Abstract
The precursor of the essential ether phospholipids is synthesized by a peroxisomal enzyme that uses a flavin cofactor to catalyze a reaction that does not alter the redox state of the substrates. The enzyme crystal structure reveals a V-shaped active site with a narrow constriction in front of the prosthetic group. Mutations causing inborn ether phospholipid deficiency, a very severe genetic disease, target residues that are part of the catalytic center. Biochemical analysis using substrate and flavin analogs, absorbance spectroscopy, mutagenesis, and mass spectrometry provide compelling evidence supporting an unusual mechanism of covalent catalysis. The flavin functions as a chemical trap that promotes exchange of an acyl with an alkyl group, generating the characteristic ether bond. Structural comparisons show that the covalent versus noncovalent mechanistic distinction in flavoenzyme catalysis and evolution relies on subtle factors rather than on gross modifications of the cofactor environment.
Keywords: peroxisomal disorder, phospholipid biosynthesis, plasmalogen, rhizomelic chondrodysplasia punctata
Textbooks typically describe a phospholipid molecule as consisting of a glycerol backbone with a polar head and two acyl chains linked via an ester bond to the glycerol sn-1 and sn-2 carbons. This description can be misleading, however, in that a significant fraction of eukaryotic and archaebacterial cell membranes contain a different group of molecules, the ether phospholipids, which represent up to 20–50% of mammalian membranes. These phospholipids’ distinctive property is the ether bond that connects an alkyl (rather than an acyl) chain to the sn-1 carbon of the glycerol moiety (1, 2). Ether phospholipids facilitate membrane fusion processes and function as effective antioxidants, signaling molecules, and storage depots for docosahexaenoic (ω-3) and arachidonic (ω-6) acids. The key fact is that these phospholipids are essential for normal growth and development (3, 4).
The initial and critical steps in the generation of ether phospholipids occur in the peroxisomes through a curious route. First, an acyl-transferase enzyme uses acyl-CoA and dihydroxyacetone phosphate (DHAP) to generate acylDHAP. This molecule is subsequently processed by alkylDHAP synthase (ADPS) in formation of the ether bond, the most crucial and chemically demanding step of the entire biosynthetic pathway (5–7). In particular, ADPS replaces the acyl group of acylDHAP with a long-chain fatty alcohol, generating alkylDHAP (Fig. 1). Defects in the foregoing metabolic steps have devastating consequences that lie at the basis of genetic diseases. In patients with a peroxisomal biogenesis disorder, such as Zellweger syndrome, peroxisomes lack matrix enzymes (so-called “ghost” organelles), and thus ether phospholipids cannot be effectively synthesized. Rhizomelic chondrodysplasia punctata is caused by more specific single-enzyme deficiencies. Patients with this autosomal recessive disorder die within the first or, at best, second decade of life (8). Disease types 2 and 3 are caused by mutations in the acyl-transferase and ADPS enzymes, whereas type 1 arises from mutations in PEX7, the protein mediating the peroxisomal import of ADPS (9–12).
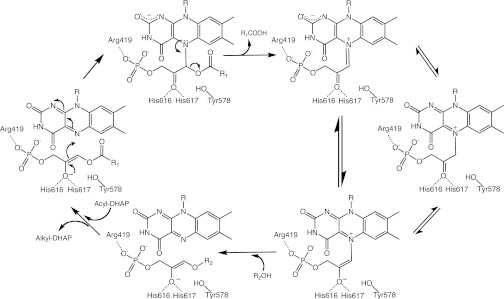
Fig. 1.
Mechanistic proposal for the ADPS reaction. The acylDHAP substrate is shown in its enolic form. The crucial covalent intermediate can have different tautomeric forms. The exact protonation states of several titratable groups purportedly involved in the reaction (Tyr578, His616-His617, N1 and N5 of the flavin, oxygens of DHAP, fatty acid product, and fatty alcohol substrate) are not firmly defined and are likely to change during the reaction. The data do not rule out adduct formation on C4a rather than on N5 of the flavin (see Fig. 4C, Right for atomic numbering); however, a C4a adduct intermediate would be difficult to reconcile with the subsequent fatty acid elimination step. The predicted negative charge of the covalent adduct can be stabilized by a network of H-bonding interactions involving protein backbone atoms, an ordered water molecule, and the N1 and O2 atoms of the flavin, as shown in Fig. S2A.
Here we report a structural and mechanistic investigation of mammalian ADPS in WT and mutated forms. The biochemical hallmark of the enzyme is that it uses a redox cofactor, flavin adenine dinucleotide (FAD), to catalyze a reaction that does not alter the redox state of the substrates (10, 13). Our findings indicate that FAD functions as a chemical trap for the substrate DHAP moiety so as to favor the acyl–alkyl exchange. They also demonstrate that pathological mutations target residues directly involved in catalysis and cofactor binding. These findings raise intriguing hypotheses about the covalent versus noncovalent mechanistic dichotomy in flavoenzyme catalysis and evolution.
Results
V-Shaped Active Site.
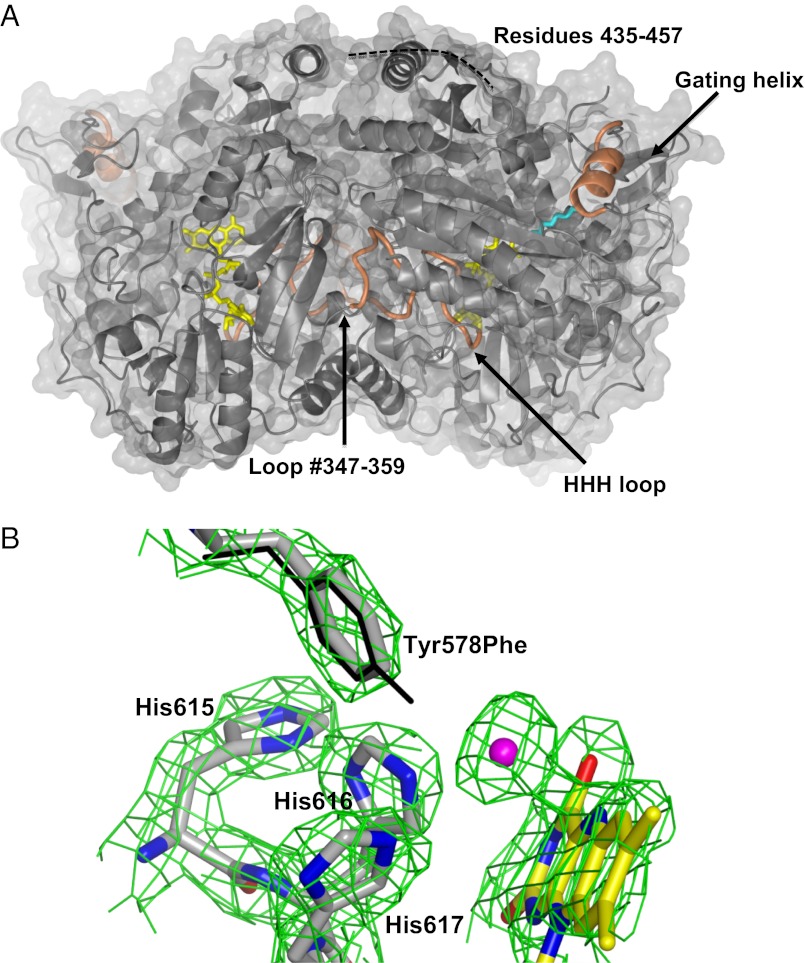
The structure of Cavia porcellus ADPS (93% sequence identical to the human enzyme) was solved at 1.9-Å resolution (Fig. 2 A and B and SI Methods). This protein is a 130-kDa dimer of identical subunits each comprising three domains (N-terminal, FAD-binding, and cap domains). The active site is located in front of the flavin ring at the interface between the FAD and cap domains and comprises residues belonging to both monomers, indicating that the dimeric assembly is essential for catalysis (Fig. 2A). Mammalian ADPS shares 33% sequence identity with the Dictyostelium discoideum enzyme, the crystal structure of which is known (14). Domain superpositions indicate a significant change in the relative domain orientation, with a 14° rotation of the D. discoideum cap domain with respect to the same domain of ADPS (Fig. S1). This change is associated to another key difference; the so-called “HHH” loop is clearly visible in the electron density of the mammalian protein, whereas it is disordered in the Dictyostelium structure (Fig. 2 A and B). This loop is named after the strictly conserved His-His-His motif (residues 615–617), which is essential for activity (14). The HHH loop of ADPS forms a sharp bend that wedges into the active site to directly contact the flavin (Figs. 2 and 3). This conformation is coupled to the ordering of residues 347–359 of a twofold-related subunit. Thus, the structure of ADPS visualizes the conformation of all elements directly involved in catalysis, a critical issue for investigation of the enzyme mechanism.
Fig. 2.
Crystal structure of ADPS. (A) The dimer with the molecular twofold axis vertical in the plane of the paper. The FAD is shown in yellow; the bound aliphatic ligand, in cyan. Loop 347–359, the “HHH loop” (615–622), and the gating α-helix (126–137) are shown in orange. # indicates that loop 347–359 is part of the active site of the twofold related subunit. ADPS is associated to the inner side of the peroxisomal membrane (13). This localization enables the liposoluble fatty alcohol substrate and fatty acid product to directly diffuse to and from the membrane (29). By analogy with the Dictyostelium ADPS structure, the site of membrane association is predicted to be on the upper side of the dimer, in the area embraced by the disordered 435–457 segment (14). (B) The final 2Fo-Fc–weighted electron density map (contoured at the 1.4 σ level) of the active site in the Tyr578Phe mutant. Protein carbons are shown in gray; flavin carbons, in yellow; oxygens, in red; nitrogens, in blue; and chloride, in magenta. The Tyr578 side chain of the WT protein is superimposed and shown with thin bonds. The only detectable change is the strong electron density peak in direct contact with the flavin N5 atom. We have interpreted this peak as a chloride ion bound to flavin, the negative charge of which is compensated for by the His side chains of the HHH loop. This observation supports the role of these His residues in stabilization of the negatively charged enolate form of the substrate (Fig. 1).
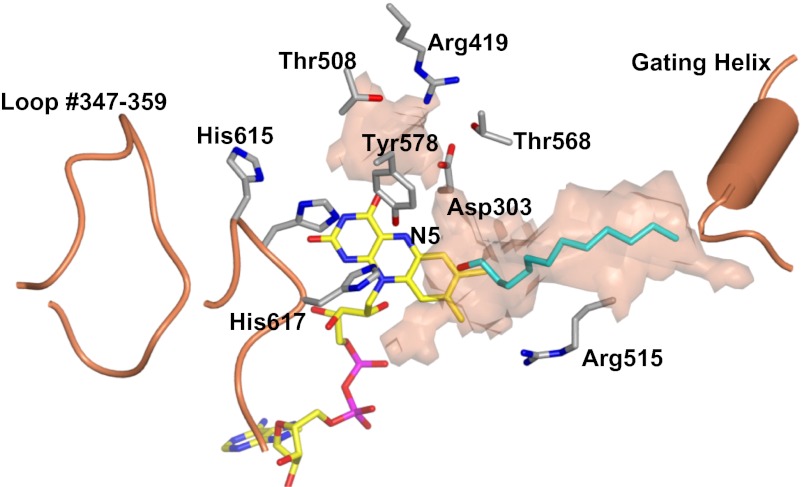
Fig. 3.
Active site of ADPS. Protein carbons are shown in gray; flavin carbons, in yellow; ligand carbons, in cyan; oxygens, in red; nitrogens, in blue; and phosphorous, in magenta. The substrate-binding site tunnel was calculated with a 1.4-Å radius probe. The semitransparent surface outlines the “probe-accessible” region of the tunnel. # indicates residues from the opposite subunit of the dimeric enzyme. The orientation is the same as that of the subunit shown in Fig. 2A.
AcylDHAP, the substrate of ADPS, comprises a fatty chain hydrophobic tail, the central three-carbon unit of DHAP, and the negatively charged phosphate group (Fig. 1). This tripartite nature perfectly matches the architecture of the substrate-binding site. ADPS has a V-shaped tunnel that runs across the cap–FAD domain interface from the gating helix to Arg419 (Fig. 3). The longest arm of the tunnel represents the binding site for the aliphatic portion of the substrate. Indeed, its electron density indicates that this segment of the active site tunnel is occupied by a long-chain molecule (possibly a PEG molecule used for crystallization). The tunnel length matches that expected for 16- to 18-carbon ligands, in line with the specificity for hexadecanoyl- and octadecanoyl-DHAP; the enzyme is inactive with shorter-chain substrates (5). The shorter arm of the V-tunnel has a more spherical shape and is lined by several hydrophilic residues, including Arg419 and two Thr residues. These features make it well suited to host the substrate phosphate group. The two arms are connected through a narrow constriction defined by His616, His617, the flavin ring, and Tyr578. Importantly, the constriction is located exactly in front of reactive locus of the cofactor, the N5 atom (Fig. 3). This arrangement of the flavin with His and Tyr side chains generates the catalytic armamentarium underlying the ether bond formation carried out by ADPS.
Pathological Mutations Targeting Functionally Crucial Residues.
We produced and studied five active-site mutations, three of them (see Fig. S2) found in patients affected by rhizomelic chondrodysplasia punctata (10, 12). Above all, the point emerging from these experiments is that all mutations completely inactivate the enzyme. We found that three of these mutations (Thr309Ile, Leu469Pro, and Arg515Leu) were defective in their ability to bind FAD, associated with poor stability of the apoenzyme (Fig. S2 A and B). The other two mutations (Arg419His and Tyr578Phe) exhibited normal stability, and their crystal structures showed no large conformational changes. However, none of the mutations was enzymatically active, and incubation with acylDHAP caused no spectral perturbation (Reactivity of the Flavin Cofactor). Arg419His targets a positively charged residue that is part of the short arm of the active site V-tunnel (Fig. 3). Crystal structure analysis showed that substitution of the long Arg side chain with a less bulky (but polar) His group causes a small (up to 1.5 Å) shift in the central portion of helix α15, located near the site of mutation (Fig. S2C). Otherwise, the protein does not exhibit any alteration with respect to the WT conformation. Similarly, the Tyr578Phe structure simply shows that the hydroxyl group of the Tyr side chain can be removed without causing any structural changes (Fig. 2B). The Arg419His and Tyr578Phe mutants are very informative, in that their lack of reactivity indicates that the mutated residues are essential for substrate binding and/or for performing an essential step of the catalytic reaction.
Reactivity of the Flavin Cofactor.
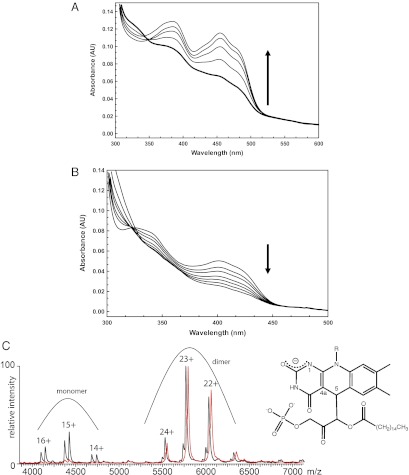
The present study is based on an improved expression system that produces a protein with enhanced stability and, above all, far less propensity for aggregation compared with previously used recombinant enzymes (13). These properties make the enzyme more amenable to biochemical studies, although it must be noted that ADPS substrates are water-insoluble and tend to cause slow protein unfolding and aggregation. The key initial observation was that incubation of the enzyme with palmitoylDHAP led to rapid bleaching of the cofactor. The absorbance spectrum indicates that the flavin becomes either two electron-reduced or covalently modified by the substrate (Fig. 4A). This bleached form decays to the original oxidized state within 10–20 min under both aerobic and anaerobic conditions, implying that the entire process is completely oxygen-independent. We found that regeneration of the oxidized enzyme resulted in the formation of two molecules, palmitic acid (detected by MS analysis of the reaction mixture) and DHAP, effectively detected using an assay based on triose phosphate isomerase (Fig. S3). Importantly, the bleached cofactor appeared to be stable only when enzyme-bound, given that protein unfolding led to the immediate formation of oxidized flavin. Again, unfolding under anaerobic condition did not prevent flavin reoxidation. These data suggest that the two electron-reduced form of the flavin probably is not the intermediate, and support the idea that the bleached form of the cofactor is a covalent adduct generated by the reaction with substrate. The adduct undergoes rapid hydrolysis in water, but is insensitive to oxygen.
Fig. 4.
Spectroscopic studies on ADPS reactivity. (A) Formation and decay of the “bleached” form of the enzyme generated by the incubation with acylDHAP. The enzyme [10 μM in 50 mM NaCl, 5% (vol/vol) glycerol, and 50 mM Tris⋅HCl (pH 8.0)] was added with 100 μM palmitoylDHAP (final). This immediately led to the bleaching of the 450-nm peak (bold line). The protein reoxidized spontaneously (at 1, 3, 10, and 30 min) and at the end of this process exhibited the spectrum of the oxidized state. Reoxidation occurs under anaerobic conditions as well. (B) Reaction of 5-deazaFAD–bound ADPS with palmitoylDHAP. The enzyme reacted slowly with the substrate to form a highly stable complex, accompanied by bleaching of the absorbance spectrum. The experimental conditions were the same as in A, except for the protein concentration (8 μM). Spectra were measured at 0, 1, 2, 5, 10, 30, and 60 min. (C) Native MS analysis of ADPS bound to 5-deazaFAD and acylDHAP. Shown is an overlay of the mass spectra of 5-deazaFAD protein incubated with palmitoylDHAP (red) and without addition of the substrate (black). The analysis indicates a shift in the molecular mass of ∼800 ± 50 Da for the dimeric enzyme (400 ± 25 Da per monomer; the expected mass of palmitoylDHAP is 406 Da). (Right) Proposed structure of the covalent adduct formed by palmitoylDHAP with 5-deazaflavin. Relevant flavin atoms are numbered.
These findings led us to explore the possibility that the covalent intermediate may result from the nucleophilic addition of acylDHAP on the cofactor (Fig. 1). Along this line, we probed ADPS for its reactivity with nitroalkanes, anionic forms of which are potentially able to nucleophilically attack the flavin (15, 16). We found that ADPS reacts with both nitroethane and nitrooctane, as indicated by the bleaching of the flavin on incubation of the enzyme with these compounds (Fig. S4 A and B). Remarkably, adduct formation caused no alteration in the active site geometry, including flavin conformation, which remained planar (Fig. S4C). These results are fully consistent with the idea that ADPS is able to react with nucleophilic reagents, giving rise to the formation of covalent adducts with the cofactor.
Flavin Analog to Trap a Catalytic Intermediate.
We attempted to extensively characterize the compound formed by incubating the enzyme with acylDHAP. The fundamental problem was that the compound decayed instantaneously on release from the protein (i.e., on protein unfolding), precluding MS analysis. The addition of cyanide, sodium borohydride, or other reducing agents did not alter the formation, stability, and decay of the intermediate. Native MS on palmitoylDHAP-incubated enzyme yielded inconclusive results, reflecting the relative instability of the intermediate, which prevented extensive buffer exchange as required by MS. For these reasons, we attempted to reconstitute the enzyme with a modified FAD in which the nitrogen in position 5 of the flavin is replaced by a carbon (5-deazaFAD). The rationale for this experiment was two-pronged: (i) This analog probes the role of the flavin N5 locus, and (ii) 5-deazaflavin is known to be reactive toward nucleophilic reagents, thereby representing a potential tool for probing the enzymatic mechanism (17–19). The 5-deazaFAD–reconstituted enzyme exhibited very low activity (3–5% compared with the native protein), likely reflecting the presence of a small fraction of FAD-bound protein, given that the same degree of activity was observed for the apoenzyme.
A most enlightening result was obtained by incubating the 5-deazaFAD ADPS with the substrate. The addition of palmitoylDHAP led to slow bleaching of the longer-wavelength absorption band in the absorbance spectrum (Fig. 4B), and, most importantly, the spectral changes lasted for several hours. In contrast, as soon as the protein was unfolded, the released 5-deazaFAD acquired the standard spectrum of the oxidized state, a process that cannot be a simple oxygen-mediated reoxidation, because oxygen does not react with this flavin analog (17). Thus, 5-deazaFAD reacts with the substrate to form a complex that is very stable as long as it remains protein-bound (SI Methods). Moreover, the observed spectral changes are fully consistent with those exhibited on formation of covalent adducts between carbon 5 of protein-bound 5-deazaflavins and nucleophilic reagents (17, 19) (Fig. 4B).
We further characterized this complex by native MS, which is a technically demanding analysis because the reconstituted enzyme preparations (although mainly dimeric and cofactor-bound) contain molecules that are monomeric and/or in the apo form. These heterogeneities cause broadening of the peaks in the acquired mass spectra, hampering accurate mass assignments. We repeated the analysis several times using two different protein preparations. The observed mass shifts (800 ± 50 Da for the dimeric enzyme) consistently indicated that an entire palmitoylDHAP molecule is bound to each protein chain (Fig. 4C). The fundamental conclusion that can be drawn from these MS experiments is that, consistent with the absorbance spectroscopy results, 5-deazaflavin forms a tight, most likely covalent, complex with the substrate to the extent that the enzymatic reaction cannot proceed any further, in line with the observed lack of activity with 5-deazaFAD.
Discussion
Taken together, our structural and biochemical data indicate that at the heart of the ADPS reaction lies the formation of a covalent adduct with the substrate to enable acyl–alkyl exchange (Fig. 1). The reaction is predicted to start with formation and/or preferential binding of the enolate form of the substrate, which can subsequently attack the flavin. A key finding supporting this model is that the pro R proton (or hydrogen) at the C1 carbon of the DHAP moiety exchanges stereospecifically with solvent during the reaction (6, 7). Tyr578 is the residue most likely responsible for the acid/base catalysis underlying substrate tautomerization. Indeed, Tyr578Phe is inactive, and incubation with the substrate does not lead to flavin bleaching, implying that the substrate is unable to react with the flavin bound to the mutant. The His residues of the HHH loop are ideally located to stabilize the negatively charged oxygen of the enolic substrate (Fig. 2B). Consistent with this, mutations of each of the three loop histidines (His615, His616, and His617) to Ala have been shown to completely abolish activity (13).
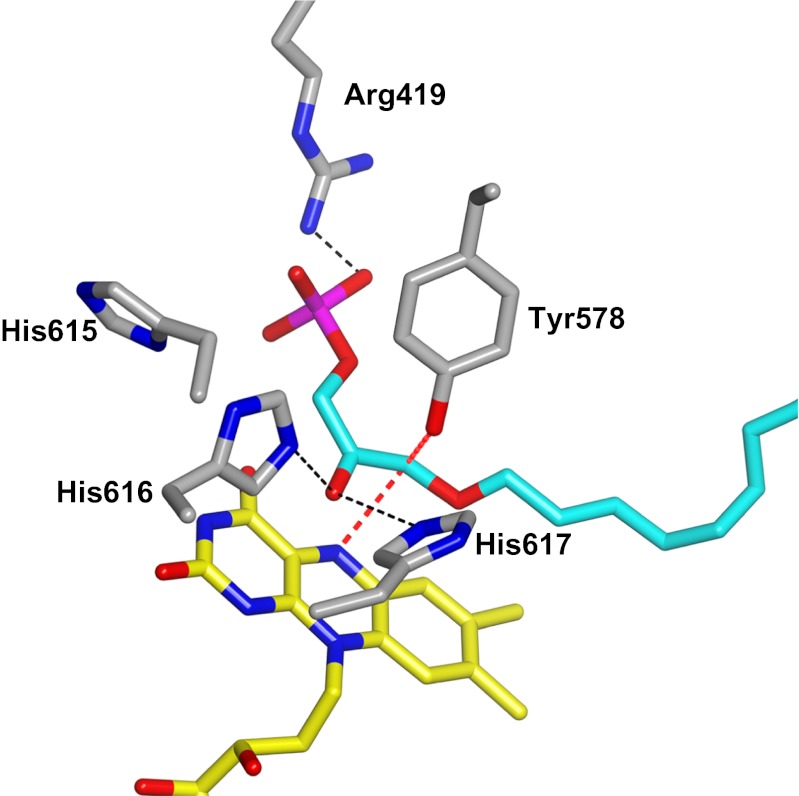
Formation of the enolate form of DHAP allows the key step of the reaction: the nucleophilic attack of C1 of DHAP onto the flavin. This step is followed by the release of the fatty acid product to form the covalent complex between hydroxyacetonephosphate and the flavin (Fig. 1). Much data support the formation of this crucial covalent intermediate. First, the intermediate exhibits spectral features fully compatible with those of a flavin covalent adduct (Fig. 4A). Second, the fatty acid product retains both carboxyl ester oxygens on cleavage from DHAP (20–22). Third, decay of the intermediate releases DHAP through an hydrolytic process that does not require oxygen or other electron acceptors. Fourth, the ADPS-bound flavin is capable of reacting with the nucleophilic nitroalkanes to form a covalent adduct on the N5 position, which can be accommodated in the active site without alterations in flavin conformation and environment (Fig. S4C). Fifth, protein-bound 5-deazaFAD is able to react with the substrate, consistent with the known reactivity of this flavin analog toward nucleophilic agents. In this case, however, the presence of a CH group instead of a nitrogen in position 5 of the flavin prevents elimination of the fatty acid and continuation of the reaction (Fig. 4C). Finally, two structural features underlying this mechanism are of special interest (Figs. 3 and 5). The binding site for the DHAP phosphate enables proper “in-phase” binding of the DHAP C1 atom with respect to the flavin, and the fine geometry of the V-shaped active site tunnel positions a constriction in direct contact with flavin N5, where the substrate–flavin covalent bond must form.
Fig. 5.
Substrate binding to ADPS. To aid data analysis, a 3D model for the bound alkylDHAP product was generated based on the prediction that the alkyl chain binds in the long arm of the catalytic tunnel, whereas the phosphate group binds in the other arm, establishing a direct electrostatic interaction with the essential Arg419. We found that modeling of the alkylDHAP product in its cis enolate form (5, 20) fits very well in the active site and requires only a small shift of Tyr578, which is relatively flexible in the crystal structures. Possible H-bond interactions are depicted as black dashed lines. The red lines indicate contacts of the substrate C1 atom with Tyr578 and flavin N5. Atoms are colored as in Fig. 3.
The fatty alcohol binds in the hydrophobic arm of the active site in place of the exiting fatty acid, which is consistent with a ping-pong mechanism (7). The hydroxyacetonephosphate–flavin intermediate does not react with either borohydride or cyanide, indicating that the enzyme stabilizes mainly the ketonic and/or enolic form of the compound rather than the imine (Fig. 1). Thus, the alcohol substrate, possibly after deprotonation by Tyr578, can react with the intermediate by simply substituting for the flavin to generate the ether-containing product. By analogy, the hydroxyacetonephosphate–flavin intermediate can be expected to react with water to release DHAP. This reaction occurs rapidly on solvent exposure (e.g., protein unfolding) or more slowly in the active site, which thus is able to protect the intermediate from unwanted reactions with the solvent.
This mechanism is most unusual for a flavoenzyme. A well-known example of covalent flavoenzyme catalysis is that of UDP-galactopyranose mutase, the reaction of which, however, requires a reduced flavin, as opposed to the catalytically competent oxidized flavin of ADPS (23). Covalent trapping of a substrate group through an imine intermediate is a catalytic “strategy” that seems rather more typical for pyridoxal phosphate-dependent proteins. Numerous flavoenzymes are known to react covalently with mechanism-based inhibitors, often through poorly understood reactions. Importantly, covalent flavin adducts are not necessarily irreversible. Covalent but reversible inhibitor formation has been reported for monoamine oxidases (24), and reversible adducts with nucleophilic reagents have been documented for enzyme-bound 5-deazaflavins (17, 19). A fascinating property of ADPS is its ability to turn this potential for covalent reactivity into a tool for catalysis. The subtle distinction between redox and covalent reactivity is illustrated by the observation that the flavin-binding site of ADPS shares many features with these sites of flavoenzyme oxidases and dehydrogenases of the vanillyl-alcohol oxidase structural family, of which ADPS is a member (25). Among the common features (Fig. S2A) are a Asp–Pro pair (Pro202–Asp203) in contact with the N5 atom of the cofactor and conserved H-bonding interactions between the flavin and protein backbone atoms (Ser319). These H bonds might help stabilize the predicted negatively charged flavin of the covalent intermediate formed by reaction with the substrate (Fig. 1 and Fig. S2A).
Another related observation is the recurrent presence in alcohol dehydrogenases of a His residue located with respect to the flavin as His617 of ADPS (26) (Fig. S5). In the dehydrogenases, the His side chain is crucial for catalysis by abstracting a proton from the substrate hydroxyl group to promote substrate dehydrogenation through hydride transfer to the cofactor. In contrast, the histidine of ADPS stabilizes the enolic form of DHAP to promote formation of a covalent intermediate with the flavin. Apparently, no drastic alterations in the geometry of the flavin site are needed to implement an unusual covalent catalysis starting from a widespread “redox” flavoenzyme scaffold.
Methods
Protein expression, purification, and crystallization, as well as crystal structure analysis, were performed following standard protocols (27), as described in SI Methods. X-ray data were collected at the European Synchrotron Radiation Facility in Grenoble, France and the Swiss Light Source in Villigen, Switzerland. Enzyme activities were assayed as described elsewhere (14, 28). Reconstitution with 5-deazaFAD took advantage of the relatively weak binding of the protein to FAD, as described in SI Methods. UV-Vis absorbance spectra were recorded with either an Agilent HP8453 diode array or a Varian Cary 100 spectrophotometer using a 100-μL cuvette with a path length of 1 cm. Native MS experiments with 5-dezaFAD–reconstituted ADPS were performed on a modified Waters Q-Tof 2 mass spectrometer. Buffer exchanges into 150 mM ammonium acetate (pH 7.5) were performed using 30-kDa molecular weight cutoff spin-filter columns (Vivaspin500; Sartorius Stedim Biotech). Samples were sprayed at a concentration of 10 μM using gold-coated borosilicate capillaries created in house using a Sutter P-97 puller and an Edwards Scancoat six sputter coater. MassLynx V4.1 (Waters) was used for experimental mass determination.
Supplementary Material
Acknowledgments
This study was supported by Telethon Grant GGP12007. A visiting professorship was supported by Fondazione Cariplo Grant 2008.3148 (to D.E.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.F.P. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4BBY, 4BC7, 4BC9, and 4BCA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215128109/-/DCSupplemental.
References
- 1.Gorgas K, Teigler A, Komljenovic D, Just WW. The ether lipid-deficient mouse: Tracking down plasmalogen functions. Biochim Biophys Acta. 2006;1763(12):1511–1526. doi: 10.1016/j.bbamcr.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 2.Wallner S, Schmitz G. Plasmalogens the neglected regulatory and scavenging lipid species. Chem Phys Lipids. 2011;164(6):573–589. doi: 10.1016/j.chemphyslip.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Lizard G, Rouaud O, Demarquoy J, Cherkaoui-Malki M, Iuliano L. Potential roles of peroxisomes in Alzheimer’s disease and in dementia of the Alzheimer’s type. J Alzheimers Dis. 2012;29(2):241–254. doi: 10.3233/JAD-2011-111163. [DOI] [PubMed] [Google Scholar]
- 4.Braverman NE, Moser AB. Functions of plasmalogen lipids in health and disease. Biochim Biophys Acta. 2012;1822(9):1442–1452. doi: 10.1016/j.bbadis.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Davis PA, Hajra AK. Stereochemical specificity of the biosynthesis of the alkyl ether bond in alkyl ether lipids. J Biol Chem. 1979;254(11):4760–4763. [PubMed] [Google Scholar]
- 6.Friedberg SJ, Gomillion M. Hydrogen exchange in the formation of dihydroxyacetone phosphate from acyl dihydroxyacetone phosphate in O-alkyl lipid synthesis in Ehrlich ascites tumor cell microsomes. J Biol Chem. 1981;256(1):291–295. [PubMed] [Google Scholar]
- 7.Brown AJ, Snyder F. Alkyldihydroxyacetone-P synthase: Solubilization, partial purification, new assay method, and evidence for a ping-pong mechanism. J Biol Chem. 1982;257(15):8835–8839. [PubMed] [Google Scholar]
- 8.White AL, Modaff P, Holland-Morris F, Pauli RM. Natural history of rhizomelic chondrodysplasia punctata. Am J Med Genet A. 2003;118A(4):332–342. doi: 10.1002/ajmg.a.20009. [DOI] [PubMed] [Google Scholar]
- 9.Braverman N, et al. A Pex7 hypomorphic mouse model for plasmalogen deficiency affecting the lens and skeleton. Mol Genet Metab. 2010;99(4):408–416. doi: 10.1016/j.ymgme.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vet EC, Ijlst L, Oostheim W, Wanders RJ, van den Bosch H. Alkyl-dihydroxyacetonephosphate synthase: Fate in peroxisome biogenesis disorders and identification of the point mutation underlying a single enzyme deficiency. J Biol Chem. 1998;273(17):10296–10301. doi: 10.1074/jbc.273.17.10296. [DOI] [PubMed] [Google Scholar]
- 11.Nimmo G, et al. Rhizomelic chrondrodysplasia punctata type 2 resulting from paternal isodisomy of chromosome 1. Am J Med Genet A. 2010;152A(7):1812–1817. doi: 10.1002/ajmg.a.33489. [DOI] [PubMed] [Google Scholar]
- 12.Thai TP, et al. Impaired membrane traffic in defective ether lipid biosynthesis. Hum Mol Genet. 2001;10(2):127–136. doi: 10.1093/hmg/10.2.127. [DOI] [PubMed] [Google Scholar]
- 13.de Vet EC, Hilkes YH, Fraaije MW, van den Bosch H. Alkyl-dihydroxyacetonephosphate synthase: Presence and role of flavin adenine dinucleotide. J Biol Chem. 2000;275(9):6276–6283. doi: 10.1074/jbc.275.9.6276. [DOI] [PubMed] [Google Scholar]
- 14.Razeto A, et al. The crucial step in ether phospholipid biosynthesis: Structural basis of a noncanonical reaction associated with a peroxisomal disorder. Structure. 2007;15(6):683–692. doi: 10.1016/j.str.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Héroux A, Bozinovski DM, Valley MP, Fitzpatrick PF, Orville AM. Crystal structures of intermediates in the nitroalkane oxidase reaction. Biochemistry. 2009;48(15):3407–3416. doi: 10.1021/bi8023042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valley MP, Tichy SE, Fitzpatrick PF. Establishing the kinetic competency of the cationic imine intermediate in nitroalkane oxidase. J Am Chem Soc. 2005;127(7):2062–2066. doi: 10.1021/ja043542f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorns MS, Hersh LB. Nucleophilic addition reactions of free and enzyme-bound deazaflavin. J Biol Chem. 1976;251(16):4872–4881. [PubMed] [Google Scholar]
- 18.Chan RL, Bruice TC. The chemistry of an electron-deficient 5-deazaflavin: 8-Cyano-10-methyl-5-deazaisoalloxazine. J Am Chem Soc. 1977;99(20):6721–6730. doi: 10.1021/ja00462a041. [DOI] [PubMed] [Google Scholar]
- 19.Jorns MS, Ballenger C, Kinney G, Pokora A, Vargo D. Reaction of enzyme-bound 5-deazaflavin with peroxides: Pyrimidine ring contraction via an epoxide intermediate. J Biol Chem. 1983;258(14):8561–8567. [PubMed] [Google Scholar]
- 20.Friedberg SJ, Weintraub ST, Singer MR, Greene RC. The mechanism of ether bond formation in O-alkyl lipid synthesis in Ehrlich ascites tumor: Unusual cleavage of the fatty acid moiety of acyl dihydroxyacetone phosphate. J Biol Chem. 1983;258(1):136–142. [PubMed] [Google Scholar]
- 21.Brown AJ, Glish GL, McBay EH, Snyder F. Alkyldihydroxyacetonephosphate synthase mechanism: 18O studies of fatty acid release from acyldihydroxyacetone phosphate. Biochemistry. 1985;24(27):8012–8016. doi: 10.1021/bi00348a026. [DOI] [PubMed] [Google Scholar]
- 22.Friedberg SJ, Satsangi N, Weintraub ST. Stereochemistry of the acyl dihydroxyacetone phosphate acyl exchange reaction. J Lipid Res. 1991;32(2):259–266. [PubMed] [Google Scholar]
- 23.Gruber TD, Westler WM, Kiessling LL, Forest KT. X-ray crystallography reveals a reduced substrate complex of UDP-galactopyranose mutase poised for covalent catalysis by flavin. Biochemistry. 2009;48(39):9171–9173. doi: 10.1021/bi901437v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binda C, et al. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proc Natl Acad Sci USA. 2003;100(17):9750–9755. doi: 10.1073/pnas.1633804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattevi A, et al. Crystal structures and inhibitor binding in the octameric flavoenzyme vanillyl-alcohol oxidase: The shape of the active-site cavity controls substrate specificity. Structure. 1997;5(7):907–920. doi: 10.1016/s0969-2126(97)00245-1. [DOI] [PubMed] [Google Scholar]
- 26.Dym O, Pratt EA, Ho C, Eisenberg D. The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme. Proc Natl Acad Sci USA. 2000;97(17):9413–9418. doi: 10.1073/pnas.97.17.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zomer AW, Michels PA, Opperdoes FR. Molecular characterisation of Trypanosoma brucei alkyl dihydroxyacetone-phosphate synthase. Mol Biochem Parasitol. 1999;104(1):55–66. doi: 10.1016/s0166-6851(99)00141-3. [DOI] [PubMed] [Google Scholar]
- 29.Forneris F, Mattevi A. Enzymes without borders: Mobilizing substrates, delivering products. Science. 2008;321(5886):213–216. doi: 10.1126/science.1151118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.