Abstract
mRNA localization and regulated translation provide a means of spatially restricting gene expression within each of the thousands of subcellular compartments made by a neuron, thereby vastly increasing the computational capacity of the brain. Recent studies reveal that local translation is regulated by stimuli that trigger neurite outgrowth/collapse, axon guidance, synapse formation, pruning, activity-dependent synaptic plasticity, and injury induced axonal regeneration. Impairments in the local regulation of translation result in aberrant signaling, physiology, and morphology of neurons, and are linked to neurological disorders. This review highlights current advances in understanding how mRNAs are translationally repressed during transport and how local translation is activated by stimuli. We address the function of local translation in the context of fragile X mental retardation.
Regulated translation at the synapse: function and molecular mechanisms
Stimulus-induced changes in gene expression are fundamental to the development and function of all organisms since they allow a shared genome to undergo specific patterns of expression to give rise to an extraordinary variety of distinct cell and tissue types. Within the nervous system, stimulus-induced changes in gene expression function to alter and refine circuit connectivity in a persistent manner. In this way, experience modifies our memories, behaviors, feelings and thoughts such that nature and nurture combine to determine who we are as individuals. Compared to non-neuronal cells, neurons, with their highly polarized morphologies, pose a distinct set of challenges, particularly in terms of the spatial regulation of gene expression. Neurons elaborate axonal and dendritic processes whose lengths often exceed the diameter of the cell soma by many orders of magnitude, and mature neurons can form up to 10,000 synaptic contacts. Stimulus-induced changes in structure and function can occur at each of these synapses, often as the result of regulated translation of localized mRNAs.
Early studies showing that ribosomes are predominantly restricted to the cell soma gave rise to a long-standing belief that all proteins were synthesized in the cell body, and then transported to distal sites. While the initial detection of ribosomes in dendrites [1] was met with some skepticism, it is now widely accepted that local protein synthesis occurs in neurons. Systematic, unbiased approaches have led to the identification of hundreds of localized mRNAs [2–5]. In invertebrate neurons [3,6], local translation of neuritically localized mRNAs underlies synapse formation [7], learning-related synaptic plasticity [8,9], and injury-induced changes in excitability [10]. In the developing vertebrate nervous system, mRNAs are detected in axonal growth cones, where they are translated during axon guidance and synaptogenesis [11–14] (see however Roche et al [15]). In mature vertebrate neurons, most studies have focused on dendritically localized transcripts whose translation is regulated by stimuli that produce changes in synaptic efficacy [16–18], although translation in axons of mature vertebrate neurons is known to be required for neural regeneration following axonal injury [19,20].
mRNA transport into dendrites has been shown to be mediated by cis-acting RNA elements typically contained within the 3’-untranslated region (UTR), which are recognized by trans-acting RNA binding proteins (see Box 1). These messenger ribonucleoproteins (mRNPs) combine with additional RNAs and proteins to form an RNA granule (see Box 2) that is actively transported by motor proteins along the cytoskeleton to its final destination [21,22]. Ongoing efforts in the field are aimed at understanding 1) how is the translation of mRNAs repressed during transport?; 2) how do activating stimuli regulate translation of specific transcripts once they have reached their destination?; and 3) how is local translation altered in diseased neurons, and what does this reveal about the function of local translation in the brain? In this review, we highlight recently gained insights into each of these questions, focusing on mRNA localization and regulated translation in mature CNS neurons (figure 1).
Box 1: RNA-binding proteins acquired in the nucleus affect the cytoplasmic fate of mRNAs.
Many proteins that bind to mRNAs in the nucleus have been shown to remain bound in the cytoplasm and to regulate mRNA localization and translation. Such proteins include heterogeneous nuclear ribonucleoproteins (hnRNPs), exon junction complexes (EJC) and a number of other nuclear RNA binding proteins.
hnRNP A2 binds an RNA element, called the A2 response element (A2RE) within the 3’-untranslated region (UTR) of the mRNA encoding myelin basic protein (MBP) and localizes MBP mRNA to the distal processes of oligodendrocytes [96]. In neurons, A2REs mediate dendritic localization of reporter transcripts, and a subset of dendritically-localized mRNAs contain A2REs, indicating a role for hnRNP A2 in dendritic mRNA targeting [97].
The EJC consists of a set of nuclear proteins that bind to pre-mRNA transcripts during splicing and that have been shown to be required for the cytoplasmic localization of oskar mRNA in Drosophila oocytes [98]. Many EJC components are present in neuronal dendrites, where they have also been shown to bind localized mRNAs [99]. When EJC components are bound within coding regions, they may recruit ribosomes to promote translation; when they are deposited downstream of coding regions, the transcripts are usually targeted for degradation via the Nonsense-Mediated Decay (NMD) pathway after the first round of translation. The mRNA encoding Arc contains two conserved introns within its 3’-UTR and thus may be rapidly degraded by NMD after the first round of translation [100]. Such regulation may ensure tight temporal control and a 'burst' of Arc protein synthesis at stimulated synapses.
The zip-code binding proteins ZBP1 and ZBP2 associate with β-actin mRNA during transcription and are required for both export and dendritic targeting of β-actin mRNA [99].
ELAV/HuD proteins, neuron-specific nuclear RNA binding proteins known to regulate mRNA stability, have been shown to associate with many localized mRNAs, including those encoding GAP43, Homer 1a, neuritin and CamKIIα [101].
LSm1, an auxiliary factor for RNA degradation, and CBP80, a (pre)mRNA binding protein have been shown to bind to dendritically localized mRNAs β-actin, eEF1α, and the IP3 receptor [102].
Box 2: Heterogeneous populations of RNA granules in dendrites.
mRNAs associate with distinct sets of proteins throughout their life cycle, from transcription to degradation, through a process of ribonucleoprotein (RNP) remodeling. The population of dendritic mRNPs is thus heterogeneous and dynamic, reflecting both the diversity of localized mRNAs and the transition between states of translational repression, activation and degradation. Understanding the composition of, and the relationship between, various dendritic RNA granules is a focus of research in the field.
RNA Transport Granules. Biochemical fractionation reveals that dendritically localized mRNAs are present in small RNPs, free of ribosomes, as well as in larger structures that contain ribosomes, kinesin and the RNA binding protein Staufen [103]. Affinity purification for kinesin interacting proteins in brain led to the identification of larger (~1000S) RNA transport granules containing localized mRNAs and several dozen proteins, including translation factors and repressors [104]. In a separate study, Elvira et al. [105] purified and characterized large RNA transport granules composed of a distinct set of localized mRNAs and proteins, indicating that distinct mRNAs are transported in distinct RNA granules.
Stress Granules (SGs) temporarily arrest mRNA translation when cells are stressed (e.g. during oxidative or metabolic stress). Once the stress is relieved, SGs dismantle and mRNA can resume normal activity [106]. SGs have been detected in dendrites, where they may form dynamically by recruitment of Pumilio proteins to RNAs and subsequent aggregation [33].
Processing Bodies (p-bodies) function to prime and process mRNAs for degradation and contain decapping enzymes, exonucleases, miRNAs and components of the RISC machinery. Many p-body markers have been detected in neuronal dendrites [107,108]. A distinct class of p-bodies, termed dendritic p-body like structures, or dlPBs, have been detected in hypothalamic and hippocampal neurons that contain many components of p-bodies but lack the exonuclease Xrm1 [109], and may thus serve as storage sites for translationally repressed mRNAs.
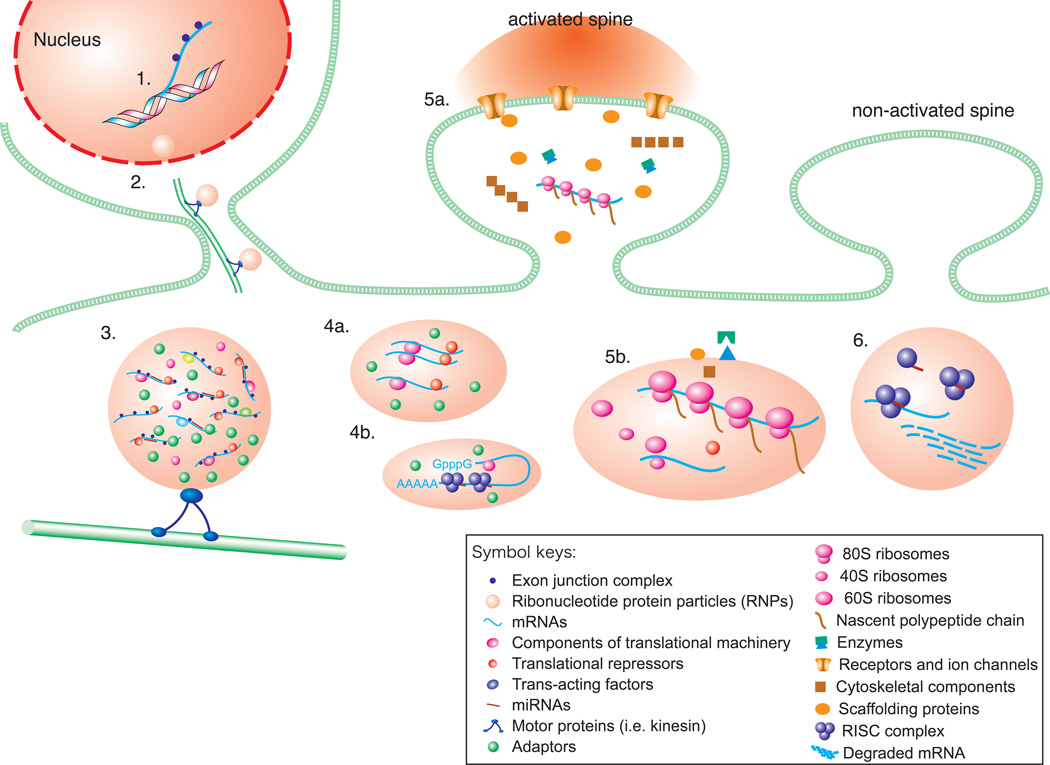
Fig. 1. Model of mRNA transport and regulated local translation in dendrites.
1. Transcription of mRNAs and co-transcriptional recruitment of transacting factors such as the exon junction complex; 2. Assembly and export of ribonucleotide protein particles (RNPs); 3. The cytoplasmic RNP is packaged into a transport granule consisting of mRNAs; components of the translational machinery, translational repressors; trans-acting factors; miRNAs, kinesin and adaptors, which are transported along microtubules by molecular motors into dendrites; 4. Mechanisms by which dendritic mRNA are thought to be maintained in a translationally repressed state are a) repression by assembly of mRNAs together with transational repressors into structures known as RNPs; and b) repression by assembly of mRNAs together with miRNAs to form the RISC complex within structures known as process-bodies (P-bodies); 5. a) Activation of neurotransmitter receptors and voltage-gated ion channels engages intracellular second messenger cascades such as the mTOR pathway, which promote translation by turning on the translational machinery and/or by removing translational repressors; b) Active translation at a stimulated synapse 80S ribosome, 40S ribosomes, 60S ribosomes, nascent polypeptide chain; newly synthesized proteins (enzymes, receptors and ion channels, cytoskeletal components, scaffolding proteins) are incorporated into the synapse; 6. Local degradation of mRNAs. miRNA-RISC-mediated local degradation of mRNAs in P-bodies (RISC complex; mRNA prior to degradation; degraded mRNA; miRNA).
How are mRNAs silenced during transport?
To spatially restrict gene expression, mRNAs must be translationally repressed until they reach their final destination. Translational repression is usually achieved by binding of trans-acting factors that prevent the mRNA from being translated. Here we consider several of best characterized such factors, eukaryotic Initiation Factor 4E (eIF4E) binding proteins, Fragile X Mental Retardation Protein (FMRP), Cytoplasmic Polyadenylation Element-Binding Protein (CPEB), Pumilio and microRNAs. Although not yet demonstrated in neurons, RNA oligomerization and formation of large silent mRNPs can also efficiently suppress translation (see below).
Repression of cap-dependent translation initiation by eIF4E binding proteins
Eukaryotic translation, the process by which mRNA is translated into proteins, consists of three main steps: initiation, elongation and termination, and in the processes of neurons, is typically cap-dependent. Cap-dependent translation refers to the form of translation in which proteins known as eukaryotic Initiation Factors (eIFs) interact with the “cap” or 7-methyl-GTP structure located at the 5’ end of the mRNA [23]. The initiation complex eIF4F is composed of eIF4E, the protein that directly binds the cap; eIF4G, which binds to Poly(A)-Binding Protein (PABP), a protein that bridges the 5’ and 3’ ends of the mRNA, and thereby promotes mRNA circularization, and eIF4A, an RNA helicase that unwinds secondary structures in the 5’UTR to allow scanning to the translation initiation site. The 40S ribosomal subunit is thought to be recruited to the mRNA by eIF3, which binds to eIF4G.
As might be expected for a complex biochemical process, the initiation phase of translation is a common target for regulation. Regulation of protein synthesis can occur by posttranslational modification of translation factors, which would modulate translation of an array of mRNAs, and/or binding of RNA regulatory proteins to the 5’ or 3’ UTRs of specific mRNAs. One of the best-characterized mechanisms involving posttranslational modification is that of mTOR-dependent phosphorylation of 4E-BP, an eIF4E binding protein. Unphosphorylated 4E-BP competes with binding of eIF4G to eIF4E, thereby blocking assembly of the eIF4F initiation complex and initiation of cap-dependent translation. In addition, some translational regulatory proteins such as Maskin, neuroguidin and CYFIP1, also inhibit cap-dependent translation through regulating binding of eIF4G to 4E. These 4E-BP-like proteins bind specific transcripts directly or indirectly via adaptor proteins, and in this way repress translation in a transcript-specific manner. mTOR-dependent phosphorylation of 4E-BP releases eIF4E, enabling it to assemble with eIF4G to form the initiation complex eIF4F.
In neurons, FMRP recruits the 4E-BP cytoplasmic FMR-interacting protein-1 (CYFIP1) to target mRNAs, repressing their translation [24]. The cytoplasmic polyadenylation element-binding protein (CPEB) associates with the 4E-BP neuroguidin and Maskin, and represses translation of a large set of CPE-containing mRNAs [25,26]. Recent work has indicated that a dendritically localized non-coding RNA, brain cytoplasmic (BC1) RNA, may regulate the interaction between CYFIP1 and FMRP [27] (but also see Iacoangeli et al [28]). Of note, BC1 also binds eIF4A, blocking its helicase activity [29].
Additional mechanisms of translational repression target later steps in translational initiation
Members of the Pumilio family of translational repressors are evolutionarily conserved RNA-binding proteins that regulate translation of specific mRNAs by binding their 3'-UTRs and promoting deadenylation [30]. In neurons, Pumilio regulates the morphology of presynaptic terminals at Drosophila neuromuscular junctions by repressing translation of eIF4E in postsynaptic structures [31], and is implicated in learning and memory, regulating the translation of synaptically localized mRNAs [32]. Pumilio2 has been detected as a component of RNA granules in dendrites of hippocampal neurons [33]. By analogy to yeast, these findings suggest that Pumilio family members may repress translation of localized mRNAs in neurons by blocking formation of the 80S ribosome.
Additionally, although not yet demonstrated in neurons, RNA oligomerization and formation of large silent mRNPs may also efficiently suppress translation of the mRNA encoding oskar, a protein required for the development of the posterior pole plasm in the Drosophila oocyte, perhaps by preventing accessibility of the translational machinery to the mRNA [34]. Of note, many components of the granule or RNP that contains oskar mRNA have been identified and seem to localize to neuronal processes, including polypyrimidine tract binding protein (PTB), barentsz and staufen [22,35].
microRNAs and transcript-specific translational repression
MicroRNAs (miRNAs) are non-coding, ~21 nt long RNAs whose mode of action involves complementary base pairing to sequences in a target mRNA, leading most commonly to translational repression of the target. Complementary binding can be partial and usually only involves the 6–8 nt long “seed site” in the miRNA. As such, each miRNA can repress the translation of a relatively large number of specific transcripts. Over half of all identified mouse miRNAs are expressed in the brain [36], and some of these are also present in dendrites and synapses [37,38]. Moreover, transcripts encoding synaptic proteins, including FMRP and PSD95, comprise the largest group of predicted targets of human miRNAs [39]. Together, these findings support the concept that miRNAs could regulate the translation of localized mRNAs in neurons [40]. Consistent with this hypothesis, Schratt and colleagues have shown that miR-134 colocalizes with and regulates translation of the mRNA encoding LIM kinase (LIMK), an actin-binding kinase that phosphorylates members of the ADF/cofilin family of actin binding and filament severing proteins, whose expression is implicated in spine morphogenesis [41]. In a more recent study, miR138 was also found to localize to dendrites, where it represses translation of the depalmitoylation enzyme APT1F and negatively regulates the size of dendritic spines [42]. Smalheiser and Lugli detected pre-miRNAs, as well as components of the miRNA processing machinery, in synaptoneurosomes [43]. In this study, the authors propose a model in which pre-miRNAs undergo activity-dependent processing into mature miRNAs at stimulated synapses to generate synapse-specific translational repression of transcripts.
How does local stimulation regulate the translation of localized mRNAs?
While translation of localized mRNAs is repressed during transport, it is likely that, upon arrival at their final destination, distinct stimuli and/or patterns of neuronal activity triggers the translation of specific subsets of localized mRNAs. How are local extracellular cues and stimuli transduced into local translational activation? How do growth cones and synapses decode the specific signal and activity patterns? Stimuli that activate mRNA translation do so both by regulating general components of the protein synthetic machinery and by regulating mRNA-specific repressors.
mTOR and S6K pathways release repression of general translation initiation
Diverse synaptic signals regulate the translation of localized RNAs. Such localized stimuli include those that trigger synapse formation, pruning, neurite outgrowth/collapse, activity-dependent synaptic plasticity and injury-induced axonal regeneration. A critical regulator of activity-dependent protein synthesis in dendrites is metabotropic glutamate receptor (mGluRs). mGluRs are G protein-coupled receptors enriched at excitatory synapses throughout the brain where they act to regulate glutamatergic neurotransmission. Group I mGluRs (mGluR1/5) activate phospholipase C, leading to intracellular Ca2+ mobilization, and the extracellular signal-regulated kinase (ERK)–mitogen-activated protein kinase (MAPK) pathway through which they modulate synapse-to-nucleus communication. Signaling by mGluR1/5 is critical to synaptic circuitry formation during development and is implicated in forms of plasticity including long-term potentiation (LTP), LTD, associative learning and cocaine addiction [44]. mGluR1/5 elicit synapse-specific modifications in synaptic strength and spine morphology by stimulating rapid local translation of dendritic mRNAs including Fmr1, which encodes FMRP [45].
The PI3 kinase/Akt (protein kinase B)/mammalian Target of Rapamycin (mTOR)/p70S6K and Ras/MAPK/p90S6K signaling pathways play central roles in activity-dependent local translation in dendrites and are critical signaling pathways downstream of group I mGluRs (figure 2) [46]. Both pathways target translational initiation by regulating the phosphorylation status of eIF4E and 4E-BPs. ERK/MAPK-dependent phosphorylation of eIF4E increases the rate of general cap-dependent translation, while mTOR-dependent phosphorylation of 4E-BPs decreases their affinity for eIF4E, releasing eIF4E, thereby promoting translational initiation[47–50]. Both pathways also induce phosphorylation of the ribosomal protein S6, which promotes translation of mRNAs with 5’ terminal oligopyrimidine tracts (5’TOPs), for example transcripts encoding ribosomal proteins. Note, however, that studies of a knock-in mouse with all S6 phosphorylation sites mutated revealed that S6 phosphorylation is dispensable for 5’TOP-dependent translation [51].
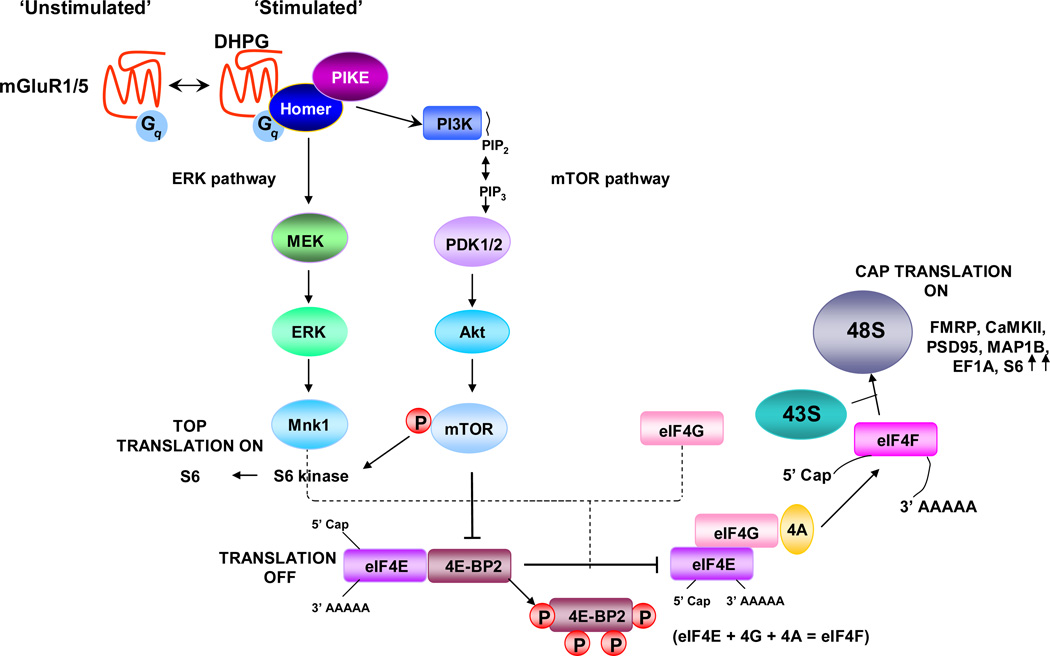
Fig. 2. Signaling pathways that regulate mTOR-dependent local protein synthesis in dendrites.
Activation of group I metabotropic glutamate receptors (mGluRs) engages the ERK/MAPK and phosphatidylinositol 3-kinase (PI3K) signaling pathways. PI3K acts via PDK1/2 to phosphorylate and activate Akt. Akt phosphorylates and activates the mammalian target of rapamycin (mTOR). Upon activation, mTOR phosphorylates and activates S6 kinase 1 (S6K1). Phosphorylation (P) of eIF4E-binding protein (4E-BP) by mTOR promotes the release of 4E-BP from the eukaryotic translation initiation factor 4E (eIF4E), enabling eIF4E to assembly with eIF4G and form the active translation initiation complex eIF4F, composed of eIF4E, eIF4A and eIF4G. eIF4F recruits the mRNA to the 43S pre-initiation complex to form the 48S initiation complex. The eIF4F complex and the poly(A) tail act synergistically to stimulate mRNA translation. ERK phosphorylates and activates MAPK-interacting serine/threonine kinase 1 (Mnk1), which phosphorylates eIF4E. S6K1 phosphorylates and activates ribosomal protein S6, stimulating TOP-dependent translation. The intracellular signaling pathways depicted here are also activated by brain-derived neurotrophic factor (BDNF) in the hippocampus and cultured cortical neurons, and by serotonin in Aplysia californica sensory neurons. m7G, 7-methyl-GTP; PIP2, phosphatidylinositol-4,5-bisphosphate; PIP3, phosphatidylinositol-3,4,5-trisphosphate. Adapted from [46].
In neurons, components of both the PI3K/Akt/mTOR and Ras/MAPK/p90S6K pathways are present at synapses where they are activated by stimuli that promote protein synthesis-dependent forms of synaptic plasticity (figure 3). In both cases, these signaling pathways are thought to promote synaptic plasticity via regulation of local protein synthesis. ERK and mTOR activation are required for late-phase LTP (L-LTP) [47,48,52–54] and mGluR-dependent long-term depression (mGluR-LTD) [47,48,52–54].
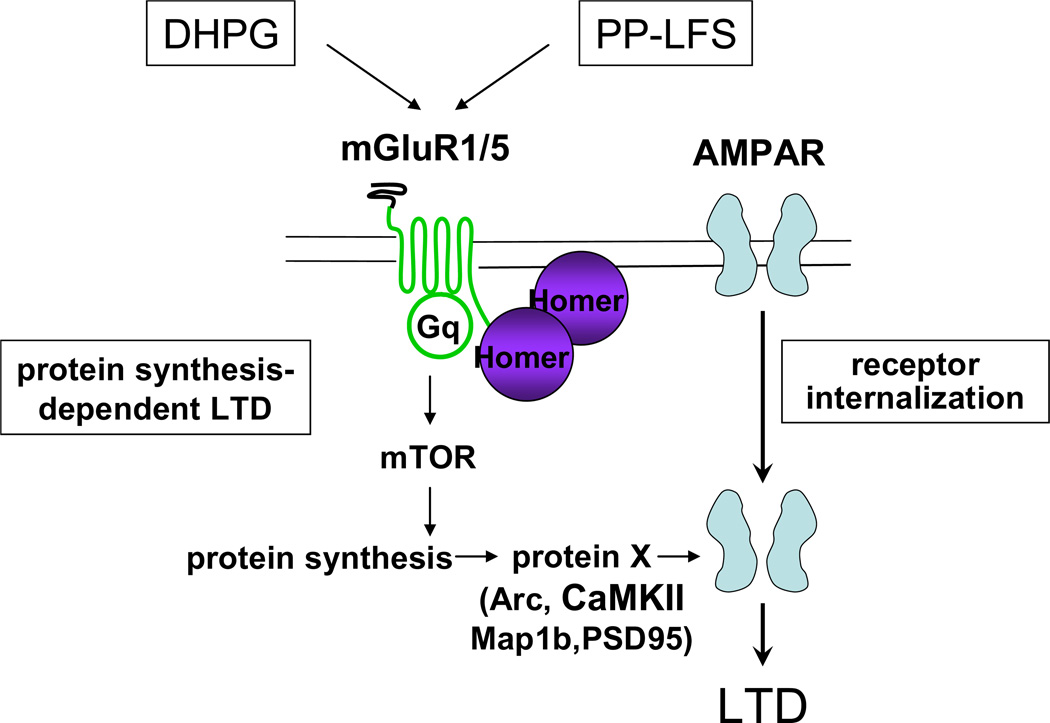
Fig. 3. mGluR-LTD at CA1 synapses of adolescent wild-type mice requires local protein synthesis.
Induction of mGluR-LTD by either bath application of the group I mGluR agonist DHPG or by paired pulse low frequency stimulation (PP-LFS) promotes the internalization of synaptic AMPARs in a group I mGluR-dependent manner, leading to a decrease in synaptic efficacy or LTD. mGluR-dependent internalization of AMPARs at CA1 synapses of adolescent wild-type mice requires mTOR-dependent local protein synthesis of an as yet unknown protein (eg., Arc, MAP1b, PSD-95, CaMKII). Correct coupling of Homer to mGluR1/5 is required for activation of mTOR, AMPAR internalization and mGluR-LTD [15, 42]. Courtesy of Huber KM.
Elongation factor eEF2 and regulation of translation elongation
Activity can also regulate local protein synthesis by targeting translational elongation. Elongation factor eEF2, which catalyzes the translocation of ribosomes along the mRNA, serves as a biochemical sensor for local translation via bidirectional activity-dependent phosphorylation [55]. Action potential-mediated network activity maintains eEF2 in a relatively dephosphorylated (active) state, whereas spontaneous neurotransmitter release (miniature synaptic transmission) promotes its phosphorylation and thus tonically suppresses local translation [55]. Following chronic silencing of activity, eEF2-becomes dephosphorylated (active) and local protein synthesis is triggered, which is required to rapidly increase synaptic expression of GluR1 and maintain synaptic homeostasis in the circuit [56].
During mGluR-LTD, which requires rapid de novo protein synthesis, eEF2 is phosphorylated by eEF2K, a Ca2+/calmodulin-dependent kinase that binds to mGluR. The group I mGluR agonist DHPG releases eEF2K to phosphorylate eEF2 and thereby inhibits elongation of translation. While this attenuates translation of most transcripts, the translation of other mRNAs is facilitated. One such mRNA encodes the Activity-regulated cytoskeletal protein Arc [57,58 ]. Arc is encoded by an immediate early gene whose mRNA is rapidly transported into dendrites [59]. The Arc protein is required for internalization of AMPA receptors during LTD [58]. Its translation is increased during DHPG-induced LTD in an eEF2-phosphorylation-dependent manner [57,58].
Stimulus-induced translational activation by release of transcript-specific repressors
Mechanisms of translational activation that impinge on general translation factors, such as those described above, regulate the translation of many transcripts. Stimulus-induced modifications of transcript-specific translational repressors in contrast can regulate translation of specific mRNAs.
Shiina and colleagues [60] identified the protein RNA Granule Protein 105 (RNG105) as a component of RNA granules that functions as a translational repressor that is released from RNA granules following BDNF stimulation, concomitant with an increase in translation of localized mRNAs such as CamKIIα. As another example, β-actin mRNA translation in axonal growth cones is repressed by a protein that binds to the “zipcode” element in its 3’UTR, zipcode binding protein 1 (ZBP1) [61]. In growth cones, ZBP1 is phosphorylated by Src kinase, which releases the protein from the mRNA, and activates β-actin translation [61].
In the case of CPEB, synaptic stimulation switches its function from a translational inhibitor (as described above) to a translational activator. Specifically, NMDA receptor stimulation activates Aurora kinase, which phosphorylates CPEB, leading to the recruitment of cleavage and polyadenylation specificity factor (CPSF), which induces poly(A)-tail elongation, release of neuroguidin from eIF4E leading to translational activation [25]. This mechanism appears to promote translation of CamKIIα mRNA, along with a number of other mRNAs, following a variety of stimuli [26].
Studies of CamKIIα translation in dendrites of Drosophila neurons during long-term memory formation have revealed a role for activity-dependent relief of miRNA-mediated translational repression [62]. Here, cholinergic stimulation was found to degrade Armitage, a component of the RISC complex that is necessary for miRNA-mediated translational silencing. Degradation of Armitage led to increased translation of CamKIIα in dendrites, presumably by relieving miRNA inhibition. Intriguingly, Armitage was degraded by the ubiquitin proteasome pathway, providing a mechanistic link between the regulation of local protein synthesis and degradation during memory [63]. While these studies suggest that regulated degradation of RISC components can function to release miRNA translational inhibition, whether and how this could occur in a miRNA-specific manner remains unknown. More recent studies in rodent hippocampal neurons revealed that the mammalian homolog of Armitage, MOV10 also undergoes activity-dependent degradation via the ubiquitin-proteasome pathway [64] MOV10 degradation was triggered by NMDA receptor activation, and promoted the translation of a panel of dendritically localized mRNAs, including those encoding Limk1 and the depalmitoylating enzyme lysophospholipase1 (Lypla1), transcripts whose dendritic translation was previously shown to be regulated by miR134 and miR138 [41,64].
An additional mechanism whereby stimulation can activate translation is by selective translation of mRNAs containing Internal Ribosomal Entry Sites (IRESs). Translation of IRES-containing mRNAs is cap-independent and IRES trans-acting factors (ITAFs) traffic the ribosome to the start site, bypassing the need to scan from the 5'-UTR. Such transcripts may be preferentially translated when cap-dependent translation is inhibited. Of note, several dendritically localized transcripts seem to contain IRESs, although no studies have shown activity-dependent, IRES-mediated translation of these localized mRNAs [65]. In Aplysia neuroendocrine bag cells, however, Sossin and colleagues have shown that translation of the mRNA encoding egg-laying hormone (ELH), an mRNA that localizes to distal neurites, undergoes activity-dependent translational activation by switching from cap-dependent to IRES-dependent translation [66].
Another way to achieve specificity of translational regulation is to regulate the dendritic/synaptic localization of mRNAs. Several transcripts undergo activity-dependent transport into dendrites, including mRNAs encoding BDNF, TrkB, eIF4E and Arc [59,67,68]. Following stimulation, Arc mRNA is not only targeted into dendrites, but it also concentrates specifically at stimulated synapses [59]. The dendritic localization of BDNF mRNA has recently been shown to be regulated by alternative 3’-polyadenylation, such that only isoforms containing the long 3’-UTR are localized to dendrites [69]. A recent study of a sensorin translational reporter in Aplysia sensory-motor neurons found that serotonin only induced translation of the reporter when it localized to sensory-motor synapses [9], indicating that the particular subcellular localization of an mRNA within the neuronal process is critical to its translational regulation.
Can distinct stimuli regulate translation of specific localized transcripts?
The discovery of hundreds of dendritically localized mRNAs, and the elucidation of mechanisms for transcript-specific translational repression and activation, suggest that distinct stimuli might regulate the translation of distinct subsets of mRNAs. Consistent with this possibility, dynamic imaging of sensorin translation in Aplysia sensory neurons revealed translational induction during serotonin-induced long-term facilitation but not during FMRFamide-induced LTD of sensory-motor synapses [9]. In contrast, however, both LTP- and LTD-inducing stimuli stimulate translation of Arc [57,58,70] and EF1α [49,71] mRNAs in dendrites of rodent hippocampal neurons. The stimuli used to elicit LTP and LTD in these studies included bath application of BDNF and DHPG, as well as high frequency tetanic stimulation. Understanding whether and how specific types and patterns of stimulation regulate translation of specific mRNAs may require monitoring translation following more physiologically relevant stimuli.
Dysregulation of local translation underlies human neurological disorders
Given the central function of local translation during the formation and function of neural circuits, it is not surprising that mutations in genes involved in local translation have been found to underlie several human neurological disorders. Of these, studies of Fragile X Mental Retardation have provided the most insight into both the physiological function of local translation and the pathophysiology of the disease.
Dysregulation of local translation in Fragile X
Fragile X syndrome is the most common heritable form of mental retardation and the most common known genetic cause of autism [44,72,73]. Fragile X syndrome is caused by loss-of-function mutations in FMRP, which is encoded by the FMR1 gene located on the X chromosome. In humans, Fragile X syndrome typically results from expansion of a CGG repeat sequence in the 5’-untranslated region and silencing of the FMR1 gene [74,75]. Patients with the Fragile X syndrome exhibit a wide-range of neurological deficits including cognitive impairment, seizures, emotional instability, sleep disorders, attention deficits, autonomic dysfunction, and autism [44,72,73]. FMRP, the gene product of the FMR1 gene, is an RNA binding protein that associates with a large array of mRNAs, many of which encode proteins important for neuronal development and plasticity. In neurons, FMRP recruits CYFIP1 to target mRNAs, repressing their translation [24]. FMRP controls activity-dependent dendritic mRNA localization, stability and translational efficiency of dendritic mRNAs in response to stimulation of mGluRs [44,72,73].
FMRP is detected in cell bodies, in dendritic shafts and branch points [76], and at the base of synaptic spines and spine heads [77]. FMRP mRNA is translated near synapses in response to neurotransmitter activation [78]; moreover, different models of experience-dependent plasticity such as whisker stimulation [79,80], visual experience [81], and exposure to enriched environment [82] also promote FMRP translation. In addition, FMRP modulates the synaptic translation of other mRNAs in a neurotransmitter-dependent manner [58,83].
Mice lacking FMRP exhibit abnormalities in dendritic spine morphology, cognitive deficits exaggerated LTD, which is protein synthesis-independent at Schaffer collateral to CA1 pyramidal cell synapses of the hippocampus and decreased LTP in the cortex [44,72,73]. Insights into the molecular cascades that link overactivated group I mGluRs to exaggerated mGluR-LTD have recently been revealed. Recent findings indicate that in wild-type neurons, FMRP represses an array of mRNAs implicated in synaptic plasticity and spine morphogenesis including PI3K-enhancer (PIKE), a direct target of FMRP (Jennifer Darnell, personal communication). In hippocampal neurons of Fmr1-deficient mice, upregulation of PI3K-enhancer PIKE, an upstream regulator of PI3K/Akt signaling, results in overactivation of PI3K/Akt and mTOR signaling in the hippocampus, as assessed by several functional read-outs, including formation of the eIF4F translation initiation complex [84]. These findings are consistent with a model whereby in the Fragile X mouse, the protein/s required for mGluR-dependent AMPA receptor internalization and mGluR-LTD are already accumulated at Schaffer collateral to CA1 synapses under basal conditions [85]. These observations provide an important functional link between overactivated mGluR signaling, aberrant protein synthesis, and exaggerated mGluR-LTD in Fmr1 KO mice [84]. Several neurotransmitter receptors are dysregulated in the absence of FMRP. For example, the Fmr1 KO mouse exhibits aberrant numbers and/or activity of mGluR5 [86,87], dopamine [88] and AMPA [89] receptor signaling.
A hallmark feature of Fragile X syndrome in humans, also observed in the Fmr1 KO mouse, is that of synaptic spine dysmorphogenesis [90]. Spines on hippocampal and cortical neurons of Fmr1 KO mice exhibit enhanced density and are thinner and longer than those of age-matched wild-type mice, resembling an immature morphology [90–93]. These observations indicate that FMRP might normally repress local translation of proteins that inhibit synapse maturation, stabilization, and elimination [90]. On the other hand, a combination of imaging and electrophysiological studies indicate that in the absence of FMRP there is a delay in establishing synaptic connections [94] and a persistent, reduction in connectivity between neurons in the somatosensory, barrel cortex of Fragile X mice [95]. These observations raise the possibility that the increased number of spines on pyramidal cells might represent a compensatory mechanism to balance [or counter] the decrease in synapse connectivity. In the somatosensory cortex, although spine dysmorphogenesis persists into adulthood [92,93], it can be rescued by exposure of Fmr1-deficient mice to an enriched environment [92].
Conclusions
Over the past decade, the study of local translation in neurons has evolved from asking questions about whether and when local translation occurs to asking more mechanistic questions about how mRNA localization and translational regulation occur. In this review, we highlighted new insights into the mechanisms whereby mRNAs are transported in a translationally repressed state to specific subcellular compartments, focusing on repression of translation by eIF4E-BPs and related proteins, and on transcript-specific translational repression by miRNAs. We then considered mechanisms whereby distinct patterns of synaptic stimulation regulate translation of localized mRNAs. Here we focused on recent studies revealing central roles for the mTOR and S6K signaling pathways in regulating translation initiation at synapses, and on studies indicating a critical role for the miRNA pathway in local translation. Finally, we concentrated our attention on the many insights into the function and molecular mechanisms of local translation that have been garnered from studies of Fragile X Mental Retardation, a form of mental retardation caused by mutations in an RNA binding protein and translational repressor, FMRP. As we hope our review of the field indicates, new technologies that allow mRNA localization and regulated translation to be analyzed, manipulated and dynamically visualized in neurons are revealing complex, finely tuned functions for local translation in neuronal structure and function. As is often the case, these studies have given rise to a new set of questions, many of which are listed in Box 3. Challenges for the future include developing approaches to address these questions, and in particular developing technologies to study local translation in intact, complex neural circuits, thereby permitting investigation into the function of local translation in the nervous system of living animals.
Box 3: Outstanding questions in the field of local translation at the synapse.
What is the nature of the cis-acting RNA localization elements that target transcripts to specific subcellular compartments within a neuron?
What are the RNA binding proteins that function to localize mRNAs within neurons and how do they mediate this localization?
What is the composition of the RNPs that localize mRNAs to dendrites? What is the relationship between RNA transport granules, P-bodies and stress granules?
What are the physiologically relevant stimuli that regulate local translation?
Do/how do distinct stimuli regulate the translation of specific subsets of transcripts?
How does the miRNA pathway contribute to local translation at synapses and thus synaptic plasticity?
Are there differences in the mechanisms of translational regulation at the synapse as compared to in the soma?
What mechanisms at synapses facilitate folding and maturation of the newly synthesized proteins?
What is the nature of the secretory pathway, which is necessary for synthesis of membrane and secreted proteins, in distal dendrites?
How does local translation of specific transcripts contribute to or alter the function of neural circuits?
Is/how is stimulus-induced transcriptional regulation in the nucleus integrated with stimulus-induced local translation at the synapse?
What are the proteins critical to mGluR-LTD that are accumulated at CA1 synapses of Fragile X mice?
Is PI3 kinase enhancer (PIKE) an FMRP target critical to aberrant protein-synthesis dependent synaptic plasticity observed in Fragile X mice?
Acknowledgements
The authors acknowledge NIH grants NS20752-23 (to RSZ) and NS045324 (to KCM), the FRAXA Foundation (to RSZ), and the F.M. Kirby Foundation (to RSZ). RSZ is the F.M. Kirby Professor in Neural Repair and Protection; KCM is the Eleanor Leslie Term Chair in Innovative Brain Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eberwine J, et al. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem Res. 2002;27:1065–1077. doi: 10.1023/a:1020956805307. [DOI] [PubMed] [Google Scholar]
- 3.Moccia R, et al. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poon MM, et al. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki T, et al. Characterization of mRNA species that are associated with postsynaptic density fraction by gene chip microarray analysis. Neurosci Res. 2007;57:61–85. doi: 10.1016/j.neures.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Gioio AE, et al. Nerve terminals of squid photoreceptor neurons contain a heterogeneous population of mRNAs and translate a transfected reporter mRNA. Eur J Neurosci. 2004;20:865–872. doi: 10.1111/j.1460-9568.2004.03538.x. [DOI] [PubMed] [Google Scholar]
- 7.Lyles V, et al. Synapse formation and mRNA localization in cultured Aplysia neurons. Neuron. 2006;49:349–356. doi: 10.1016/j.neuron.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 8.Miniaci MC, et al. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59:1024–1036. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang DO, et al. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weragoda RM, et al. Memory-like alterations in Aplysia axons after nerve injury or localized depolarization. J Neurosci. 2004;24:10393–10401. doi: 10.1523/JNEUROSCI.2329-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung KM, Holt CE. Live visualization of protein synthesis in axonal growth cones by microinjection of photoconvertible Kaede into Xenopus embryos. Nat Protoc. 2008;3:1318–1327. doi: 10.1038/nprot.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin A, Holt C. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebeo J, et al. Requirement for Protein Synthesis at Developing Synapses. J Neurosci. 2009;29:9778–9793. doi: 10.1523/JNEUROSCI.2613-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Hengst U, et al. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11:1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roche FK, et al. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;29:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 18.Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr.Opin.Neurobiol. 2009 doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willis DE, Twiss JL. The evolving roles of axonally synthesized proteins in regeneration. Curr Opin Neurobiol. 2006;16:111–118. doi: 10.1016/j.conb.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Toth CC, et al. Locally synthesized calcitonin gene-related Peptide has a critical role in peripheral nerve regeneration. J Neuropathol Exp Neurol. 2009;68:326–337. doi: 10.1097/NEN.0b013e31819ac71b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiebler MA, Bassell G. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Napoli I, et al. The fragile X syndrome protein represses activitydependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 25.Jung MY, et al. Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol Cell Biol. 2006;26:4277–4287. doi: 10.1128/MCB.02470-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Zalfa F, et al. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J Biol Chem. 2005;280:33403–33410. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- 28.Iacoangeli A, et al. On BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci U S A. 2008;105:734–739. doi: 10.1073/pnas.0710991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin D, et al. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol. 2008;28:3008–3019. doi: 10.1128/MCB.01800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstrohm AC, et al. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 31.Menon KP, et al. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 32.Chen G, et al. Identification of synaptic targets of Drosophila pumilio. PLoS Comput Biol. 2008;4:e1000026. doi: 10.1371/journal.pcbi.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vessey JP, et al. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J Neurosci. 2006;26:6496–6508. doi: 10.1523/JNEUROSCI.0649-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chekulaeva M, et al. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 35.Ma S, et al. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim Biophys Acta. 2007;1773:912–923. doi: 10.1016/j.bbamcr.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kye MJ, et al. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13:1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lugli G, et al. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.John B, et al. Human MicroRNA targets. Plos Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konecna A, et al. What are the roles of microRNAs at the mammalian synapse? Neurosci Lett. 2009 doi: 10.1016/j.neulet.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 41.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 42.Siegel G, et al. A functional screen implicates microRNA-138- dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smalheiser NR, Lugli G. microRNA Regulation of Synaptic Plasticity. Neuromolecular Med. 2009 doi: 10.1007/s12017-009-8065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronesi JA, Huber KM. Metabotropic glutamate receptors and fragile x mental retardation protein: partners in translational regulation at the synapse. Sci Signal. 2008;1:pe6. doi: 10.1126/stke.15pe6. [DOI] [PubMed] [Google Scholar]
- 45.Greenough WT, et al. Synaptic regulation of protein synthesis and the fragile X protein. Proc.Natl.Acad.Sci.USA. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat.Rev.Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 47.Kelleher RJ, 3rd, et al. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 48.Hou L, Klann E. Activation of the phosphoinositide 3-kinase- Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2004;24:6352–6361. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsokas P, et al. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J Neurosci. 2005;25:5833–5843. doi: 10.1523/JNEUROSCI.0599-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banko JL, et al. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26:2167–2173. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 52.English JD, Sweatt JD. A requirement for the mitogen-activated protein kinase cascade in hippocampal long term potentiation. J Biol Chem. 1997;272:19103–19106. doi: 10.1074/jbc.272.31.19103. [DOI] [PubMed] [Google Scholar]
- 53.Gallagher SM, et al. Extracellular signal-regulated protein kinase activation is required for metabotropic glutamate receptor-dependent long-term depression in hippocampal area CA1. J Neurosci. 2004;24:4859–4864. doi: 10.1523/JNEUROSCI.5407-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang SJ, et al. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci U S A. 2002;99:467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutton MA, et al. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 56.Sutton MA, et al. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 57.Park S, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waung MW, et al. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steward O, et al. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 60.Shiina N, et al. A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J Neurosci. 2005;25:4420–4434. doi: 10.1523/JNEUROSCI.0382-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ross AF, et al. Characterization of a beta-actin mRNA zipcodebinding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashraf SI, et al. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 63.Fonseca R, et al. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006;52:239–245. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 64.Banerjee S, et al. A Coordinated Local Translational Control Point at the Synapse Involving Relief from Silencing and MOV10 Degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 65.Pinkstaff JK, et al. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc Natl Acad Sci U S A. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costa-Mattioli M, et al. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tongiorgi E, et al. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moon IS, et al. Neuronal activation increases the density of eukaryotic translation initiation factor 4E mRNA clusters in dendrites of cultured hippocampal neurons. Exp Mol Med. 2009 doi: 10.3858/emm.2009.41.8.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.An JJ, et al. Distinct role of long 3' UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Messaoudi E, et al. Sustained Arc/Arg3.1 synthesis controls longterm potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang F, et al. The mRNA for elongation factor 1alpha is localized in dendrites and translated in response to treatments that induce long-term depression. J Neurosci. 2005;25:7199–7209. doi: 10.1523/JNEUROSCI.1779-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bassell G, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng Y, et al. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995;268:731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 75.Garber K, et al. Transcription, translation and fragile X syndrome. Curr.Opin.Genet.Dev. 2006;16:270–275. doi: 10.1016/j.gde.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 76.Ferrari F, et al. The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol.Cell Neurosci. 2007;34:343–354. doi: 10.1016/j.mcn.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 77.Feng Y, et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol.Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 78.Weiler IJ, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc.Natl.Acad.Sci.USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Todd PK, et al. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci U.S.A. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bureau I. The development of cortical columns: role of Fragile X mental retardation protein. J Physiol. 2009;587:1897–1901. doi: 10.1113/jphysiol.2008.167155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gabel LA, et al. Visual experience regulates transient expression and dendritic localization of fragile X mental retardation protein. J Neurosci. 2004;24:10579–10583. doi: 10.1523/JNEUROSCI.2185-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Irwin SA, et al. Fragile X mental retardation protein levels increase following complex environment exposure in rat brain regions undergoing active synaptogenesis. Neurobiol.Learn.Mem. 2005;83:180–187. doi: 10.1016/j.nlm.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 83.Weiler IJ, et al. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc Natl Acad Sci U.S.A. 2004;101:17504–17509. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma A, et al. Dysregulation of mTOR signaling in the Fragile X mouse. J.Neurosci. 2009 doi: 10.1523/JNEUROSCI.3696-09.2010. acceptable, pending minor revisions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bear MF, et al. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 86.Giuffrida R, et al. A reduced number of metabotropic glutamate subtype 5 receptors are associated with constitutive homer proteins in a mouse model of fragile X syndrome. J Neurosci. 2005;25:8908–8916. doi: 10.1523/JNEUROSCI.0932-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dolen G, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H, et al. FMRP acts as a key messenger for dopamine modulation in the forebrain. Neuron. 2008;59:634–647. doi: 10.1016/j.neuron.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 89.Pilpel Y, et al. Synaptic ionotropic glutamate receptors and plasticity are developmentally altered in the CA1 field of Fmr1 knockout mice. J Physiol. 2009;587:787–804. doi: 10.1113/jphysiol.2008.160929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat.Rev.Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 91.Nimchinsky EA, et al. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Restivo L, et al. Enriched environment promotes behavioral and morphological recovery in a mouse model for the fragile X syndrome. Proc.Natl.Acad.Sci.USA. 2005;102:11557–11562. doi: 10.1073/pnas.0504984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galvez R, Greenough WT. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am.J Med.Genet.A. 2005;135:155–160. doi: 10.1002/ajmg.a.30709. [DOI] [PubMed] [Google Scholar]
- 94.Bureau I, et al. Circuit and plasticity defects in the developing somatosensory cortex of FMR1 knock-out mice. J Neurosci. 2008;28:5178–5188. doi: 10.1523/JNEUROSCI.1076-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gibson JR, et al. Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J Neurophysiol. 2008;100:2615–2626. doi: 10.1152/jn.90752.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoek KS, et al. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- 97.Shan J, et al. Binding of an RNA trafficking response element to heterogeneous nuclear ribonucleoproteins A1 and A2. J Biol Chem. 2000;275:38286–38295. doi: 10.1074/jbc.M007642200. [DOI] [PubMed] [Google Scholar]
- 98.Palacios IM, et al. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 99.Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7:1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- 100.Giorgi C, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 101.Tiruchinapalli DM, et al. Activity-dependent expression of ELAV/Hu RBPs and neuronal mRNAs in seizure and cocaine brain. J Neurochem. 2008;107:1529–1543. doi: 10.1111/j.1471-4159.2008.05718.x. [DOI] [PubMed] [Google Scholar]
- 102.di Penta A, et al. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. The Journal of Cell Biology. 2009;184:423–435. doi: 10.1083/jcb.200807033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 104.Kanai Y, et al. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 105.Elvira G, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 106.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 107.Barbee SA, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zeitelhofer M, et al. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci. 2008;28:7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cougot N, et al. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci. 2008;28:13793–13804. doi: 10.1523/JNEUROSCI.4155-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]