Abstract
Ribulose-1,5-Biphosphate-carboxylase-oxygenase (RuBisCO) has a crucial role in carbon fixation but a slow catalytic rate, a problem overcome in some plant lineages by physiological and anatomical traits that elevate carbon concentrations around the enzyme. Such carbon-concentrating mechanisms are hypothesized to have evolved during periods of low atmospheric CO2. Hornworts, the sister to vascular plants, have a carbon-concentrating mechanism that relies on pyrenoids, proteinaceous bodies mostly consisting of RuBisCO. We generated a phylogeny based on mitochondrial and plastid sequences for 36% of the approximately 200 hornwort species to infer the history of gains and losses of pyrenoids in this clade; we also used fossils and multiple dating approaches to generate a chronogram for the hornworts. The results imply five to six origins and an equal number of subsequent losses of pyrenoids in hornworts, with the oldest pyrenoid gained ca. 100 Mya, and most others at <35 Mya. The nonsynchronous appearance of pyrenoid-containing clades, the successful diversification of pyrenoid-lacking clades during periods with low [CO2], and the maintenance of pyrenoids during episodes of high [CO2] all argue against the previously proposed relationship between pyrenoid origin and low [CO2]. The selective advantages, and costs, of hornwort pyrenoids thus must relate to additional factors besides atmospheric CO2.
Keywords: independent origins
The enzyme most important in photosynthesis is Ribulose-1,5-Biphosphate-carboxylase-oxygenase, or RuBisCO. This enzyme has a low CO2 attraction and catalytic rate, which plants compensate for by increasing their CO2 affinity, synthesizing large quantities of the enzyme, or increasing the CO2 concentration near it (1–3). Among green organisms (algae and land plants), CO2-concentrating mechanisms (CCMs, also referred to as carbon-concentrating mechanisms) have evolved in numerous lineages to increase the CO2:O2 ratio inside their photosynthetic cells. These mechanisms are C4 photosynthesis, Crassulacean acid metabolism, and the pyrenoids found in algae and hornworts (4, 5). Pyrenoids are proteinaceous bodies consisting of up to 90% RuBisCO (6, 7). In algae, CO2 levels near the pyrenoid are raised up to 180 times above the concentration in the rest of the cell, enhancing photosynthesis in aquatic environments where CO2 diffusion is limited (4). The extent to which pyrenoids are crucial to algal CCMs, however, has been controversial because, although all pyrenoid-containing algae have CCMs, not all algae expressing CCMs have pyrenoids (8). Some algal lineages without pyrenoids have passive or active ways of accumulating CO2 and they may rely on carbon anhydrases to create a pool of internal dissolved carbon (DiC) (8), suggesting alternative adaptive advantages of pyrenoids besides RuBisCO concentration and more efficient CO2 reduction.
Hornworts, the sister group to vascular plants (9, 10), have pyrenoids in approximately 100 of their 200 species (11) and are the only embryophytes with such structures. The ultrastructure of hornwort pyrenoids ranges from compact pyrenoids, as found in species of Notothylas, to highly dissected pyrenoids in some Nothoceros and Phymatoceros (12). Additionally, hornwort plastids have a stacked arrangement of thylakoid membranes (grana) that results in the spatial separation of photosystems and increases the efficiency of light capture in terrestrial environments (13). Hornwort grana consist of stacks of short thylakoids and lack end membranes. Therefore, unlike other land plants, hornwort grana are devoid of the membrane “sacs” that enclose intrathylakoid spaces. Presumably, the perpendicular channel thylakoid system in hornwort plastids serves to isolate biochemical processes (13).
Organic isotope discrimination supports a function in CO2 concentration for hornwort pyrenoids (14–18). Mass spectrometry analyses show that hornworts with pyrenoids (e.g., Phaeoceros and Notothylas) have lower compensation points (11–13 vs. 64 ppm) and larger pools of DiC (suggesting an accumulation of inorganic CO2) than the C3 liverworts, the organisms chosen as the reference point, a comparison across a vast phylogenetic distance.
Despite their important function, molecular phylogenies suggest losses of pyrenoids in some hornworts: for example, in the common ancestor of the genera Megaceros (approximately four species) and Phaeomegaceros (10 species) (11, 12). However, these phylogenies only included 11 of the 12 genera of hornworts and 37 species. Taken together, the morphological heterogeneity of hornwort pyrenoids and the losses implied by the previous studies focusing on hornwort phylogenetics raise the question of whether pyrenoid presence indeed is the ancestral state in hornworts, as long thought (18–20), or whether they instead evolved in parallel within hornworts. Answering this question also bears on the issue of which conditions might have favored evolution of pyrenoids.
Based on a fossil-calibrated phylogeny that includes 77 species (32–38% of 200–240 species from all 12 genera), we trace herein the evolution of pyrenoids in hornworts. Our main questions are: (i) How many times have pyrenoids (and putatively a CCM) been gained and lost in hornworts, and (ii) What is the timing of the origin of hornwort pyrenoids? Under the hypothesis that algal pyrenoids evolved at times of low atmospheric CO2 concentrations (21–23), one might expect to find a clustering of their evolution in hornworts during times of especially low CO2 levels and few losses during decreasing CO2 levels.
Results and Discussion
Independent Gains and Losses of the Pyrenoids.
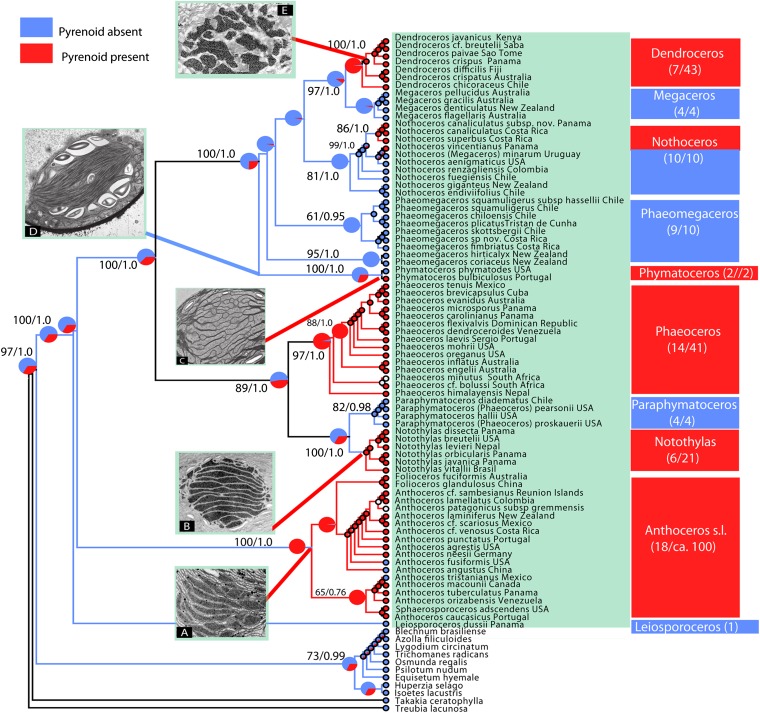
Hornwort pyrenoids, structures associated with a CCM, evolved five to six times independently (Fig. 1). The remaining land plants, as well as Leiosporoceros dussii, the sister species to all other hornworts, lack pyrenoids, suggesting that pyrenoidless plastids are the ancestral condition in hornworts and that pyrenoids first evolved within the hornwort lineage. Our maximum-likelihood (ML) and Bayesian reconstructions (Fig. 1 and Table S1) support the presence of pyrenoids in the Anthoceros-Folioceros-Sphaerosporoceros clade (Anthoceros sensu lato) [maximum-likelihood probability (ML prob.) pyr = 0.99; Bayesian prob. pyr = 0.95] with pyrenoid losses in Anthoceros angustus, Anthoceros fusiformis, Anthoceros tristanianus, and potentially Anthoceros patagonicus subsp. gremmenii. Among those taxa, the plastids of A. fusiformis and A. angustus contain a starch-free area that resembles a pyrenoid but does not concentrate RuBisCO (13). For the Notothyladaceae (Notothylas, Phaeoceros, and Paraphymatoceros), we inferred ancestral lack of pyrenoids with low certainty (ML prob. pyr = 0.53; Bayesian prob. pyr = 0.52), and for Phaeoceros (ML prob. pyr = 0.95; Bayesian prob. pyr = 0.95) and Notothylas (ML prob. pyr= 0.99; Bayesian prob. pyr = 0.99) with high certainty. Paraphymatoceros, the sister genus to Notothylas, is pyrenoidless (ML prob. pyless= 0.92; Bayesian prob. pyless = 0.99). In Phymatocerotaceae, which comprise only two species, one species contains a pyrenoid in the plastid (the European Phymatoceros bulbiculosus); the other lacks pyrenoids (the Californian Phymatoceros phymatodes). In Dendrocerotaceae, the ancestral condition is the absence of pyrenoids (ML prob. pyless = 0.81; Bayesian prob. pyless = 0.99), with three independent reversals to pyrenoid presence: two in Nothoceros and one in the most recent common ancestor (MRCA) of Dendroceros (ML prob. pyr = 0.97; Bayesian prob. pyr = 0.98) (Fig.1).
Fig. 1.
Gains and losses of the pyrenoid (and putative CCM) in hornworts inferred under ML and an asymmetrical two-parameter Markov model. A phylogram showing genetic branch lengths is supplied as Fig. S1. Values at branches show ML bootstrap values and Bayesian posterior probabilities. Pyrenoid presence is shown in red, pyrenoid absence in light blue, and the pie diagrams represent the probability of either character. Pyrenoids evolved from five to six times, namely in Anthoceros sensu lato, Notothyladaceae (Phaeoceros and Notothylas), P. bulbiculosus, some Neotropical Nothoceros, and in Dendroceros. Transmission electronic micrographs show the main pyrenoid ultrastructural types. (A) Anthoceros-type. Sphaerosporoceros adscendens. Present in Anthoceros sensu lato and Phaeoceros, which have a darkly stained pyrenoid is traversed by multiple thylakoids, giving the appearance of multiple pyrenoids. (B) Notothylas-type. Notothylas breutelii. The pyrenoid is typically compact and darkly stained, with single thylakoids traversing the structure. (C) Nothoceros-type. Nothoceros canaliculatus. A lightly stained pyrenoid at the ultrastructural level and less conspicuous in living material; found in P. bulbiculosus and Nothoceros species. (D) Pyrenoidless condition. Phaeomegaceros hirticalyx. Pyrenoidless plastid with abundant short central grana and mostly peripheral starch. (E) Dendroceros-type. Dendroceros crispatus. The pyrenoid is composed of subunits of irregular shape, with numerous electron-opaque inclusions with thylakoids, including grana, interrupting the pyrenoid.
Physiological data suggest that pyrenoid presence in hornworts is related to carbon concentration, analogous to algae with pyrenoids (14–18; but see ref. 8 for algae with CCMs despite lacking pyrenoids) (Table S2). Hornworts without pyrenoids have the RuBisCO enzyme distributed throughout the stroma, similar to the condition in the remaining land plants (6). Hornworts with pyrenoids have a larger DiC pool and lower photorespiration than liverworts, which rely on the C3 pathway (17, 18). Work on Chlamydomonas has shown pyrenoid formation to be controlled by nuclear genes, such as the CIA6 gene family (24) and the nuclear-encoded small subunit of RuBisCO (rbcS) (25). Whether the genes underlying hornwort pyrenoid formation are the same is not yet known.
Multiple independent transitions to pyrenoids in hornworts are supported by ultrastructural data. There are at least four pyrenoid types. In the Notothylas-type, pyrenoids are typically compact and stain darkly with osmium, with single thylakoids traversing them. In the Anthoceros type, found in Phaeoceros and Anthoceros (including Folioceros and Sphaerosporoceros), pyrenoids are darkly stained and traversed by multiple thylakoids, giving the appearance of multiple units. The isotope Δ13C discrimination in Notothylas- and Anthoceros-type plastids is quite similar (7.2–10.0 0/00), although the Phaeoceros (10.3–14.0 0/00) species seem to have a higher DiC pool than Notothylas (17). The Nothoceros-type pyrenoids are lightly stained at the ultrastructural level and less conspicuous in living material; this type is found in Phymatoceros bulbiculosus and Nothoceros species (12) (Fig. 1 and Table S2). The pyrenoid in these species is similar to the Anthoceros/Phaeoceros type, but differs in the level of osmium staining and the degree of dissection. The degree of staining may be an indication of less protein in the pyrenoid (26). Finally, the Dendroceros-type pyrenoid is present in the strictly epiphytic genus Dendroceros (43 species) and is composed of subunits of irregular shape, with numerous electron-opaque inclusions with thylakoids, including grana, interrupting the pyrenoid (13) (Fig. 1). Dendroceros is also the only hornwort to have a desiccation tolerance mechanism; its complex pyrenoid might relate to the drought-stressed epiphytic habitats it inhabits.
The parallel evolution of hornwort pyrenoids is analogous to the repeated evolution of complex anatomical and biochemical pathways in C4 plants (27–29). In angiosperms, C4 photosynthesis has evolved independently in 66–68 lineages, providing one of the most compelling cases of convergent evolution (27). Pyrenoids in algae are likely to have evolved several times as well (21), occurring in chloroplasts from a single endosymbiotic event (green and red algae, but not glaucophytes) and also in chloroplasts from several endosymbiotic events (cryptophytes, haptophytes, dinoflagellates, euglenophytes, and chlorarachniophyta) (4, 30). All these data fit with our finding that hornwort pyrenoids evolved and were lost several times during the past 100 million years, a period during which CO2 concentrations underwent large fluctuations.
Timing of Pyrenoid Acquisition in Hornworts.
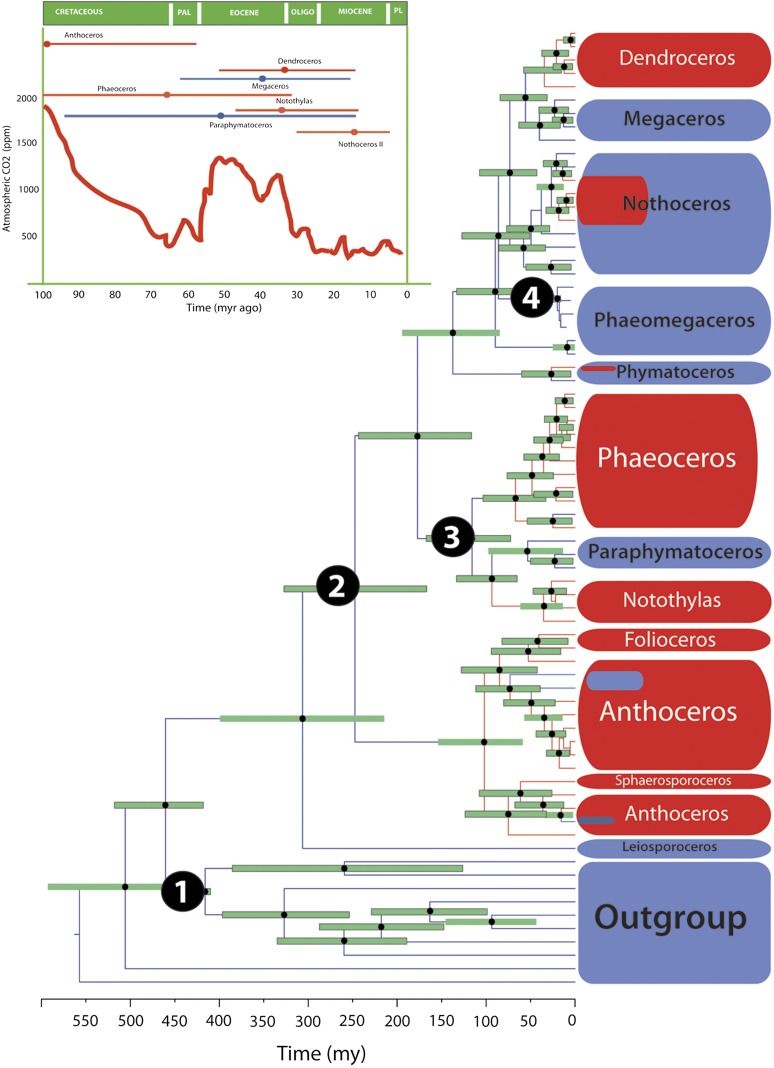
Fig. 2 provides a time tree for hornworts, with the chronogram tested for its sensitivity to model assumptions by using an array of substitution models and calibration priors (Materials and Methods). The fossil-calibrated clocks yielded a hornwort rbcL rate of 5 × 10−4 (2–8 × 10−4, 95% confidence range) substitutions per site per million years, five-times faster than the rbcL rate of 1 × 10−4 reported from 38 woody seed plants (31) but identical to the plastid rate reported from 37 land plants (32). The MRCA of all hornworts dates back to the Carboniferous (303.80 Mya), but with the exception of Anthoceros sensu lato (101.27 Mya), all genus-level crown groups are recent (from 8.8 to 62.2 Mya). Such recent crown group ages in a clade of fossil-documented presence in the Carboniferous mirror the situation in cycads, a group with similar species number and mostly Miocene and Oligocene genera (33). Probably hornwort diversification is another example of an ancient clade (in one sense a “living fossil”), having radiated mostly in the last 60 million years (Fig. 2 and Table S3).
Fig. 2.
Chronogram for hornworts and their outgroups obtained from mitochondrial and plastid sequences modeled under a relaxed uncorrelated log-normal clock. Node heights represent mean ages and bars the 95% highest posterior density intervals. Numbers represent the calibrations (Material and Methods): 1: Vascular plant divergence, 2: Anthoceros spore type A; 3: Notothylites nirulai fossil; and 4: fossil assigned to Phaeomegaceros sp. (Inset) Average values of atmospheric CO2 (ppm) levels during the last 100 Mya (56, 57). In the Inset, highest posterior density age inferred for the pyrenoid-bearing clades: Anthoceros, Phaeoceros, Notothylas, Nothoceros, and Dendroceros (red) and their pyrenoidless sister clades, Paraphymatoceros and Megaceros (blue).
The oldest pyrenoid-containing clade, Anthoceros sensu lato, originated ca. 101 Mya, coincidentally with a drastic drop in atmospheric CO2 in the Late Cretaceous (Fig. 2, Inset). The other four pyrenoid-containing clades are much younger, Phaeoceros (ca. 66 Mya), Notothylas (ca. 35 Mya), Dendroceros (ca. 34 Mya), and some Neotropical Nothoceros (ca. 18 Mya). Whether episodic changes in the CO2 levels promoted the success of hornwort lineages with pyrenoids at the expense of pyrenoid-lacking lineages is open to question. Pyrenoidless clades, such as Paraphymatoceros and Phaeomegaceros, also diversified during periods with low CO2 atmospheric levels (Fig. 2), and pyrenoid-containing hornwort clades originated (or rediversified) at greatly different times (Fig. 2). This finding does not fit the hypothesis that pyrenoid evolution relates to times of low CO2 concentration (see ref. 34 for a critical discussion of the evidence supporting this hypothesis for algae).
In angiosperms, the evolution of C4 pathways, another CCM, has been seen as causally related to low atmospheric CO2 levels (28, 35). Clearly, however, correlation does not establish causality (cf. 35), and the fact that C4 mechanisms have evolved more or less continuously over the past 60 Mya (27); Fig. 3 (27) argues against their link with particular CO2 concentrations. Episodes of positive selection on RuBisCO in Haptophyta (including coccolithophores) and Bacillariophyta (diatoms) have also been seen as linked to low CO2 levels; however, the inferred times of strong selection are based on small taxon samples and cover such broad time windows as to cast doubt on this suggestion [they are between 375 and 285 Mya for Haptophyta and 142–97 Mya, 114–92 Mya, and 73–56 Mya for diatoms (22)]. In hornworts, investigations of periods of positive selection on the plastid rbcL gene are hampered by the presence of RNA editing; in A. angustus, posttranscriptional change of individual bases occurs in approximately half the functional plastid genes (36).
Evolutionary Implications.
Hornwort clades with pyrenoids tend to have higher species numbers than their pyrenoidless sister clades (Fig. 1): Anthoceros (18 of approximately 83 species sampled), Phaeoceros (14 of 41 species), Folioceros (2 of 17), Notothylas (6 of 21), and Dendroceros (7 of ≤43). Most of the species in these genera contain visible pyrenoids (37, 38). The following pyrenoidless genera are interspersed among their pyrenoid-containing sister genera: Paraphymatoceros (4 of 4) -Notothylas (6 of 21); Megaceros (4 of 4)-Dendroceros (7 of 43). Losses of the pyrenoid occurred ca. 40 Mya (Megaceros), ca. 53 Mya (Paraphymatoceros), ca. 19 Mya (Phaeomegaceros I), and ca. 8 Mya (Phaeomegaceros II) (Fig. 2).
The relatively frequent gains and losses of pyrenoids in hornworts suggest that RuBisCO concentration between thylakoids may have benefits and costs unrelated to CO2 concentration. Pyrenoids have sometimes been seen as an adaptation to low conductance of CO2 in wet habitats, rather than low atmospheric gas levels (18). The hornwort pyrenoid may also play a role in the biogenesis of photosystem II, as is the case in Chlamydomonas pyrenoids (39), but experimental studies using hornwort pyrenoids are lacking. A promising model species would be Phaeoceros laevis, the pyrenoid of which consists of approximately 25 small units either distributed throughout the chloroplast or forming an aggregate central mass (30).
Materials and Methods
Taxon Sampling.
We sampled 77 species of hornworts; Table S4 provides a list of the sampled species with taxonomic author names, geographic origin of material, herbarium vouchers, and GenBank accession numbers for all sequences. Determination of plant material relied mostly on comparison type material, but two of the sampled species are not yet formally described. Morphological data do not support the genera Hattorioceros (J. Haseg.) J. Haseg. and Mesoceros Piippo. The single species of Hattorioceros was set apart by its very small spores (less than 20 μm in diameter) without a trilete mark (40). These spores are most likely from fungi, probably ascomycetes, that develop within the sporophyte. Mesoceros was described from Yunnan and Papua New Guinea (41). Both its species are based on plants with features intermediate between Anthoceros (black spores) and Phaeoceros (solid thallus, few antheridia per chamber with irregular jacket cells) (41). The generic type species is Mesoceros mesophoros, and the isotype of this name consists of a package containing a mixed collection of an Anthoceros species and a Phaeoceros species. As outgroups, we included up to 11 species representing early land plants (liverworts, mosses) and seedless vascular plants (lycophytes, ferns).
Isolation of DNA, Amplification, and Sequencing.
DNA isolation followed standard protocols. To deduce phylogenetic relationships, we used the exon 2 of the mitochondrial nad5, excluding an intron that is unique to Leiosporoceros, Anthoceros, Folioceros, and Sphaerosporoceros, and the plastid gene rbcL. Total DNA from fresh, silica-dried, or herbarium material was extracted with the Nucleo-Spin plant kit according to the manufacturer’s protocol (Macherey–Nagel) or using a modified cetyltrimethyl ammonium bromide protocol. Primers and standard PCR protocols are listed in ref. 12.
PCR products were cleaned using ExoSap-it (Affymetrix), and sequencing reactions were run on an ABI 3130 capillary sequencer (Applied Biosystems, Perkin-Elmer), following the manufacturers’ protocols. Sequence editing and alignment were carried out in Sequencher 4.7 (Gene Codes). Alignments for this study have been deposited in TreeBASE (submission no. S13448).
Phylogenetic Analysis.
Individual and combined phylogenetic analyses were performed under ML optimization and the GTR + CAT substitution model, using RAxML 7.2.8 (42) with the RAxML online interface (43). Statistical support was assessed via 100 ML bootstrap replicates that used the same substitution model.
Bayesian analyses were conducted in MrBayes 3.2 (44), using the default two runs and four chains (one cold and three heated), with uniform priors on most parameters. Model parameters were unlinked, posterior probabilities of tree topologies were estimated from both partitions, and trees were sampled every 1,000th generation. Burn-in and convergence were assessed by comparing the bipartitions across the two runs. Runs were terminated when the maximum SD across all bipartitions was less than 0.0314, as we expected the average SD of split frequencies to approach zero, reflecting that both chains are increasingly similar. Convergence was usually achieved in MrBayes after 4 × 106 generations. We used 50% majority-rule consensus trees to assess posterior probabilities for nodes of interest.
Ancestral Character Reconstruction of the Pyrenoid.
Each tip in the tree was scored for pyrenoid presence (score 0) or absence (score 1), based on observations made on the sequenced material (live, preserved in alcohol specimens) and compared with the relevant literature. Where possible, spores collected from ripe capsules were grown in agar (for culture methods, see ref. 45). Light microscopy was used to confirm pyrenoid presence, and data on pyrenoid type were obtained from transmission electronic micrographs (12, 13, 26). Ancestral reconstruction relied on ML as implemented in Mesquite using the Markov one- and two-parameter models (46) and the highest likelihood tree from RAxML.
Ancestral character-state reconstructions were also performed using a reverse-jump Markov chain Monte Carlo approach (47), as implemented in BayesTraits (48). Reconstructions of ancestral states were restricted to nodes with a posterior probability of ≥70% in our Bayesian sample of 5,000 trees (burn-in excluded). Most of the nodes of interest were found in 75–100% of the Bayesian trees. The rates at which model parameters were permitted to change during the Markov chain (ratedev) were set so that the acceptance rate of the proposed changes globally ranged between 20% and 40%. Rate coefficients and ancestral character states were sampled every 1,000 generations, and the chain was run for 50 × 106 generations. The ancestral state probability at a particular node was derived by averaging the posterior state probabilities.
Dating Analyses.
Dating relied on Bayesian divergence time estimation as implemented in BEAST 1.7.2 (49). Identical sequences and sequences differing only in nucleotide ambiguities were removed, yielding a matrix of 72 species, 62 hornwort species, and 10 outgroups. We carried out two runs either using the simple JC + Γ model of substitution or the more complex GTR + Γ model, identified as the best-fit in jModelTest (50). A Yule tree prior was used as the tree model. Runs were repeated under either a strict-clock model or a relaxed-clock model, the latter with rate variation across branches uncorrelated and log-normally distributed. The Markov chain Monte Carlo chains were run for 30 million in the strict-clock model, and for 100 million generations in the relaxed-clock model, with parameters sampled every 3,000th and 10,000th generation, respectively. Tracer 1.50 was used to assess effective sample sizes for all estimated parameters and to judge the percentage of burn-in for tree constructions. Trees were combined in TreeAnnotator 1.6.1, and maximum-clade credibility trees with mean node heights were visualized using FigTree 1.3.1.
The fossil record in hornworts is sparse, and earliest reliable fossils are only from the Upper Cretaceous (Table S5). Based on a review of the relevant literature, we selected three fossils for calibration (below). We also constrained the MRCA of vascular plants to 416 (± 3) Mya, using a normal distribution based on the fossil record (51) (Table S5). We used the age of Anthoceros spore type A from the Baqueró Formation, Argentina (118.56 ± 3.7 Mya) (52) to constrain the stem node of Anthoceros and the rest of the hornworts. The spore exine is ornamented with warts in the area of contact of the four meiotic products (proximal face) and the fossil has three proximal warts characteristic of two extant Neotropical species, Anthoceros tuberculatus and A. tristanianus. We used the age of Notothylites nirulai from the Deccan Intertrappean beds of Mohgaonka, India (Maastrichtian, 65–70 Mya) (53) to constrain the age of the stem node of Notothylas and Phaeoceros (including Paraphymatoceros). The petrified fossil of an entire plant has similar thallus size, sporophyte size, lack of stomata in sporophyte, and elater shape as seen in extant Notothylas. As our fourth calibration, we used a fossil assigned to Phaeoceros sp. from the Uscari Formation, Costa Rica (Lower Miocene 15–23 Mya) (54). The spore fossil has six depressions in its distal face and closely resembles extant Phaeomegaceros fimbriatus in size and ornamentation.
Dating analyses relied on either exponential or γ-priors on the three fossil time constraints. Exponential or γ-priors were centered on the fossil age. We also carried out runs that removed one calibration fossil at a time, leaving only the vascular plant constraint. An additional analysis was conducted using a mean substitution rate across mitochondrial, nuclear, and plastid genomes of 3 × 10−4 substitutions per site per million years (55).
Supplementary Material
Acknowledgments
We thank B. Buck, D. C. Cargill, J. Faubert, C. García, T. Hedderson, A. Ibarra, J. Larraín, W. Manzke, and Y. Queralta for plant material; L. L. Forrest and B. Goffinet in Storrs and M. Silber in Munich for laboratory support; and K. Renzaglia for electron micrographs. This study was supported in part by Deutsche Forschungsgemeinschaft Grant RE-603/14-1 and National Science Foundation Award DDIG-0910258 (to J.C.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.R. is a guest editor invited by the Editorial Board.
Database deposition: The sequences reported in this paper have been deposited in the TreeBASE database, treebase.org (accession no. S13448).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213498109/-/DCSupplemental.
References
- 1.Leakey ADB, Lau JA. Evolutionary context for understanding and manipulating plant responses to past, present and future atmospheric [CO2] Philos Trans R Soc Lond B Biol Sci. 2012;367(1588):613–629. doi: 10.1098/rstb.2011.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitney SM, Houtz RL, Alonso H. Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol. 2011;155(1):27–35. doi: 10.1104/pp.110.164814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson I, Backlund A. Structure and function of Rubisco. Plant Physiol Biochem. 2008;46(3):275–291. doi: 10.1016/j.plaphy.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Badger MR, et al. The diversity of coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can J Bot. 1998;76(6):1052–1071. [Google Scholar]
- 5.Raven JA, Cockell CS, De La Rocha C. The evolution of inorganic carbon concentrating mechanisms in photosynthesis. Philos Trans R Soc Lond B Biol Sci. 2008;363(1504):2641–2650. doi: 10.1098/rstb.2008.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaughn KC, et al. The pyrenoid is the site of ribulose I, 5-bisphosphate carboxylase/oxygenase accumulation in the hornwort (Bryophyta: Anthocerotae) chloroplast. Protoplasma. 1990;156:117–129. [Google Scholar]
- 7.Borkhsenious ON, Mason CB, Moroney JV. The intracellular localization of ribulose-1,5-bisphosphate Carboxylase/Oxygenase in Chlamydomonas reinhardtii. Plant Physiol. 1998;116(4):1585–1591. doi: 10.1104/pp.116.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giordano M, Beardall J, Raven JA. CO2 concentrating mechanisms in algae: Mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol. 2005;56:99–131. doi: 10.1146/annurev.arplant.56.032604.144052. [DOI] [PubMed] [Google Scholar]
- 9.Qiu YL, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA. 2006;103(42):15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y, Graham SW. Inferring the higher-order phylogeny of mosses (Bryophyta) and relatives using a large, multigene plastid data set. Am J Bot. 2011;98(5):839–849. doi: 10.3732/ajb.0900384. [DOI] [PubMed] [Google Scholar]
- 11.Renzaglia KS, et al. Bryophyte phylogeny: Advancing the molecular and morphological frontiers. Bryologist. 2007;110(2):179–213. [Google Scholar]
- 12.Duff RJ, Villarreal JC, Cargill DC, Renzaglia KS. Progress and challenges toward developing a phylogeny and classification of the hornworts. Bryologist. 2007;110(2):214–243. [Google Scholar]
- 13.Vaughn KC, et al. The anthocerote chloroplast: A review. New Phytol. 1992;120(2):169–190. [Google Scholar]
- 14.Smith EC, Griffiths H. The occurrence of the chloroplast pyrenoid is correlated with the activity of a CO2-concentrating mechanism and carbon isotope discrimination in lichens and bryophytes. Planta. 1996a;198(1):6–16. [Google Scholar]
- 15.Smith EC, Griffiths H. A pyrenoid-based carbon concentrating mechanism is present in terrestrial bryophytes of the class Anthocerotae. Planta. 1996b;200:203–212. [Google Scholar]
- 16.Smith EC, Griffiths H. The role of carbonic anhydrase in photosynthesis and the activity of the carbon concentrating mechanism in bryophytes of the class Anthocerotae. New Phytol. 2000;145(1):29–37. [Google Scholar]
- 17.Hanson D, Andrews TJ, Badger MR. Variability of the pyrenoid-based CO2 concentrating mechanisms in hornworts (Anthocerotophyta) Funct Plant Biol. 2002;29(2/3):407–416. doi: 10.1071/PP01210. [DOI] [PubMed] [Google Scholar]
- 18.Meyer M, Seibt U, Griffiths H. To concentrate or ventilate? Carbon acquisition, isotope discrimination and physiological ecology of early land plant life forms. Philos Trans R Soc Lond B Biol Sci. 2008;363(1504):2767–2778. doi: 10.1098/rstb.2008.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crum H. Structural Diversity of Bryophytes. Ann Arbor, MI: Univ of Michigan Herbarium; 2001. [Google Scholar]
- 20.Shaw J, Renzaglia KS. Phylogeny and diversification of bryophytes. Am J Bot. 2004;91(10):1557–1581. doi: 10.3732/ajb.91.10.1557. [DOI] [PubMed] [Google Scholar]
- 21.Raven JA, Edwards D. 2004. in The Evolution of Plant Physiology. From Whole Plant to Ecosystems, eds Hemsley AR, Poole I (Elsevier, London, UK), pp 17–41.
- 22.Young JN, Rickaby REM, Kapralov MV, Filatov DA. Adaptive signals in algal Rubisco reveal a history of ancient atmospheric carbon dioxide. Phil Trans R Soc Lond B Biol Sci. 2012;367(1588):483–492. doi: 10.1098/rstb.2011.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Badger MR, Hanson DT, Price GD. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct Plant Biol. 2002;29(2/3):161–173. doi: 10.1071/PP01213. [DOI] [PubMed] [Google Scholar]
- 24.Ma Y, Pollock SV, Xiao Y, Cunnusamy K, Moroney JV. Identification of a novel gene, CIA6, required for normal pyrenoid formation in Chlamydomonas reinhardtii. Plant Physiol. 2011;156(2):884–896. doi: 10.1104/pp.111.173922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genkov T, Meyer M, Griffiths H, Spreitzer RJ. Functional hybrid Rubisco enzymes with plant small subunits and algal large subunits: Engineered rbcS cDNA for expression in Chlamydomonas. J Biol Chem. 2010;285(26):19833–19841. doi: 10.1074/jbc.M110.124230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burr FA. Phylogenetic transitions in the chloroplast number of the Anthocerotales I. The number and ultrastructure of the mature plastids. Am J Bot. 1970;57(1):97–110. [Google Scholar]
- 27.Sage RF, Sage TL, Kocacinar F. Photorespiration and the evolution of C4 photosynthesis. Annu Rev Plant Biol. 2012;63:19–47. doi: 10.1146/annurev-arplant-042811-105511. [DOI] [PubMed] [Google Scholar]
- 28.Christin PA, et al. Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr Biol. 2008;18(1):37–43. doi: 10.1016/j.cub.2007.11.058. [DOI] [PubMed] [Google Scholar]
- 29.Edwards EJ, et al. C4 Grasses Consortium The origins of C4 grasslands: Integrating evolutionary and ecosystem science. Science. 2010;328(5978):587–591. doi: 10.1126/science.1177216. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths DJ. The pyrenoid. Bot Rev. 1970;36(1):29–58. [Google Scholar]
- 31.Albert VA, et al. Functional constraints and rbcL evidence for land plant phylogeny. Ann Miss Bot Garden. 1994;81(3):534–567. [Google Scholar]
- 32.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19(1):101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 33.Nagalingum NS, et al. Recent synchronous radiation of a living fossil. Science. 2012;334(6057):796–799. doi: 10.1126/science.1209926. [DOI] [PubMed] [Google Scholar]
- 34.Raven JA, Giordano M, Beardall J, Maberly SC. Algal evolution in relation to atmospheric CO2: Carboxylases, carbon-concentrating mechanisms and carbon oxidation cycles. Philos Trans R Soc Lond B Biol Sci. 2012;367(1588):493–507. doi: 10.1098/rstb.2011.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roalson EH. C(4) photosynthesis: Differentiating causation and coincidence. Curr Biol. 2008;18(4):R167–R168. doi: 10.1016/j.cub.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 36.Kugita M, Yamamoto Y, Fujikawa T, Matsumoto T, Yoshinaga K. RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res. 2003;31(9):2417–2423. doi: 10.1093/nar/gkg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asthana AK, Srivastava SC. Indian hornworts (A taxonomic study) Bryophyt Bibl. 1991;42:1–158. [Google Scholar]
- 38.Singh DK. Notothylaceae of India and Nepal (A morpho-Taxonomic Revision) India: Bishen Singh Mahendra Pal Singh; 2002. [Google Scholar]
- 39.Uniacke J, Zerges W. Stress induces the assembly of RNA granules in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol. 2008;182(4):641–646. doi: 10.1083/jcb.200805125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasegawa J. Hattorioceros striatisporus (Hasegawa) Hasegawa newly found in Fiji. Bryol Res. 2000;7(9):273–275. [Google Scholar]
- 41.Piippo S. Bryophyte Flora of the Huon Peninsula, Papua New Guinea. LIV. Anthocerotophyta. Acta Bot Fennica. 1993;148:27–51. [Google Scholar]
- 42.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 43.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web-Servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. Accessed May, 2012. [DOI] [PubMed] [Google Scholar]
- 44.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villarreal A JC, Renzaglia KS. Structure and development of Nostoc strands in Leiosporoceros dussii (Anthocerotophyta): A novel symbiosis in land plants. Am J Bot. 2006;93(5):693–705. doi: 10.3732/ajb.93.5.693. [DOI] [PubMed] [Google Scholar]
- 46.Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol. 2001;50(6):913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- 47.Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am Nat. 2006;167(6):808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- 48.Pagel M, Meade A. 2004. BayesTraits. Package available form www.evolution.rdg.ac.uk. Accessed May 2012.
- 49.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 51.Clarke JT, Warnock RCM, Donoghue PCJ. Establishing a time-scale for plant evolution. New Phytol. 2011;192(1):266–301. doi: 10.1111/j.1469-8137.2011.03794.x. [DOI] [PubMed] [Google Scholar]
- 52.Archangelsky S, Villar de Seone L. Estudios palinógicos de la formación Baqueró (Cretácico), provincia de Santa Cruz, Argentina. Ameghiniana. 1996;33(3):7–19. [Google Scholar]
- 53.Chitaley SD, Yawale NR. On Notothylites nirulai gen. et sp. nov. a petrified sporogonium from the Deccan-Intertrappean beds of Mohgaonkalan M. P. (India) Botanique. 1980;9(1/4):111–118. [Google Scholar]
- 54.Graham A. Miocene communities and paleoenvironments of Southern Costa Rica. Am J Bot. 1987;72(10):1501–1518. [Google Scholar]
- 55.Shaw AJ, et al. Peatmoss (Sphagnum) diversification associated with Miocene Northern Hemisphere climatic cooling? Mol Phylogenet Evol. 2010;55(3):1139–1145. doi: 10.1016/j.ympev.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 56.Beerling DJ, Royer DL. Convergent Cenozoic CO2 history. Nat Geosci. 2011;4(7):418–420. [Google Scholar]
- 57.Berner RA, Kothavala Z. GEOCARB III: A revised model of atmospheric CO2 over Phanerozoic time. Am J Sci. 2001;301:182–204. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.