Abstract
The relatively short read lengths associated with the most cost-effective DNA sequencing technologies have limited their use in de novo genome assembly, structural variation detection, and haplotype-resolved genome sequencing. Consequently, there is a strong need for methods that capture various scales of contiguity information at a throughput commensurate with the current scale of massively parallel sequencing. We propose in situ library construction and optical sequencing on the flow cells of currently available massively parallel sequencing platforms as an efficient means of capturing both contiguity information and primary sequence with a single technology. In this proof-of-concept study, we demonstrate basic feasibility by generating >30,000 Escherichia coli paired-end reads separated by 1, 2, or 3 kb using in situ library construction on standard Illumina flow cells. We also show that it is possible to stretch single molecules ranging from 3 to 8 kb on the surface of a flow cell before in situ library construction, thereby enabling the production of clusters whose physical relationship to one another on the flow cell is related to genomic distance.
Keywords: molecular biophysics, transposase, jumping reads
Massively parallel, short read sequencing technologies are inherently limited with respect to several key goals, including the resequencing of segmental duplications and structurally complex regions of the human genome, the resolution of haplotype information in diploid and polyploid genomes, and the de novo assembly of complex genomes. Further reductions in the cost-per-base of sequencing will do little to advance us toward these goals. Rather, equivalently parallel methods of obtaining contiguity information at different scales are required. For example, the fact that the original de novo assemblies of the human and mouse genomes achieved a high quality (1, 2), despite an order-of-magnitude less sequence coverage than lower quality assemblies based on short reads alone (3–5), is primarily a consequence of the inclusion of a broad spectrum of complementary sources of contiguity information, including (i) long primary read lengths; (ii) mate-paired reads from plasmids, fosmids, and BACs; (iii) hierarchical clone-by-clone sequencing; and (iv) genetic maps.
Even as new approaches to DNA sequencing mature and surpass current technology, it may remain the case that the best technologies in terms of cost-per-base are read length limited. If so, how will we obtain contiguity information? One possibility is the supplementation of low-cost, short-read sequence with contiguity information obtained by other technologies. Approaches to generate this information fall into six categories: (i) long-range “jumping read” protocols enable one to obtain read pairs separated by a controlled distance. These methods are currently the gold standard in the field and have been used to achieve some of the goals outlined above. For example, 3-kb insert mate pairs were used to reveal extensive structural variation in the human genome (6); 10-kb inserts were used to detect more complex structural variation in epithelial cancer genomes (7); and 40-kb fosmid inserts enabled the de novo assembly of the mouse genome with an N50 scaffold length comparable to the draft assembly (8). These experimental results are consistent with early computer simulations demonstrating improved assembly quality with increasing insert sizes from 1.2 to 10 kb (9). However, all current “jumping read” protocols use circularization or in vivo steps, such that these methods are laborious, limited in efficiency, and have a maximum insert size of 40 kb due to the use of fosmid vectors. (ii) Bar coding and sequencing of clone dilution pools (or their in vitro equivalent) can yield haplotype information on a genome-wide scale (10–12). However, the resolution of the method is limited to the types of fragments (e.g., fosmid or whole-genome amplification products) and number of pools that one can efficiently process. (iii) In vitro protocols for molecular tagging enable the hierarchical assembly of locally derived reads. However, current methods are limited to ∼1-kb “subassemblies” (13). (iv) Optical mapping using restriction enzymes or fluorescent probes has been successful in generating long-range contiguity maps for de novo genome assembly (14–17). However, these processes are limited by false-positive and -negative cut sites due to star activity, inefficient cleavage, or mishybridization, necessitating multiple optical maps from the same region to generate a consensus map. Furthermore, the nonuniform distribution of restriction enzyme recognition sites can limit the amount of useful information derived from repetitive or low complexity regions. (v) Optical sequencing on stretched single DNA molecules has yielded up to 3 bp of contiguous sequence information from multiple locations along the same molecule (18). Because reads are generated directly from single molecules, issues of sample quantity and PCR bias are largely avoided. However, significant technical hurdles (e.g., read length, instrumentation) must be overcome before optical sequencing on single molecules can be realized as a widely useful and robust technology. (vi) Long-read single molecule sequencing using polymerases (19), nanopores (20), or transmission electron microscopy. The Pacific Biosciences RS sequencing platform is limited to read lengths of a few kilobases and suffers from a high intrinsic error rate, whereas the nanopore- and transmission electron microscopy-based methods are still in early development.
Here, we propose in situ library construction and optical sequencing within the flow cells of massively parallel sequencing instruments as an efficient path toward a single technology that simultaneously captures contiguity information and primary sequence at diverse scales. The basic premise is to exploit the physical properties of DNA (random coiling or stretching due to flow or electric current), in situ library construction [via in vitro transposition of adaptors to high–molecular-weight (HMW) DNA within a flow cell], and the fully developed aspects of a widely available massively parallel sequencing instrument (amplification, sequencing-by-synthesis, imaging, and data processing) to generate multiple spatially related reads whose physical separation is either known or can be inferred from the relative coordinates at which the reads originate on the flow cell. In one approach, we take advantage of the random coil configuration adopted by DNA in solution to spatially confine the ends and generate paired-end reads in close proximity. In a second approach, instead of allowing the DNA to adopt a random coil while both ends hybridize, we apply an electric field to stretch the molecules that only have a single end hybridized. This results in the production of clusters whose physical relationship to one another on the flow cell is related to genomic distance.
Results
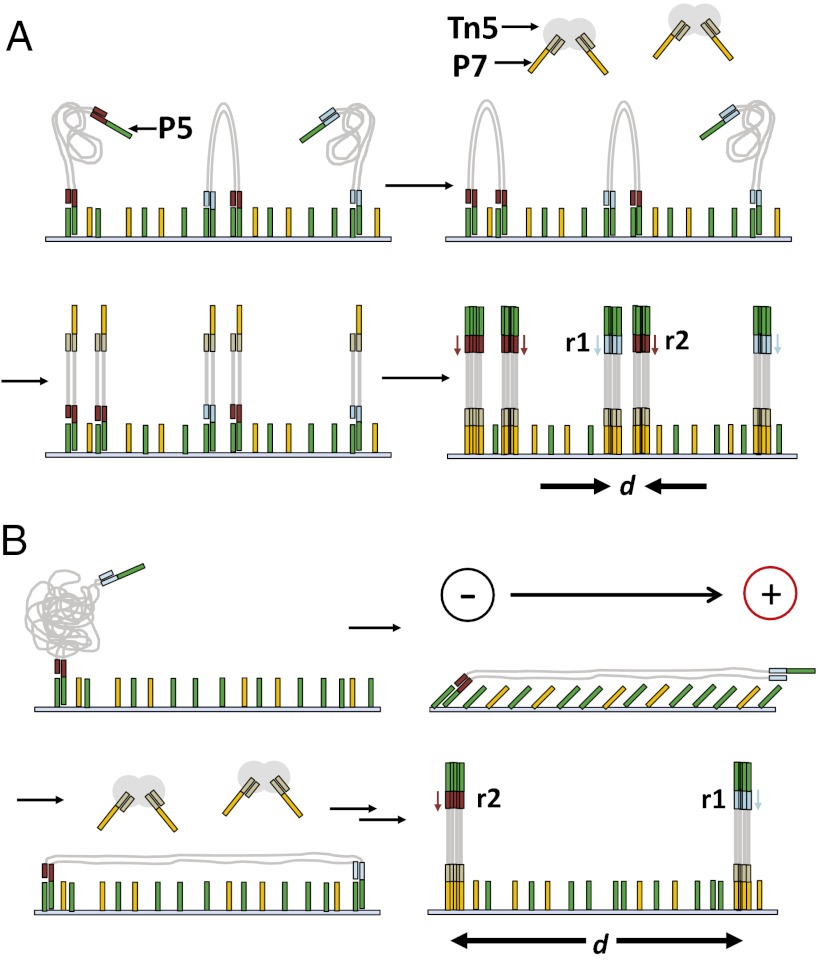
We first sought to develop a method for the in situ library preparation of coiled molecules (Fig. 1A). This required that we generate HMW DNA libraries containing single-stranded flow cell-compatible 3′-tails. Briefly, we physically sheared genomic DNA from Escherichia coli, size selected it for 1- to 8-kb molecules, and then repaired the ends (Materials and Methods). Next, we self-annealed two hairpin adaptors (A and B; Table S1) containing three uracil bases near the loop of the hairpin (Fig. S1) and blunt ligated the adaptors to the size-selected DNA. The loop portion of A and B was designed to be complementary to the same flow cell primer (P5), whereas the double-stranded portion of each hairpin was unique. We removed unligated genomic DNA and adaptors with treatment by exonuclease III and VII to yield an enriched population of molecules with hairpin adaptors on both ends. We then treated the molecules with uracil DNA glycosylase and endonuclease VIII to open the hairpin loop and release single-stranded flow cell complementary 3′-tails. As two hairpin adaptors were used, the resulting population of adaptor-flanked HMW DNA molecules was expected to include A-A (25%), B-B (25%), and A-B (50%) species in terms of their 3′-tails.
Fig. 1.
Sample preparation for in situ library construction. (A) Surface-mediated bridge PCR performs poorly for inserts > 1 kb, which limits the Illumina platform’s ability to generate native long paired-end reads from HMW DNA. To circumvent this, HMW templates were modified with flow cell-compatible adaptors (P5) (Fig. S1) and hybridized under stationary flow. When one template end hybridizes, it spatially confines the other end, thereby increasing the probability that it will also hybridize in close physical proximity. The immobilized templates are subsequently subjected to in situ transposition with transposomes loaded with sequences corresponding to the second flow cell adaptor (P7). Without a transposition event, each template molecule contains only one of the two required flow cell adaptors required to generate a cluster. For templates that are transposed, this process generates two low–molecular-weight templates that are both capable of cluster formation. After bridge PCR amplification, it is anticipated that 50% of the templates will produce two overlapping or closely located clusters that each contain shotgun sequence derived from one or the other end of the HMW molecule. Reads from either end can be deconvolved by using the two different sequencing primers (shown in red and blue). (B) HMW DNA molecules that are annealed at one end can be stretched using an electric field before the other end anneals. In situ transposition then generates cluster pairs derived from the same parent molecule that are separated by a distance proportional to the length of the parent. With these two approaches, we are able to use the information provided by the spatial coordinates at which clusters are generated to infer long-range contiguity.
We hybridized both ends of these molecules to standard Illumina flow cell surfaces at a dilute concentration using a slightly modified thermal cycling protocol (Materials and Methods). Next, we added to each lane a Tn5 transposase that had been loaded with a custom adaptor compatible with hybridization to the other flow cell primer (P7). This randomly fragmented and added adaptors to the surface-hybridized HMW molecules, thereby generating low–molecular-weight sequencing-ready templates. To improve base calling, we backfilled each lane with a human control library before obtaining two separate single-end 36-bp reads (SE36) on an Illumina GAIIx. For the stretching experiments (Fig. 1B), we modified the thermal cycling protocol and buffer conditions during the hybridization step to facilitate the application of an electric field immediately after the hybridization step (Materials and Methods).
Reconstructing Contiguity Information.
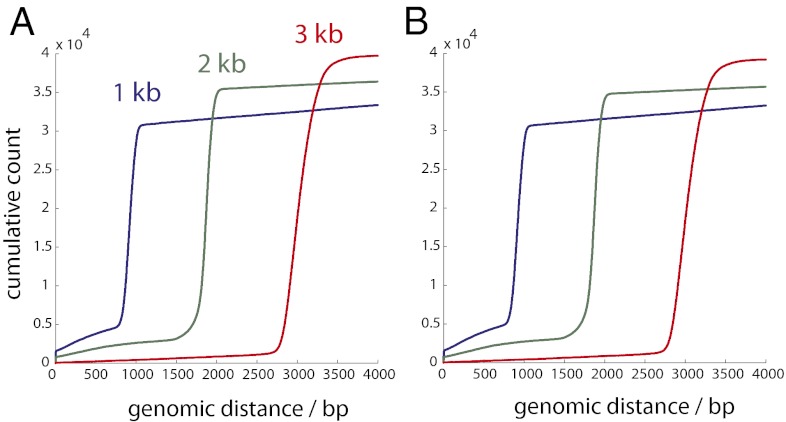
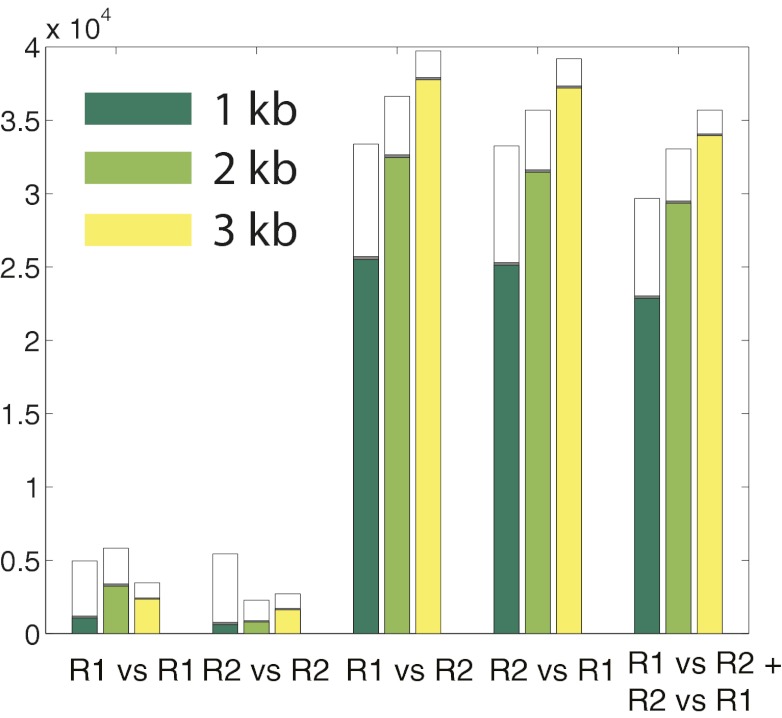
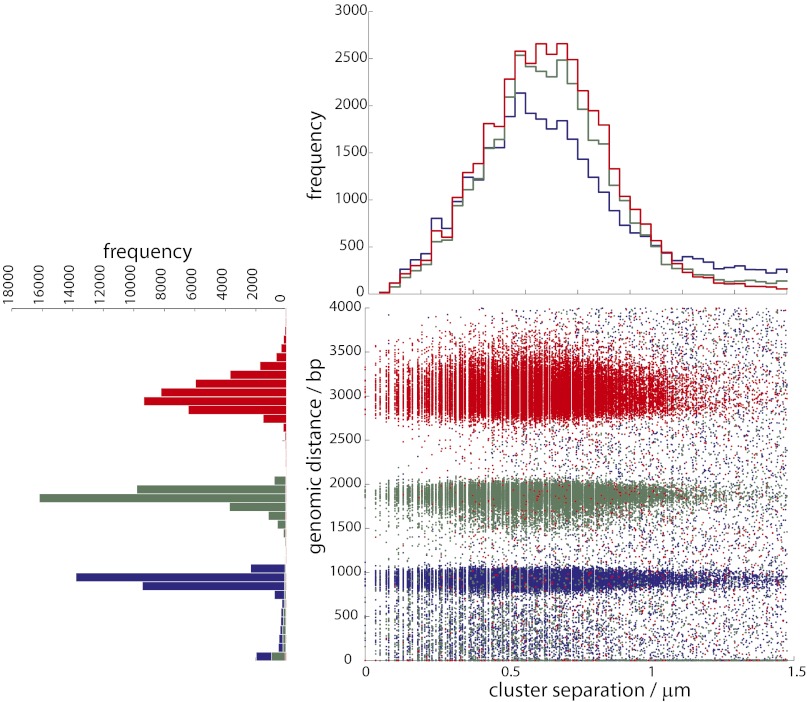
We obtained an average of 3.5 M reads mapped to E. coli in each of three lanes (Table S2). For the 1-, 2-, and 3-kb libraries, we first sought to determine how many clusters in read 1 had a related nearest neighbor in read 2 that mapped to a nearby genomic location. We performed a nearest-neighbor search to identify cluster pairs <1.5 μm apart between reads 1 and 2 (i.e., corresponding to A-B species). For the 3-kb library, we found a total of 696,169 nearest-neighbor pairs with both reads mapped to E. coli. Of these, 38,329 of 696,169 (5.5%) pairs were within 20% of the expected genomic separation (Fig. 2 and Tables S3 and S4) and 38,150 of 38,329 (99%) pairs were in the correct orientation based on the design of the in situ library construction (Fig. 3). By using two primer sequences to serially obtain reads, we were able to deconvolve related cluster pairs that were physically separated by a few nanometers (essentially overlapping) to >1.0 μm (Fig. 4). The mean genomic distances (± SD) were 985 ± 401, 1,846 ± 274, and 2,995 ± 361 bp for the 1-, 2-, and 3-kb libraries, respectively. The mode physical separation distance for the 1-kb pairs was 0.44 μm, and for the 2- and 3-kb pairs it was 0.67 μm, with the tail of the distribution showing some cluster pairs separated by >1.0 μm. These physical separation distances are higher than expected based on a freely jointed chain model of DNA tethered to a surface (Fig. S2) but can be explained as an artifact of cluster formation (SI Note 2). Applying a more restrictive filter that requires mutual exclusivity (i.e., the nearest neighbor of cluster x is y and that of y is x) reduces the number of candidate pairs by up to 10% but does not yield any noticeable gain in specificity (Fig. 3). It was also possible to identify related nearest-neighbor clusters within a single read (i.e., corresponding to A-A or B-B species), but this was limited to cluster pairs that were spatially separated by at least ∼0.9 μm (Fig. S3).
Fig. 2.
For the 1-, 2-, and 3-kb libraries (blue, green, and red), we identified the nearest-neighbor E. coli mapped pairs that were within 1.5 μm of each other and 4,000 bp genomic distance by comparing (A) read 1 against read 2 and (B) read 2 against read 1. The cumulative number of cluster pairs is plotted against the numerically sorted genomic distance for each nearest-neighbor pair.
Fig. 3.
Nearest-neighbor cluster pair data for the 1-, 2-, and 3-kb libraries for different nearest-neighbor searches with E. coli mapped reads. Each search consisted of a reference read and a comparison read. The nearest neighbor for each cluster in the reference read was identified in the comparison read (e.g., for R1 vs. R1, the nearest neighbor for every cluster in read 1 was identified in read 1). The R1 vs. R2 + R2 vs. R1 search represents only pairs that were exclusive nearest neighbors (i.e., the nearest neighbor of X is Y and that of Y is X). The white bars are the total number of cluster pairs with <1.5-μm physical separation and <4,000-bp genomic separation; the gray bars are the number of pairs within the targeted size range for that library size (800–1,200, 1,600–2,400, and 2,400–3,600 bp, respectively); and the colored bars are pairs that also have reads on opposite strands.
Fig. 4.
Scatter plot and histograms showing the relationship between genomic distance and cluster separation. Every cluster in read 1 that had a nearest neighbor in read 2 within 1.5 μm and 4,000 bp genomic distance was identified for the three libraries (blue, 1 kb; green, 2 kb; red, 3 kb).
To estimate our false-positive rate, we measured how often two reads from different, unrelated flow cell tiles would be selected as nearest neighbors. Within this set of NN pairs, we calculated how many pairs were separated by the expected genomic distance for a given library (Tables S3 and S4). For the 3-kb E. coli mapped reads, we found 641,587 NN pairs across different tiles that were within 1.5 µm, but only 155 of 641,587 (0.02%) NN pairs had both reads within the expected mapping distance and in the correct orientation.
We hypothesize that only 5.5% of NN pairs were related due to the low probability for HMW DNA to adopt an appropriate conformation that favors both ends annealing to a surface. The 3D probability distribution for the end-to-end vector of a DNA molecule with one end tethered to a surface indicates that the free end has a much higher probability of being far away from the surface than close to it (SI Note 1). This problem is exacerbated with increasing DNA length. When only one end of a molecule hybridizes and the molecule undergoes transposition, it will generate a singleton read without a related nearest neighbor.
There were a large number of low-quality reads for all three libraries did not map to human, E. coli, transposase mosaic, or adaptor sequences (Table S2). In the 3-kb library, for example, 2,539,680 of 2,755,611 (92%) unmapped reads did not pass a Q30 quality score filter and 1,901,371 of 2,755,611 (69%) had the lowest possible average raw quality score (Fig. S4A). We suspect that they may be due to issues that the cluster finding algorithm has when dealing with mixed, large, and oddly shaped clusters. When we considered all reads and recalculated the nearest-neighbor pairs, 47% of the pairs both mapped to human (internal standard control library), 16% had one read mapped to E. coli and one unmapped read, 10% had both unmapped, and 7% had both mapped to E. coli. For the pairs that had one unmapped and one mapped read, only 6% had an unmapped read with an average raw quality score >30, whereas 78% had the lowest possible raw quality score (Fig. S4B). Although the source of these unmapped reads is unclear, they can largely be filtered out based solely on quality score.
In Situ Stretching and Tagging of HMW Molecules.
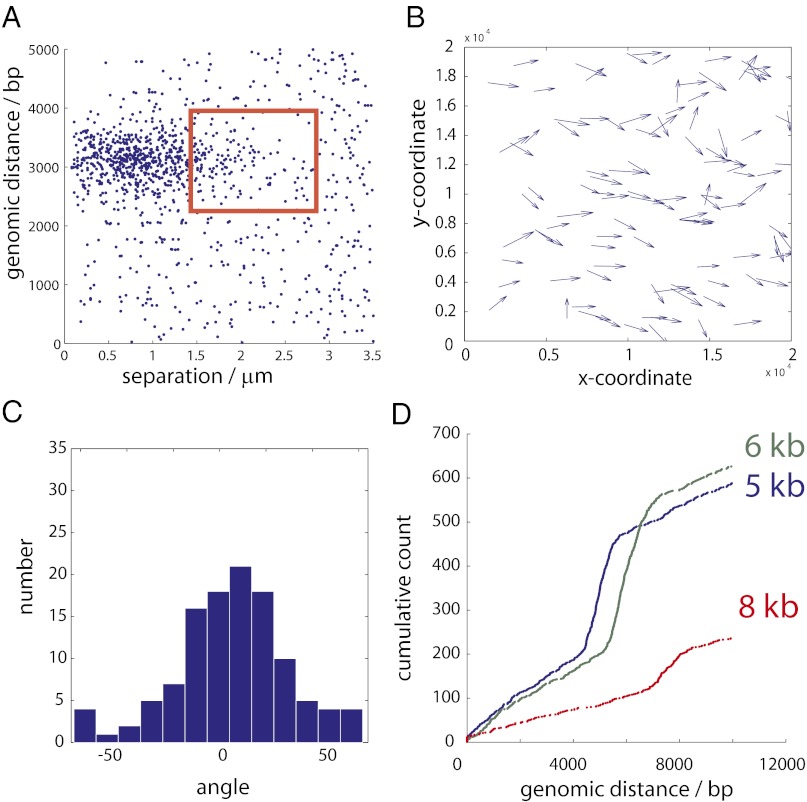
We surmised that it might be possible to (i) improve the hybridization efficiency and (ii) eventually introduce insert size information into the physical separation distance between paired clusters by performing in situ stretching before library construction. Using the 3-kb E. coli library, we attempted to perform in situ stretching within the Illumina flow cell by applying an electric field immediately after the hybridization step. In the absence of an applied electric field, the angle θ between each cluster pair was calculated with respect to the axis of current flow in the chamber. As expected, the angles were uniformly distributed and not related to the distance between paired clusters (Fig. S5). In the presence of the electric field, there were 784 cluster pairs that had a genomic separation of 2,600–3,400 bp and <3.5 μm physical separation. Of these, 669 pairs were separated by <1.5 μm and 115 were separated by 1.5–3.0 μm (Fig. 5A). The distribution of angles for cluster pairs within the 1.5–3.0 μm group significantly deviated from a uniform distribution (χ2 goodness-of-fit test, P = 3.8 × 10−10) and pairs were oriented parallel to the electric field (Fig. 5B). The majority (78%) of the cluster pairs separated by 1.5–3.0 μm had an angle of −45° < θ < +45° with respect to the electric field (Fig. 5C), suggesting that these molecules were stretched before the free ends hybridized to the surface.
Fig. 5.
(A) Scatter plot showing the genomic distance vs. cluster separation distance for the stretched 3-kb library. Cluster pairs that had 2,400–3,600 bp genomic separation and 1.5–3 µm physical separation are enclosed in the red box. (B) Vector plot of the stretched cluster pair orientations from every tile superimposed onto a single “virtual” tile. Each arrow represents a single cluster pair from the red box in A, and the arrow’s length is proportional to the distance between the clusters (not to scale). Electrical current flow is parallel to the x-axis. (C) Histogram of the angle between stretched cluster pairs. 0° corresponds to being oriented in parallel with the direction of current flow; ±90° corresponds with being perpendicular to the current flow. (D) Five-, 6-, and 8-kb libraries (blue, green, and red) were stretched during hybridization. We identified the nearest-neighbor pairs that were within 3.0 μm and 10,000 bp genomic distance by comparing read 1 against read 2. The cumulative number of cluster pairs is plotted against the numerically sorted genomic distance for each nearest-neighbor pair.
Next, we attempted to stretch 5-, 6-, and 8-kb E. coli libraries. These libraries had previously demonstrated low hybridization efficiencies and poor nearest-neighbor pairing with the random coil hybridization approach (Fig. S6). When the field was applied, we detected up to 342 nearest-neighbor pairs within the expected genomic distance for each library (Fig. 5D). Although applying a field significantly reduced the total number of related cluster pairs (from 5.5% of all mapped reads to 0.3% or less), these results demonstrate that in situ stretching and sequencing of HMW DNA can be accomplished within native flow cells. Because the contour length of a 5-kb molecule is 1.7 μm and the average cluster diameter is ∼3 pixels (∼1 μm), we were unable to draw any conclusions on how genomic distance related to physical separation for libraries of this size. We anticipate that longer DNA fragments will produce clusters that are sufficiently spatially separated such that the distance will be related to the length of the parent molecule.
Discussion
We have successfully demonstrated that in situ library preparation of HMW DNA molecules can enable the capture of long-range sequence information up to 8 kb apart on a commercially available sequencing platform. The methods described here hold great promise to overcome the limitations of other contiguity-determining methods by taking advantage of existing sequencing hardware and a single-step enzyme-based in situ library preparation. However, two significant hurdles remain before it can be adopted for widespread use: (i) the efficiency of generating physically related clusters needs to be improved, and (ii) the source of the unmapped reads needs to be identified and mitigated.
There are five key factors that affect the generation of related nearest-neighbor clusters: (i) the production of a clean HMW library with uniform single-stranded flow cell compatible 3′-adaptors. Control experiments without hairpin adaptors resulted in complete degradation of the library, suggesting that ExoIII/VII treatment is highly effective at eliminating any library molecules that do not have two adaptors. Additional control experiments have shown that USER treatment is efficient at uracil excision for making adaptors single stranded, so the current library construction methods are robust. (ii) The hybridization of both ends to the flow cell surface. Although localizing both ends of a molecule near a surface is not favored due to entropic arguments, it is more favorable than the circularization of a same-length single molecule due to the fact that each end can hybridize to any one of thousands of flow cell adaptors. Future work to improve efficiency may be directed at introducing alternating electric current or flow to improve single-stranded tail accessibility during hybridization, or an alternative approach of tethering each 3′-tail to a magnetic bead and using a magnet to draw them to the surface. There is also room to optimize the DNA library concentration, buffer conditions, annealing temperature, and incubation time, and the potential to increase throughput by eliminating the in-lane control library. (iii) The uniform and nondestructive in situ transposition into bridged molecules. We used a coarse range of transposase concentrations and incubation times to identify a working level of activity; future efforts may be directed at optimizing Tn5 loading conditions and in situ activity on surfaces. (iv) The generation of clusters using a nonstandard method. Although there did not appear to be a correlation between physical separation distance and read quality (Fig. S7), it is difficult to assess how our in situ method is influenced by the standard cluster finding and base-calling algorithms. In the method described here, we have limited control of the final length of the related templates after transposition (one could be 200 bp and the other could be 800 bp) and clusters are intentionally seeded in very close proximity. This could result in large, oddly shaped mixed clusters that are not optimized for current algorithms, thereby giving rise to unmapped reads. Additionally, closely spaced molecules with A-A or B-B tails will also give rise to mixed, unmapped reads. (v) The average cluster density in the lane. These experiments were performed at a dilute library concentration (∼10% of standard flow cell cluster densities) to trivialize our nearest-neighbor finding algorithm. As cluster density increases, however, a nearest-neighbor search becomes less likely to find the correct related read pair. If we assume an optimal cluster density and ideal hybridization conditions, we predict that each Illumina GA lane is capable of generating up to 1.5 million NN pairs that are spatially isolated to allow for identification (SI Note 3). For the approach that includes in situ stretching, there are numerous variables that could be optimized to improve pairing efficiency: hybridization buffer conditions, field strength, length of time the field was applied, temperature, and DNA length. It may also be beneficial to adopt a different set of adaptor sequences and introduce a required “bridge” oligonucleotide during stretching, such that only one DNA end can hybridize to the surface until that time (21).
To move beyond 40-kb fosmid jumping reads and enable paired read information on the scale of 100 kb to 1 Mb and beyond, technologies compatible with massively parallel sequencing that do not involve circularization or in vivo manipulation are required. Such methods will be crucial to facilitate routine resolution of complex structural variation, complete haplotype phasing, and accurate de novo assembly of whole genomes. Although in situ library construction and optical sequencing is currently in an early stage of development, with further improvements it offers a promising pathway forward to solve many of these contiguity problems that are not well addressed with current methods. We ultimately envision that the methods described here will lead to the generation of reads with spatially separated coordinates such that their physical relationship is correlated with genomic distance. This could enable the optical sequencing of multiple, ordered reads from many single HMW molecules on existing massively parallel sequencing hardware.
Materials and Methods
Library Construction.
We diluted 10 µL of E. coli type B genomic DNA (10 µg/µL; USB; part 14380) to 200 µL in water and then physically sheared it for 20 s on a Bioruptor (Diagenode). We loaded the entire volume across 12 lanes on a 1% agarose gel and ran it at 100 V for 2 h. We size selected appropriate bands (1–8 kb), purified the DNA (1–3 kb libraries were purified on the Boreal Genomics Aurora or with a Qiagen QIAquick Gel Extraction Kit; 5–8 kb libraries were purified exclusively on the Aurora), and end-repaired the molecules (End-It; Epicentre). This process gave ∼1 µg of end-repaired material for each library size in a total volume of 30 µL. Next, we self-annealed hairpin adaptors (Integrated DNA Technologies) (Table S1) by adding 5 µL of 100 µM stock oligonucleotide to 20 µL of 5× SSC, heating to 95 °C for 5 min, and slowly cooling to room temperature at 0.1 °C/s. We ligated the hairpins adaptors to the size-selected genomic DNA using QuickLigase (NEB) overnight at room temperature [20 µL of end-repaired DNA, 40 µL of QuickLigase buffer, 4 µL of each hairpin adaptor (25 µM), and 12 µL of QuickLigase = 80 µL total volume]. After ligation, we removed unligated genomic DNA and adaptors by adding 1 µL of exonuclease III (NEB) and 0.5 µL of exonuclease VII (Epicentre) and incubating the reaction at 37 °C for 3 h. Finally, we removed the uracil bases by adding 2 µL of USER (NEB) and incubating the reaction at 37 °C for 30 min to generate single-stranded flow cell complementary 3′-tails. We performed a final size selection on a 1% agarose gel run at 100 V for 2 h and quantified the libraries on a Qubit (Invitrogen).
Transposome Loading.
We obtained synthetic DNA oligonucleotides containing transposase mosaic, primer sites, and flow cell adaptor sequence from IDT (Table S1). We annealed the adaptors by mixing 5 µL of each adaptor (100 µM stock) with 40 µL of TE, heating to 95 °C, and slowly cooling to 4 °C. To load the transposase, we mixed 10 µL of stock Tn5 transposase (EzTn5; Epicentre) with 2.5 µL of the double-stranded adaptors (11 µM), 2.5 µL of deionized water, and 5 µL of glycerol. We incubated the solution at room temperature for 20 min and then diluted it with 24 µL of high–molecular-weight buffer (Nextera Kit; Epicentre) and 76 μL of dH2O to give a 1× solution.
In Situ Flow Cell Library Construction and Sequencing.
We wrote a custom cluster generation protocol on a standard Illumina Cluster Station to accommodate template and transposome loading. First, we primed the flow cell with hybridization buffer and heated it to 96 °C at a rate of 1 °C/s. At 96 °C, we loaded a standard Illumina sequencing library into a control lane and hybridization buffer into the other seven lanes. After a 2-min incubation, we lowered the temperature to 65 °C at 0.05 °C/s to hybridize the control library. Next, we removed the tubing on the manifold for the control lane on both the input and output sides of the flow cell. We then added the E. coli libraries to each lane (∼10 pM final concentration diluted in hybridization buffer) at a rate of 15 µL/min for 2.5 min, followed by slowly cooling the flow cell at 0.02 °C/s to a final temperature of 40 °C. After a 5-min incubation, we heated the flow cell at 1 °C/s to 55 °C. We then added the loaded transposomes (0.03×; 1× diluted in HMW buffer) at 15 µL/min to the lanes containing E. coli DNA. We incubated the flow cell at 55 °C for 5 min to allow transposition to take place and then cooled it to 40 °C. Next, we installed a new manifold on the cluster station and injected Illumina wash/amplification buffer across the entire flow cell. We synthesized the first strand with Bst DNA polymerase (NEB) in 1× ThermoPol buffer complemented with 1 mM dNTPs at 65 °C for 5 min and then 74 °C for 5 min. We then backfilled each E. coli lane with a standard human control library to act as an internal control and to improve base calling. We generated clusters with 35 cycles of bridge amplification and obtained two separate single-end 36-bp (SE36) reads on Illumina Genome Analyzer IIx with RTA 1.8 and SBS, version 5.
For the stretching experiments, we loaded template libraries into the flow cell at 75 °C and slowly cooled the chamber at 0.1 °C/s to 55 °C. Next, we connected a 1.5-mL Eppendorf tube containing stretching buffer (5× SSC and 200 mM KCl) to the input and output port of each lane with a short piece of manifold tubing; the output Eppendorf was also connected to the pump with another piece of tubing (fluid flow went from input Eppendorf → flow cell → output Eppendorf → pump). We immersed electrodes in each Eppendorf tube, sealed the output Eppendorf shut with rubber cement, and flowed 120 µL of stretching buffer across each lane. To stretch the DNA, we applied a 28 V/cm electric field across each lane for 2 s. We then flushed 120 µL of wash buffer through the chamber before in situ transposition and sequencing.
Data Collection and Analysis.
We extracted the X–Y coordinates of every cluster from read 1 and read 2 from the fastq files using a custom Perl script. First, we used these data to calculate the image offsets using the normxcorr2 function in MATLAB and corrected the X–Y coordinates for read 2 accordingly. We then mapped read 1 and read 2 separately to either the E. coli genome alone or the E. coli genome, the human genome (HG19), and adaptor/mosaic sequences using the Burrows–Wheeler Aligner (BWA). We determined the identities of nearest-neighbor clusters between read 1 and read 2 using a custom Perl script.
Supplementary Material
Acknowledgments
J.J.S. was funded by a Helen Hay Whitney Foundation postdoctoral fellowship, J.B.H. was funded by a National Research Service Award, and A.A. was funded by a National Science Foundation graduate research fellowship. Funding was also supplied by National Human Genome Research Institute Grant 1R01HG006283-01.
Footnotes
Conflict of interest statement: J.J.S., C.L., J.B.H., A.A., and J.S. have applied for a patent relating to the method described in this study.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202680109/-/DCSupplemental.
References
- 1.Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409(6822):860–921, and errata (2001) 411(6838):720 and (2001) 412(6846):565.
- 2.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8(1):61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillmer AM, et al. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res. 2011;21(5):665–675. doi: 10.1101/gr.113555.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams LJ, et al. Paired-end sequencing of Fosmid libraries by Illumina. Genome Res. 2012 doi: 10.1101/gr.138925.112. 10.1101/gr.138925.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roach JC, Boysen C, Wang K, Hood L. Pairwise end sequencing: A unified approach to genomic mapping and sequencing. Genomics. 1995;26(2):345–353. doi: 10.1016/0888-7543(95)80219-c. [DOI] [PubMed] [Google Scholar]
- 10.Kitzman JO, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol. 2011;29(1):59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat Biotechnol. 2011;29(1):51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters BA, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487(7406):190–195. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiatt JB, Patwardhan RP, Turner EH, Lee C, Shendure J. Parallel, tag-directed assembly of locally derived short sequence reads. Nat Methods. 2010;7(2):119–122. doi: 10.1038/nmeth.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz DC, et al. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262(5130):110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 15.Zhou S, et al. Validation of rice genome sequence by optical mapping. BMC Genomics. 2007;8(1):278. doi: 10.1186/1471-2164-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou S, et al. A single molecule scaffold for the maize genome. PLoS Genet. 2009;5(11):e1000711. doi: 10.1371/journal.pgen.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riehn R, et al. Restriction mapping in nanofluidic devices. Proc Natl Acad Sci USA. 2005;102(29):10012–10016. doi: 10.1073/pnas.0503809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramanathan A, et al. An integrative approach for the optical sequencing of single DNA molecules. Anal Biochem. 2004;330(2):227–241. doi: 10.1016/j.ab.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 19.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 20.Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.