between Tbx1, Lhx2, and Myf5 embedded within the PM network, affecting early pharyngeal muscle specification and patterning. Thus, Lhx2 plays an important role in PM progenitor cells, consistent with its roles in the specification of other stem/progenitor cell populations, such as telencephalic progenitors (41), retina progenitors (42), hematopoietic progenitors (43), and hair follicle progenitors (21).
Fig. 6.
PM progenitors form a regulatory network that coordinates early cardiogenesis and craniofacial myogenesis, (A) A summary of the genetic interactions of the PM transcriptional network and its impact on cardiogenesis and myogenesis. (B) A proposed model for a domain-specific subdivision of DGS-like anomalies in mouse models into rostral (heart and craniofacial) and caudal (arch artery) phenotypes. The model is based on the progressive alignment of the pharyngeal arches with the heart tube during its looping stages (50). The corresponding mouse knockout phenotypes are shown along these two domains.
In addition, we identified a genetic link between Tcf21, Tbx1, and Lhx2 in the PM transcriptional circuit. Genetic perturbation of these factors resulted in specific DGS-like phenotypes. We demonstrated, using single- and double-knockout experiments, that Lhx2 removal has specific cardiac phenotypes, and it enhances the severity of both craniofacial muscles and heart phenotypes of Tbx1 mutants. This finding suggests that both genes work in the same genetic pathway. Hence, Lhx2 can be included within the growing list of transcription factors that have been found to play important roles in second-heart field development, based on the cardiac phenotypes of single and compound mutations in these genes (44).
Although human TCF21 and LHX2 do not map to chromosome 22q11.2, the shared morphological defects and link to Tbx1 suggest that these genes might be genetic modifiers of DGS. Genetic variations in the ISL1 locus in human were shown to be linked to an increased risk for congenital heart defects (45). Could LHX2 and TCF21 contribute to the variations in cardiovascular phenotype seen in DGS patients? To draw genotype-phenotype correlations in such patients, a genome-wide association study, as well as a candidate gene approach, is currently underway. Results from this study could shed light on whether common DNA variants alter the degree of expressivity of the syndrome.
One of the enigmatic features of DGS is that it varies in its penetrance from patient to patient. Importantly, some DGS patients do not display either a deletion or a mutation in the Tbx1 locus (46). Changes in the levels of Tbx1, loss and gain, lead to a dose-dependent spectrum of DGS malformations (47, 48). Therefore, Tbx1 levels must be precisely regulated in order for the pharyngeal apparatus and its derivatives to properly form. Our study adds to the understanding of how Tbx1 levels could be fine-tuned by interactions with other PM transcription factors (Fig. 6A).
Tbx1 is expressed in both rostral and caudal PM cells. It has been shown that cranial PM cells enter the arterial pole of the heart to populate the right ventrical and OFT, and caudal PM cells contribute to the myocardium at the base of the great arteries (49). Previous studies addressing DGS etiology reported various cardiac anomalies, including both aortic arch and cardiac defects, for the following knockout models: Fgf8 and Six1/Eya1 (35), VegfA (50), and retinoic acid-related genes (51). We suggest that Lhx2 and Tcf21, expressed in the cranial PM, function as domain-specific modifiers of the Tbx1 pathway, as judged by the uncoupling of the aortic arch phenotype from that of the outflow tract (Fig. 6B). In sum, our study sheds light on the developmental principles underlying the etiology of congenital birth defects.
Our findings imply that the heart and pharyngeal muscles show varying degrees of sensitivity to early perturbations of the PM. For example, although the pharyngeal muscle phenotypes of Lhx2 and Tcf21 mutants are largely restored, albeit with patterning/hypoplastic defects, the cardiac defects are beyond repair. Detailed analyses of pharyngeal muscles in mouse and zebrafish DGS models (or in human patients) have not been well-characterized. Facial asymmetry, for example, is a rare symptom observed in babies only when they cry, known as “asymmetric crying faces,” is caused by the absence or hypoplasia of a pharyngeal muscle at the corner of the mouth. This defect has been shown to be associated with cardiovascular anomalies in DGS babies (52). Therefore, it would be important to better characterize the linkage between craniofacial muscle patterning and cardiovascular defects.
Regulatory networks of transcription factors have been found in diverse organisms, from bacteria to humans. The network architectures of the transcription factors function to enhance the stability of gene expression and functional outputs. The PM network characteristics that we (present study) and others (16, 17, 28, 33) have gradually uncovered in recent years seem to be hierarchical, and involve extensive cis-regulatory interactions. We propose that the overall biological outputs of the PM network (e.g., cardiogenesis and myogenesis) and precise signal strengths are network properties, rather than properties specific to individual PM members.
Experimental Procedures
Mice.
The following mouse transgenic lines and their genotyping have been previously described: Myf5Cre (53), Rosa26YFP (54), Pitx2−/− (55), Myf5nlacZ (32), MesP1Cre (34), Lhx2sKO (20), Lhx2cKO (22), Tcf21−/− (56), and Tbx1−/− (11). All animal experiments were performed in accordance with the Weizmann Institute of Science regulations for animal care and handling.
FACS, Microarrays, Staining, qPCR, and ChIP.
Interlimb somites and pharyngeal arches were dissected from E9.5, E10.5, and E11.5 Myf5Cre;RosaYFP mouse embryos. RNA was purified amplified, and hybridized to Affymetrix arrays (detailed in SI Experimental Procedures). X-Gal staining, histology, immunohistochemistry, and whole-mount in situ hybridization were performed as previously reported (29). Antibodies are listed in SI Experimental Procedures. cDNA or immunoprecipitated DNA was analyzed by qPCR using SYBR Green methodology, as recommended by the manufacturer. Primers used are listed in Table S2. ChIP was done according to ref. 57. Minor modifications and antibodies used are detailed in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Kfir-Baruch Umansky for his technical help and advice. This work was supported by grants from the European Research Council, the Israel Science Foundation, the United States–Israel Binational Science Foundation, the German Israeli Foundation, the Association Française Contre les Myopathies, the Kirk Center for Childhood Cancer and Immunological Disorders, the Jeanne and Joseph Nissim Foundation for Life Sciences Research, and a donation from the Jack Gitlitz Estate (all to E.T.); National Institutes of Health-National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR054406 (to C.K.); a Studentship from The Institute Of Cancer Research (to J.W.C.); and Ministry of Science and Innovation Grant BFU2011-22928 (to J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208690109/-/DCSupplemental.
References
- 1.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468(7326):911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzahor E, Evans SM. Pharyngeal mesoderm development during embryogenesis: Implications for both heart and head myogenesis. Cardiovasc Res. 2011;91(2):196–202. doi: 10.1093/cvr/cvr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grifone R, Kelly RG. Heartening news for head muscle development. Trends Genet. 2007;23(8):365–369. doi: 10.1016/j.tig.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Tzahor E. Heart and craniofacial muscle development: A new developmental theme of distinct myogenic fields. Dev Biol. 2009;327(2):273–279. doi: 10.1016/j.ydbio.2008.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Sambasivan R, Kuratani S, Tajbakhsh S. An eye on the head: The development and evolution of craniofacial muscles. Development. 2011;138(12):2401–2415. doi: 10.1242/dev.040972. [DOI] [PubMed] [Google Scholar]
- 6.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6(11):826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 7.Hutson MR, Kirby ML. Neural crest and cardiovascular development: A 20-year perspective. Birth Defects Res C Embryo Today. 2003;69(1):2–13. doi: 10.1002/bdrc.10002. [DOI] [PubMed] [Google Scholar]
- 8.Srivastava D. Developmental and genetic aspects of congenital heart disease. Curr Opin Cardiol. 1999;14(3):263–268. doi: 10.1097/00001573-199905000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Baldini A. Dissecting contiguous gene defects: TBX1. Curr Opin Genet Dev. 2005;15(3):279–284. doi: 10.1016/j.gde.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Yamagishi H, Srivastava D. Unraveling the genetic and developmental mysteries of 22q11 deletion syndrome. Trends Mol Med. 2003;9(9):383–389. doi: 10.1016/s1471-4914(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 11.Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410(6824):97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 12.Yagi H, et al. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362(9393):1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 13.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27(3):286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 14.Merscher S, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104(4):619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 15.Sambasivan R, Tajbakhsh S. Skeletal muscle stem cell birth and properties. Semin Cell Dev Biol. 2007;18(6):870–882. doi: 10.1016/j.semcdb.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Kelly RG, Jerome-Majewska LA, Papaioannou VE. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum Mol Genet. 2004;13(22):2829–2840. doi: 10.1093/hmg/ddh304. [DOI] [PubMed] [Google Scholar]
- 17.Shih HP, Gross MK, Kioussi C. Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proc Natl Acad Sci USA. 2007;104(14):5907–5912. doi: 10.1073/pnas.0701122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landry JR, et al. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood. 2005;106(8):2680–2687. doi: 10.1182/blood-2004-12-4755. [DOI] [PubMed] [Google Scholar]
- 19.Thomas T, et al. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125(16):3005–3014. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- 20.Porter FD, et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124(15):2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 21.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312(5782):1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangale VS, et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319(5861):304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135(2):193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- 25.Liao J, et al. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol. 2008;316(2):524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu JR, et al. Control of facial muscle development by MyoR and capsulin. Science. 2002;298(5602):2378–2381. doi: 10.1126/science.1078273. [DOI] [PubMed] [Google Scholar]
- 27.Moncaut N, et al. Musculin and TCF21 coordinate the maintenance of myogenic regulatory factor expression levels during mouse craniofacial development. Development. 2012;139(5):958–967. doi: 10.1242/dev.068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong F, et al. Pitx2 promotes development of splanchnic mesoderm-derived branchiomeric muscle. Development. 2006;133(24):4891–4899. doi: 10.1242/dev.02693. [DOI] [PubMed] [Google Scholar]
- 29.Harel I, et al. Distinct origins and genetic programs of head muscle satellite cells. Dev Cell. 2009;16(6):822–832. doi: 10.1016/j.devcel.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nathan E, et al. The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development. 2008;135(4):647–657. doi: 10.1242/dev.007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carvajal JJ, Cox D, Summerbell D, Rigby PW. A BAC transgenic analysis of the Mrf4/Myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Development. 2001;128(10):1857–1868. doi: 10.1242/dev.128.10.1857. [DOI] [PubMed] [Google Scholar]
- 32.Tajbakhsh S, Rocancourt D, Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 1996;384(6606):266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- 33.Sambasivan R, et al. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev Cell. 2009;16(6):810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Saga Y, et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126(15):3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 35.Guo C, et al. A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. J Clin Invest. 2011;121(4):1585–1595. doi: 10.1172/JCI44630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yun S, et al. Lhx2 links the intrinsic and extrinsic factors that control optic cup formation. Development. 2009;136(23):3895–3906. doi: 10.1242/dev.041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res. 2009;104(8):933–942. doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- 38.Nowotschin S, et al. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133(8):1565–1573. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- 39.Ai D, et al. Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol. 2006;296(2):437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acharya A, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139(12):2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou SJ, Perez-Garcia CG, Kroll TT, O’Leary DD. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci. 2009;12(11):1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tétreault N, Champagne MP, Bernier G. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev Biol. 2009;327(2):541–550. doi: 10.1016/j.ydbio.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 43.Dahl L, Richter K, Hägglund AC, Carlsson L. Lhx2 expression promotes self-renewal of a distinct multipotential hematopoietic progenitor cell in embryonic stem cell-derived embryoid bodies. PLoS ONE. 2008;3(4):e2025. doi: 10.1371/journal.pone.0002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelly RG. The second heart field. Curr Top Dev Biol. 2012;100:33–65. doi: 10.1016/B978-0-12-387786-4.00002-6. [DOI] [PubMed] [Google Scholar]
- 45.Stevens KN, et al. Common variation in ISL1 confers genetic susceptibility for human congenital heart disease. PLoS ONE. 2010;5(5):e10855. doi: 10.1371/journal.pone.0010855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scambler PJ. 22q11 deletion syndrome: A role for TBX1 in pharyngeal and cardiovascular development. Pediatr Cardiol. 2010;31(3):378–390. doi: 10.1007/s00246-009-9613-0. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Baldini A. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum Mol Genet. 2008;17(1):150–157. doi: 10.1093/hmg/ddm291. [DOI] [PubMed] [Google Scholar]
- 48.Liao J, et al. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet. 2004;13(15):1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- 49.Lescroart F, et al. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development. 2010;137(19):3269–3279. doi: 10.1242/dev.050674. [DOI] [PubMed] [Google Scholar]
- 50.Stalmans I, et al. VEGF: A modifier of the del22q11 (DiGeorge) syndrome? Nat Med. 2003;9(2):173–182. doi: 10.1038/nm819. [DOI] [PubMed] [Google Scholar]
- 51.Roberts C, Ivins S, Cook AC, Baldini A, Scambler PJ. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of DiGeorge Syndrome in the chick. Hum Mol Genet. 2006;15(23):3394–3410. doi: 10.1093/hmg/ddl416. [DOI] [PubMed] [Google Scholar]
- 52.Stewart HS, Clayton-Smith J. 22q11 deletion: A cause of asymmetric crying facies. Arch Dis Child. 1996;75(1):89. doi: 10.1136/adc.75.1.89-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tallquist MD, Weismann KE, Hellström M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127(23):5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- 54.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin CR, et al. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401(6750):279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 56.Quaggin SE, et al. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126(24):5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- 57.Hilton T, Gross MK, Kioussi C. Pitx2-dependent occupancy by histone deacetylases is associated with T-box gene regulation in mammalian abdominal tissue. J Biol Chem. 2010;285(15):11129–11142. doi: 10.1074/jbc.M109.087429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.