Abstract
The general transcription factor II B (TFIIB) plays a central role in both the assembly of the transcription complex at gene promoters and also in the events that lead to transcription initiation. TFIIB is phosphorylated at serine-65 at the promoters of several endogenous genes, and this modification is required to drive the formation of gene promoter–3′ processing site contacts through the cleavage stimulation factor 3′ (CstF 3′)-processing complex. Here we demonstrate that TFIIB phosphorylation is dispensable for the transcription of genes activated by the p53 tumor suppressor. We find that the kinase activity of TFIIH is critical for the phosphorylation of TFIIB serine-65, but it is also dispensable for the transcriptional activation of p53-target genes. Moreover, we demonstrate that p53 directly interacts with CstF independent of TFIIB phosphorylation, providing an alternative route to the recruitment of 3′-processing complexes to the gene promoter. Finally, we show that DNA damage leads to a reduction in the level of phospho-ser65 TFIIB that leaves the p53 transcriptional response intact, but attenuates transcription at other genes. Our data reveal a mode of phospho-TFIIB-independent transcriptional regulation that prioritizes the transcription of p53-target genes during cellular stress.
The general transcription factor TFIIB plays a central role in the assembly of the transcription complex at the gene promoter (1–3). It has emerged in recent years that TFIIB also engages in contact with factors that are involved in transcription termination and facilitates the formation of loops between the gene promoter and terminator (4–6). The highly conserved B-finger/reader region of TFIIB plays critical role(s) in transcription initiation, promoter–terminator contacts, and also transcriptional activation, suggesting that these events are linked (1).
We recently reported that TFIIB is phosphorylated at ser-65 at the promoters of several endogenous genes and that this event is required for productive transcription (7). Moreover, phosphorylation of TFIIB ser-65 directly augments the interaction between TFIIB and the CstF complex and facilitates promoter–3′ processing site contacts. Whether or not these events are universally required for the transcription of genes by RNA polymerase II (RNAPII) is not known.
Recent years have seen the emergence of multiple and distinct pathways to transcription (8–11). In this regard, the posttranslational modifications of the RNAPII carboxyl-terminal domain (CTD), specifically phosphorylation, and the enzymes responsible have revealed new gene-specific pathways in transcription control. This is particularly evident at p53-responsive genes, which require distinct events to ensure that the target genes involved in stress-response pathways are robustly and rapidly induced under suboptimal cellular conditions (12–16).
In this study we demonstrate that the phosphorylation of TFIIB ser-65 is not universally required for transcription, and that target genes under the control of p53 can bypass this event. This involves the circumvention of the requirement for TFIIB phosphorylation to recruit the CstF complex to the p21 promoter. We provide evidence that TFIIB is dephosphorylated during genotoxic stress, which correlates with the general attenuation of transcription and prioritizes the transcription of p53-target genes.
Results
p53-Target Genes Do Not Require the Integrity of TFIIB Ser-65.
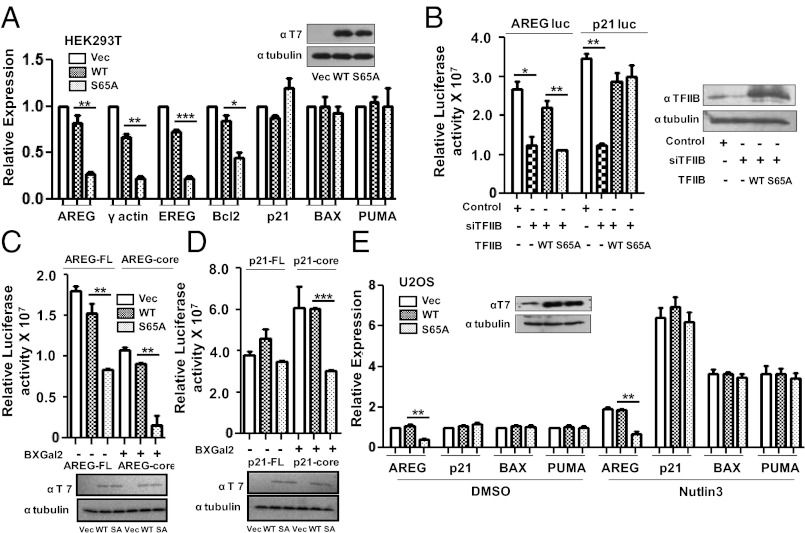
Our recent study demonstrated that the phosphorylation of TFIIB at ser-65 is essential for the productive transcription of several genes including GAPDH, γ-actin, β-tubulin, and amphiregulin (AREG) (7). We have now analyzed a larger cohort of genes to determine the requirement for the integrity of TFIIB ser-65. pCDNA3 or the same vector driving expression of either wild-type TFIIB or the phosphorylation-defective mutant TFIIB derivative S65A were transfected into HEK293T cells and 48 h later cDNA prepared and analyzed by quantitative real-time polymerase chain reaction (qPCR) for expression of the target genes shown (Fig. 1A). It was surprising to find that transcription of the p21, BAX, and PUMA genes was not significantly impaired by the expression of TFIIB S65A, although the expression of AREG, γ-actin, epiregulin (EREG), and Bcl2 were significantly inhibited. Taken together with our previous study (7), these data suggest that, although phosphorylation of TFIIB ser-65 is required for transcription of a significant proportion of genes, it is not universally required.
Fig. 1.
The integrity of TFIIB ser65 is not critical for the expression of p53-target genes. (A) qPCR analysis of AREG, γ-actin, EREG, Bcl2, p21, BAX, and PUMA gene expression was carried out after 48 h of transfection with pCDNA3 vector, T7-tagged WT TFIIB and TFIIB S65A in HEK293T cells. Western analysis with anti-T7 antibody confirmed equivalent expression of WT TFIIB and TFIIB S65A. (B) Luciferase activity of FL promoter constructs of AREG and p21 was measured in cells transfected with pSUPER shTFIIB along with WT or S65A TFIIB. Transfection with empty pSUPER vector and shTFIIB are controls. Error bars denote SD of three independent experiments. Western blotting analysis was performed with anti-TFIIB and anti-β tubulin antibodies. (C) Luciferase assay was performed after pCDNA3 vector, T7-tagged WT TFIIB and TFIIB S65A transfection with FL promoter or core promoter constructs of AREG and (D) p21 promoter constructs. Error bars denote SD of three independent experiments. Western blotting analysis was as above. (E) qPCR analysis of AREG, p21, BAX, and PUMA gene expression was carried out in U2OS cells transfected with pCDNA3 vector, T7-tagged WT TFIIB and TFIIB S65A for 40 h followed by 8 h of treatment with 10 μM Nutlin3 or DMSO. Western analysis was as above.
We next sought to determine whether the differential requirement for TFIIB ser-65 in AREG and p21 transcription would also be apparent with luciferase reporters under the control of the respective promoter regions. An RNAi-based endogenous replacement strategy was used to deplete endogenous cellular TFIIB and simultaneously express either wild-type TFIIB or TFIIB S65A (which harbor a silent mutation to render them refractory to the RNAi; ref. 17). In agreement with the effects observed in Fig. 1A, AREG promoter-luciferase activity was significantly reduced when TFIIB S65A was the sole source of TFIIB. In contrast, TFIIB S65A was able to support transcription of the p21 promoter-luciferase reporter (Fig. 1B). TFIIB can engage in sequence-specific DNA contact with the core promoter (1). We therefore considered the possibility that the differential requirement for the integrity of TFIIB ser-65 for transcription of the AREG and p21 genes may be due to core promoter sequence. Thus, we compared the activities of luciferase constructs containing either the full-length (FL) AREG and p21 promoters (as above) or the core promoter sequences alone of AREG (−56 to +4) and p21 (−56 to +3) genes (both driven by the same acidic activator GAL4-RII). The relative luciferase activities of the both FL and the core promoter of AREG gene were significantly inhibited by the expression of TFIIB S65A (Fig. 1C, compare lanes 2–3 and 5–6). In contrast to the p21 FL promoter-luciferase reporter, however, the p21 core promoter-luciferase construct was inhibited by the expression of TFIIB S65A compared with their respective activities in the presence of wild-type TFIIB (Fig. 1D, compare lanes 2–3 and 5–6). Thus, the region upstream of the core promoter of p21 gene confers the tolerance to TFIIB ser-65 substitution in transcription.
So far our data suggest that the basal transcription of p53-target genes is not repressed by TFIIB S65A. We next tested the effect of TFIIB S65A on the regulation of p53-target genes under conditions in which p53 is stabilized by Nutlin3 in U2OS cells. Nutlin3 treatment resulted in marked activation of p53-target genes p21, Bcl-2–associated X protein (BAX), and p53 upregulated modulator of apoptosis (PUMA), which was not affected by TFIIB S65A, and AREG expression was still inhibited (Fig. 1E). Thus, both basal and activated transcriptions of p53-target genes are not significantly affected by TFIIB S65A.
Presence of Functional p53 Bypasses the Requirement for TFIIB Serine-65.
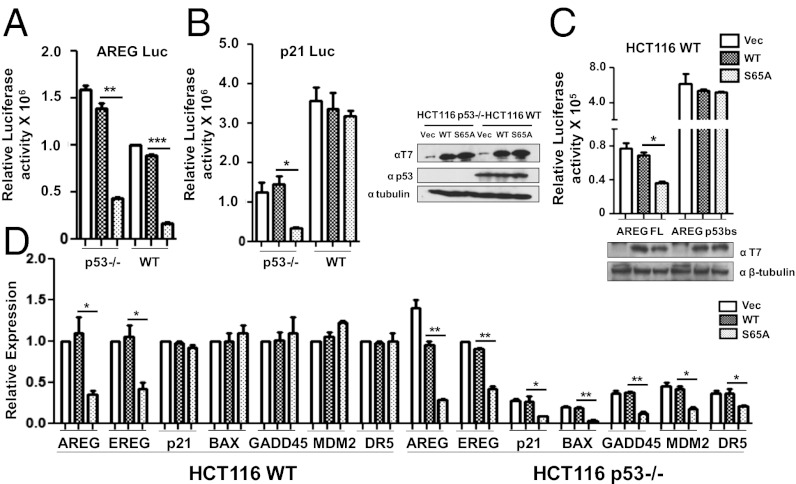
Our data so far suggest that p53 may play a role in bypassing the requirement of TFIIB ser-65 in the productive transcription of the p21 gene. Hence we used HCT116 cells (p53 WT) and the isogenic HCT116 p53-null cells (p53−/−) to analyze the luciferase activities of AREG and p21 FL promoters in presence of either wild-type TFIIB or TFIIB S65A. AREG FL promoter activity was inhibited by the expression of TFIIB S65A in both p53-null and HCT116WT cells (Fig. 2A). In contrast, p21 FL promoter activity was inhibited by the expression of TFIIB S65A in the absence of p53 in HCT116 p53−/− cells, but not in p53 WT cells (Fig. 2B). RNAi-mediated replacement of endogenous TFIIB with the TFIIB derivatives had similar effect on the endogenous expression of AREG and p21 genes in HCT116WT cells (Fig. S1A). To confirm that direct binding of p53 to the promoter confers resistance to TFIIB S65A, we generated an AREG promoter-luciferase construct under the control of four tandem upstream p53-binding sites (AREG-p53bs). The relative luciferase activity of AREG-p53bs was compared with the original AREG FL promoter in the presence of wild-type TFIIB or TFIIB S65A. The results demonstrate that the addition of p53-binding sites rendered the AREG promoter resistant to the inhibitory effect of TFIIB S65A in HCT116WT cells but not in p53-null cells (Fig. 2C and Fig. S1B). These data provide strong evidence that binding of functional p53 to its cognate site at the target gene promoter is sufficient to activate transcription in the presence of TFIIB S65A. In addition, we also tested other p53-target genes and found that along with p21, BAX, and PUMA, the expression of growth arrest and DNA-damage-inducible protein A (GADD45A), murine double minute 2 (MDM2), and death receptor 5 (DR5) was not affected by TFIIB S65A in HCT116WT cells but were repressed in presence of TFIIB S65A in p53−/− cells (Fig. 2D). These results suggest that a majority of p53-target genes bypass the requirement for phospho-ser65 TFIIB during transcriptional activation that is dependent on p53 expression.
Fig. 2.
p53 can bypass the requirement for the integrity of TFIIB Ser65. (A) Luciferase assay was performed in HCT116 p53−/− and HCT116WT cell lines with FL promoter constructs of AREG and (B) p21 after pCDNA3 vector, T7-tagged WT TFIIB and TFIIB S65A transfection. Western blotting was carried out with anti-T7, anti-p53, and anti-β tubulin antibodies. (C) Luciferase assay was performed with AREG p53bs and original AREG FL promoter constructs in the presence of wild-type TFIIB and mutant S65A in HCT116WT cells. Western blotting was done with anti-T7 antibody with β-tubulin. (D) qPCR analysis of candidate gene expression was performed after transfection with pCDNA3 vector, WT TFIIB and TFIIB S65A in HCT116WT and p53−/− cells.
TFIIB Kinase and Its Role in Transcription.
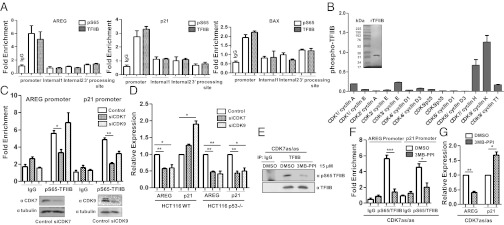
Our results so far suggest that p53-target genes do not require the phosphorylation of TFIIB ser-65. Even so, ChIP analysis with phospho-ser-65 antibodies revealed that TFIIB assembled at the p21 and BAX promoters is still phosphorylated (Fig. 3A and Fig. S2). We therefore sought to determine the protein kinase(s) responsible for the phosphorylation of TFIIB in vivo. We reported before that the cyclin-dependent kinase (CDK) inhibitors: olomoucine and 6-dichloro-1-h-ribofuranosyl-benzimidazole (DRB) inhibited the phosphorylation of TFIIB at ser-65 both in vitro and in vivo (7). To identify the protein kinase involved in TFIIB phosphorylation, recombinant TFIIB was purified, and an in vitro kinase assay was performed with a panel of different functional CDK/cyclin combinations. The activity of the different kinases toward TFIIB was normalized with a standard substrate (Fig. 3B). It is interesting to note that CDK7/cyclin H and CDK9/cyclin K were the most active TFIIB kinases compared with the other CDK/cyclin pairs.
Fig. 3.
TFIIH phosphorylates TFIIB Ser-65. (A) ChIP assay was performed with anti-TFIIB and pSer65-TFIIB antibodies and the promoter sequences, two different internal regions and the 3′-processing sequence of AREG, p21, and BAX genes, were analyzed for enrichment. (B) Recombinant TFIIB was purified (Left) and in vitro filter-binding kinase assay performed with a panel of different CDK/cyclin combinations. The values of TFIIB phosphorylation are plotted relative to a standard substrate for each kinase (Right). (C) ChIP analysis was carried out with anti-pS65-TFIIB antibody with HEK 293T cells transfected with either siRNAs against CDK7 and CDK9 or control siRNA for 48 h to monitor the levels of phosphorylated TFIIB at the AREG and p21 promoters. qPCR data of four independent transfections are plotted as fold enrichment relative to 18S DNA. Error bars denote SD. Western blotting analyses with either anti-CDK7 or anti-CDK9 antibodies confirmed knockdown at the protein level compared with β-tubulin loading control (Bottom). (D) qPCR analysis of AREG and p21 gene expression was performed after transfection with siRNAs against CDK7 and CDK9 in HCT116WT and p53−/− cells. (E) TFIIB was immunoprecipitated from CDK7 (as/as) HCT116 cells after 12 h of 15 μM 3MB-PPI or control DMSO treatment and immunoblotted using anti-pS65-TFIIB and anti-TFIIB antibodies. (F) ChIP analysis was with anti-pS65-TFIIB and anti-TFIIB antibodies in CDK7 (as/as) cells treated with 15 μM 3MB-PPI for 12 h. The normalized level of phospho-S65TFIIB (over TFIIB ChIP signal) at the AREG and p21 promoters is plotted as fold enrichment relative to 18S DNA. Error bars denote SD. (G) qPCR analysis of AREG and p21 gene expression was performed after 12 h of 15 µM 3MB-PPI in CDK7 (as/as) cells.
We next used RNAi-mediated knockdown of CDK7 and CDK9 to determine the effect on the phosphorylation of TFIIB ser-65 at the AREG and p21 promoters. ChIP assays with anti-pS65-TFIIB antibodies revealed that knockdown of CDK7 significantly reduced the level of phospho-S65 TFIIB at both the promoters. In contrast CDK9 ablation did not affect the phosphorylation of TFIIB at the AREG promoter but a small reduction in the ChIP signal was observed at the p21 promoter (Fig. 3C).
As demonstrated above, TFIIB phosphorylation at ser-65 is essential for the expression of the AREG gene, but not the p21 gene. We therefore tested the effect of CDK7 or CDK9 depletion on transcription in HCT116WT and p53−/− cells. Ablation of either CDK7 or CDK9 reduced expression of the AREG gene in HCT116WT cells but induced p21 transcription (Fig. 3D). However, consistent with the effects of TFIIB S65A expression, knockdown of either CDK7 or CDK9 inhibited the expression of both AREG and p21 in p53-null cells. These observations were confirmed by using the CDK inhibitor DRB at concentrations of 50 and 200 μM, which stimulated p21 expression in HCT116WT cells, but inhibited p21 transcription in p53-null cells. In contrast, transcription of the AREG gene was inhibited by DRB (50 and 200 μM) in both the cell lines (Fig. S3 A and B).
We next used an alternative approach to further confirm a role for CDK7 in the phosphorylation of TFIIB ser-65. The analog-sensitive cell line CDK7(as/as) HCT116 allows the kinase activity of CDK7 to be specifically inhibited by treating the cells with 3MB-PPI (18). We found that, upon inhibition of the CDK7 kinase activity using (15 μM) 1-(tert-Butyl)-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine (3MB-PPI) (Fig. S3C), the level of phospho-ser65-TFIIB was significantly reduced as detected by IP-immunoblotting (Fig. 3E). In addition, ChIP assays demonstrated a marked reduction in the level of phospho-ser65-TFIIB (relative to general TFIIB) at gene promoters (Fig. 3F). Furthermore, AREG transcription was repressed, but we observed a moderate increase in p21 expression (Fig. 3G). These results provide independent evidence for a role of TFIIH (CDK7) in TFIIB ser-65 phosphorylation.
TFIIB Phosphorylation Is Reduced upon Treatment of Cells with Doxorubicin.
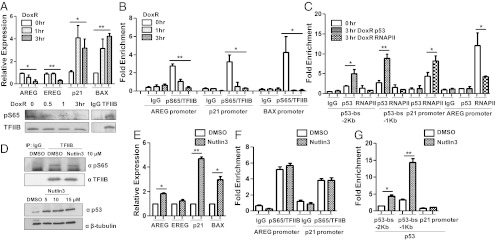
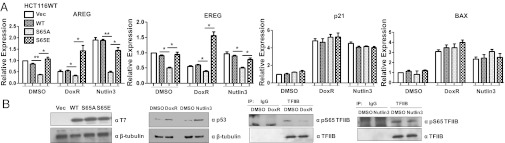
We next determined the role of TFIIB Ser-65 under conditions in which p53 is activated by cellular stress using the drug doxorubicin (DoxR). We performed these experiments at time points before significant cell death occurred to ensure that the transcriptional responses were accurately measured (Fig. S4). The expression of candidate genes was analyzed in HCT116WT cells treated with DoxR for up to 3 h. AREG and EREG expression were reduced by 3 h posttreatment, and p21 and BAX expression were rapidly induced. Moreover, immunoblotting of TFIIB immunoprecipitates demonstrated that the general level of phospho-ser65-TFIIB was reduced following 3 h of DoxR treatment (Fig. 4A). We next used ChIP to confirm the status of TFIIB ser-65 phosphorylation (normalized to the general TFIIB signal) at the AREG and p21 promoters after DoxR treatment. It is interesting to note that the phospho-Ser65-TFIIB ChIP signal significantly reduced at the AREG, p21, and BAX promoters upon DoxR treatment (Fig. 4B). DoxR also induced p53 and RNAPII recruitment to the p21 promoter, but RNAPII recruitment to the AREG promoter was reduced (Fig. 4C). Comparable effects were observed when we analyzed another DNA damaging agent, cisplatin (Fig. S5). Taken together, these data suggest that the reduction of TFIIB ser-65 phosphorylation following DNA damage plays a role in the general down-regulation of gene transcription. The net effect would be to prioritize transcription of genes that do not require TFIIB ser-65 phosphorylation, which includes the p53-target genes.
Fig. 4.
The level of phospho-TFIIB ser65 is reduced upon treatment with DNA-damaging agents. (A) qPCR analysis of AREG, EREG, p21, and BAX gene expression was performed at different time points of 5 µg/mL DoxR treatment in HCT116WT cells (Upper). TFIIB was immunoprecipitated from HCT116WT cells after different time points of 5 µg/mL DoxR treatment and immunoblotted using anti-pS65-TFIIB and anti-TFIIB antibodies (Lower). (B) ChIP analysis was carried out with anti-pS65-TFIIB and anti-TFIIB antibodies in HCT116WT cells treated with 5 µg/mL DoxR after different time points. The normalized level of phospho-S65TFIIB (over TFIIB ChIP signal) at the AREG, p21, and BAX promoters is plotted as fold enrichment relative to 18S DNA. (C) ChIP analysis was also carried out with anti-p53 and anti-RNAPII antibodies in HCT116WT cells treated with 5 µg/mL DoxR after 3 h and compared with untreated cells. The levels of p53 and RNAPII at the upstream promoter sequence (p53-binding site [bs] at −2 and −1 Kb) and core promoter region of p21 and core promoter of AREG is plotted as fold enrichment relative to 18S DNA. (D) TFIIB was immunoprecipitated from HCT116WT cells after 8 h of 10 μM Nutlin3 or control DMSO treatment and immunoblotted using anti-pS65-TFIIB and anti-TFIIB antibodies (Upper). HCT116WT cells were treated with the indicated concentrations of Nutlin3 for 8 h and immunoblotted with anti-p53 antibody and β-tubulin antibodies (Lower). (E) qPCR analysis of the indicated mRNAs was performed after 8 h of 10 μM Nutlin3 treatment in HCT116WT cells (F) ChIP analysis was carried out with anti-pS65-TFIIB and anti-TFIIB antibodies in HCT116WT cells treated with 10 μM Nutlin3 for 8 h. The normalized level of phospho-S65TFIIB (over TFIIB ChIP signal) at the AREG and p21 promoters is plotted as fold enrichment relative to 18S DNA. (G) As in F, Chip assay was done with anti-p53 antibody to analyze its occupancy at the indicated regions of p21 promoter. Error bars denote SD.
We next tested whether TFIIB ser-65 dephosphorylation is specifically DNA damage-dependent or whether it is due to the activation of p53. We therefore stimulated p53 function by using Nutlin3, an antagonist of Mdm2 that disrupts the p53–Mdm2 association. Western blotting analysis of TFIIB immunoprecipitates demonstrated that the general level of phospho-ser65-TFIIB was not significantly affected following 8 h of Nutlin3 treatment, even though these conditions led to the stabilization of p53 (Fig. 4D). The expression of p21 and BAX was induced by Nutlin3, but AREG and EREG genes were not inhibited (but, in fact, slightly elevated) in HCT116WT cells treated with Nutlin3 for 8 h (Fig. 4E). ChIP analysis confirmed that TFIIB ser-65 phosphorylation (normalized to the general TFIIB signal) was not reduced at the AREG or p21 promoters following Nutlin3 treatment (Fig. 4F), but p53 occupancy at the p21 promoter was induced (Fig. 4G). These results show that p53 activation alone does not influence the phosphorylation status of TFIIB in the absence of a DNA damage signal.
Dephosphorylation of TFIIB Plays a Role in DNA-Damage–Dependent Transcription Repression.
So far our studies suggest that TFIIB is dephosphorylated in response to DNA damage and that this correlates with the general transcriptional attenuation and the specific induction of p53-dependent target genes. Our previous study suggested that the phospho-mimetic mutant TFIIB S65E does not inhibit transcription and is functionally comparable to the wild-type TFIIB (7). We next used the TFIIB S65E mutant to determine whether the transcriptional down-regulation of AREG after DNA damage requires TFIIB ser-65 dephosphorylation. The effect of TFIIB S65E was compared with wild-type TFIIB and TFIIB S65A on the transcriptional response of AREG and EREG genes after treatment of HCT116WT cells with DoxR or Nutlin3 for 6 h. The data show that expression of TFIIB S65E does indeed prevent transcriptional inhibition of AREG and EREG genes under DNA-damaging conditions in the presence of DoxR (Fig. 5A). In the case of Nutlin3 treatment, expression of TFIIB S65E maintained AREG and EREG expression similar to that observed with WT TFIIB, and p21 and BAX expression was significantly induced following the drug treatments. Western blotting analysis of TFIIB immunoprecipitates from HCT116WT cells confirmed that TFIIB was predominantly dephosphorylated in DoxR-treated cells but not in Nulin3-treated cells, and both treatments induced p53 (Fig. 5B). These data suggest that the dephosphorylation of TFIIB ser-65 plays a causative effect in AREG and EREG silencing after DNA damage.
Fig. 5.
Dephosphorylation of TFIIB plays a role in DNA-damage dependent transcription repression. (A) qPCR analysis of AREG, EREG, p21, and BAX gene expression was performed with HCT116WT cells transfected with pCDNA3 vector, T7-tagged WT, S65A and S65E TFIIB for 40 h followed by 6 h treatment with 5 µg/mL DoxR, 10 μM Nutlin3, and control DMSO. (B) Western blotting was carried out in transfected cells with anti-T7 and anti-β-tubulin antibodies. p53 induction was analyzed in the 5 µg/mL DoxR and 10 μM Nutlin3-treated cells using anti-p53 antibody. Western blotting was also performed with TFIIB immunoprecipitates from HCT116WT cells treated with either 5 µg/mL DoxR or 10 μM Nutlin3 along with the control DMSO for 6 h and immunoblotted using anti-pS65-TFIIB and anti-TFIIB antibodies.
Differential Requirement of TFIIB-Ser65 in the Recruitment of 3′-Processing Complexes.
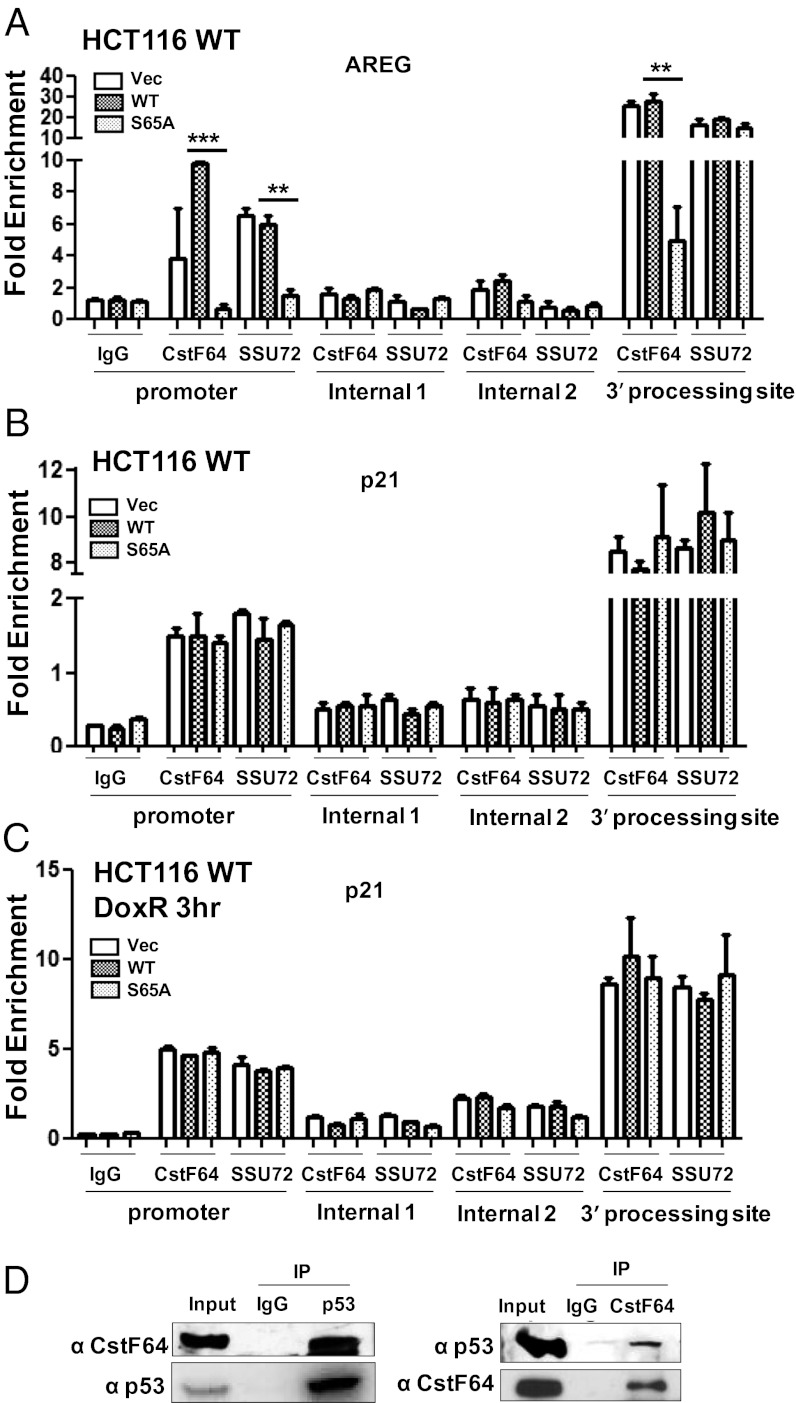
Several studies have linked TFIIB with transcription reinitiation events, where TFIIB interacts with the components of transcription termination and polyadenylation complexes cleavage and polyadenylation specificity factor (CPSF) and CstF, which localize to both the gene-promoter and 3′-processing site (4–6, 19). We have shown that phosphorylation of TFIIB ser-65 is crucial for its interaction with CstF-64 and its recruitment to the gene-promoter and 3′-processing site (7). Therefore, we next analyzed the requirement for TFIIB ser-65 phosphorylation in the recruitment of CstF and CPSF to the p21 gene in HCT116WT and p53−/− cells. Consistent with our previous report, the recruitment of CstF-64 but not of Ssu72 to the AREG 3′-processing region, and of both factors to the promoter region was compromised by the expression of TFIIB S65A (Fig. 6A). In contrast, at the p21 gene there was no significant defect in CstF-64 and Ssu72 occupancy at either the promoter or 3′-processing regions in HCT116WT cells in presence of TFIIB S65A (Fig. 6B). ChIP analysis was also performed with CstF-64 and Ssu72 antibodies in HCT116WT cells transfected with vector, WT, or TFIIB S65A followed by treatment with DoxR for 3 h. The results showed that the 3′-processing factors were efficiently recruited to both the promoter and 3′-processing regions of the p21 gene under conditions of transcriptional activation following DNA damage (Fig. 6C). However, in HCT116 p53−/− cells, the overexpression of TFIIB S65A resulted in reduced occupancy of both CstF-64 and Ssu72 at the p21 promoter (Fig. S6A). Similarly, CstF-64 occupancy at the BAX promoter was compromised in p53-null cells but not in WT cells (Fig. S6 B and C). Thus, in the absence of p53, the p21, and BAX genes require phospho-ser65-TFIIB for the recruitment of CstF to both the gene promoter and 3′-processing sites.
Fig. 6.
Differential requirement of TFIIB ser65 for the recruitment of 3′-processing factor at the p21 promoter. (A) ChIP assays to determine recruitment of CstF-64 and SSU72 at the promoter and 3′-processing regions of the AREG and (B) the p21 gene in HCT116WT cells transfected with pCDNA3, WT TFIIB or TFIIB S65A for 48 h. (C) Similar ChIP assays were performed at p21 gene in transfected HCT116WT cells treated with 5 µg/mL DoxR for 3 h. Two internal regions (1, 2) of the AREG and p21 genes were analyzed for any nonspecific enrichment in CstF-64 and SSU72 ChIP DNA. qPCR data of three independent transfections are plotted as fold enrichment relative to 18S DNA. Error bars denote SD. (D) Coimmunoprecipitation assay was performed with nuclear extracts prepared from HCT116WT cells using anti-p53 (Left) and anti-CstF-64 antibodies (Right). Immunoprecipitates were probed with the indicated antibodies.
Our results suggest that p53 can override the requirement for phospho-ser65-TFIIB in the recruitment of CstF and CPSF to the gene promoter. As mentioned above, phosphorylation of TFIIB ser-65 is required for its interaction with CstF. Recently, p53 has been reported to associate with the CstF 3′ end processing factor (20). We therefore studied the interaction of p53 with the CstF complex through coimmunoprecipitation analysis. Immunoblotting of the p53-immunoprecipitates with anti-CstF-64 antibodies revealed that p53 complexes with CstF, and the reverse immunoprecipitation experiments further confirmed the interaction (Fig. 6D). Taken together, our results suggest that p53-target genes might bypass the phosphorylation-dependent TFIIB–CstF interaction by interaction between p53 and components of the CstF complex.
Discussion
Several studies have uncovered alternate pathways associated with the transcriptional regulation of p53-target genes, and especially the p21 gene, in resting as well as under DNA-damaging conditions (14–16). Diverse stress signals have been shown to elicit varied responses with respect to p21 expression that is mediated by p53 together with the components of the basal transcription machinery. Indeed, TFIIB has been implicated in the regulation of transcription reinitiation at the p21 promoter, which may follow either p53-dependent or p53-independent pathways (12). Moreover, basal levels of p53 can influence the recruitment of TFIIB at the p21 promoter and facilitate pre-initiation complex (PIC) formation in unstressed cells (13). The data we present here suggests that TFIIB phosphorylation plays a role in the selective expression of p53-target genes during cellular stress.
It is interesting to note the promoter of the p21 gene is marked by the presence of nonproductively initiated or stalled RNAPII and hence its transcription is significantly regulated at the elongation step (12, 15). p53 has been shown to stimulate elongation through its interaction with FACT (facilitates chromatin transcription) and TFIIH (21, 22) and was reported to interact with CstF–3′ processing complex (20). Our data suggest that the latter interaction might functionally substitute TFIIB phosphorylation. We also note that phosphorylated TFIIB is present at the promoters of p53-target genes. Hence, in absence of p53, phosphorylation of TFIIB ser-65 is required for the transcription of p21 and BAX, along with the majority of genes that we have tested. However, upon DNA damage, p53 plays a dominant role in activating its target genes as TFIIB is dephosphorylated, which correlates with general transcription attenuation. It is likely that other activators may form similar contacts with the 3′-processing machinery (23) and thus it will be interesting to determine if they can also bypass the requirement for TFIIB phosphorylation.
Ser-65 of TFIIB resides within the B-finger/B-reader of TFIIB that projects into the catalytic center of RNAPII at the transcription initiation site (2, 3). Because TFIIB ser-65 is buried within the active center of RNAPII it may not be available to engage in interactions with a kinase or other transcription factors at the PIC. Our studies thus far have not determined when TFIIB phosphorylation occurs during the transcription cycle. Our previous publication (7) suggested that the phosphorylation of TFIIB ser-65 is required for a function that occurs after the phosphorylation of the RNAPII CTD at ser-5, which coincides with transcription initiation. The TFIIB N terminus is ejected from the RNAPII catalytic center shortly after transcription initiation (5–8 nt), thus exposing the region for phosphorylation and/or interaction with 3′-processing factors. It is possible that TFIIB ser-65 phosphorylation occurs at this point, perhaps to stimulate further rounds of transcription through establishing contact with the 3′-processing complexes. Alternatively, phosphorylation of TFIIB could occur outside the PIC to generate “transcriptionally competent” TFIIB that is then recruited to the promoter.
Our results add to the growing body of evidence that suggests alternate RNAPII CTD kinases may be required at some genes, that is particularly evident with the p21 gene (15, 24). Indeed, the role of CDK7 in the phosphorylation of TFIIB correlates with the differential requirement for CDK7 and TFIIB phosphorylation at the AREG gene versus the p21 gene. Further work will be required to determine how the different CDKs and other protein kinases regulate RNAPII CTD and also TFIIB phosphorylation in gene transcription under normal as well as stress conditions.
Methods
Cell Culture.
HEK 293T and U2OS cells were grown in DMEM, HCT116WT and p53−/− cells were grown in RPMI-1640 medium, and CDK7 (as/as) HCT116 cells were grown in McCoy's 5A medium supplemented with 10% (vol/vol) FBS at 37 °C. Details in SI Methods.
Western Blotting.
Western blotting analysis was performed as described before (7) using antibodies listed in Table S1.
ChIP Assay.
ChIP assay was performed with HEK293T, HCT116WT, and HCT116 p53−/− cells as described previously (7). Statistical significance was analyzed by Student t test (P < 0.05). The sequences of the qPCR primers used in the study are listed in Table S2.
Supplementary Material
Acknowledgments
We thank Prof. David Goodrich for providing the HCT116WT and p53−/− cell lines and Dr. Stephane Larochelle and Prof. Robert Fisher for CDK7 (as/as) HCT116 cell line. This work was funded by the National Institute of General Medical Sciences Grant 1R01GM098609.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207483109/-/DCSupplemental.
References
- 1.Deng W, Roberts SG. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116(5):417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- 2.Kostrewa D, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462(7271):323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327(5962):206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23(22):2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvo O, Manley JL. Strange bedfellows: Polyadenylation factors at the promoter. Genes Dev. 2003;17(11):1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 6.Hampsey M, Singh BN, Ansari A, Lainé JP, Krishnamurthy S. Control of eukaryotic gene expression: Gene loops and transcriptional memory. Adv Enzyme Regul. 2011;51(1):118–125. doi: 10.1016/j.advenzreg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Fairley JA, Roberts SG. Phosphorylation of TFIIB links transcription initiation and termination. Curr Biol. 2010;20(6):548–553. doi: 10.1016/j.cub.2010.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Alessio JA, Wright KJ, Tjian R. Shifting players and paradigms in cell-specific transcription. Mol Cell. 2009;36(6):924–931. doi: 10.1016/j.molcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339(2):225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11(11):761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shandilya J, Roberts SG. The transcription cycle in eukaryotes: From productive initiation to RNA polymerase II recycling. Biochim Biophys Acta. 2012;1819(5):391–400. doi: 10.1016/j.bbagrm.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12(4):1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 13.Gomes NP, Espinosa JM. Differential regulation of p53 target genes: It’s (core promoter) elementary. Genes Dev. 2010;24(2):111–114. doi: 10.1101/gad.1893610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laptenko O, Beckerman R, Freulich E, Prives C. p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proc Natl Acad Sci USA. 2011;108(26):10385–10390. doi: 10.1073/pnas.1105680108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes NP, et al. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20(5):601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 17.Elsby LM, O’Donnell AJ, Green LM, Sharrocks AD, Roberts SG. Assembly of transcription factor IIB at a promoter in vivo requires contact with RNA polymerase II. EMBO Rep. 2006;7(9):898–903. doi: 10.1038/sj.embor.7400767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larochelle S, et al. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Mol Cell. 2007;25(6):839–850. doi: 10.1016/j.molcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27(5):806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Nazeer FI, et al. p53 inhibits mRNA 3′ processing through its interaction with the CstF/BARD1 complex. Oncogene. 2011;30(27):3073–3083. doi: 10.1038/onc.2011.29. [DOI] [PubMed] [Google Scholar]
- 21.Léveillard T, et al. Functional interactions between p53 and the TFIIH complex are affected by tumour-associated mutations. EMBO J. 1996;15(7):1615–1624. [PMC free article] [PubMed] [Google Scholar]
- 22.Keller DM, et al. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell. 2001;7(2):283–292. doi: 10.1016/s1097-2765(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 23.El Kaderi B, Medler S, Raghunayakula S, Ansari A. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J Biol Chem. 2009;284(37):25015–25025. doi: 10.1074/jbc.M109.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glover-Cutter K, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29(20):5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.