Abstract
Noroviruses interact with histo-blood group antigen (HBGA) receptors in a strain-specific manner probably detecting subtle structural differences in the carbohydrate receptors. The specific recognition of types A and B antigens by various norovirus strains is a typical example. The only difference between the types A and B antigens is the acetamide linked to the terminal galactose of the A but not to the B antigen. The crystal structure of the P dimer of a GII-4 norovirus (VA387) bound to types A and B trisaccharides has elucidated the A/B binding site on the capsid but did not explain the binding specificity of the two antigens. In this study, using site-directed mutagenesis, we have identified three residues on the VA387 capsid that are sterically close to the acetamide and are required for binding to A but not B antigens, indicating that the acetamide determines the binding specificity between the A and B antigens. Further mutational analysis showed that a nearby open cavity may also be involved in binding specificity to HBGAs. In addition, a systematic mutational analysis of residues in and around the binding interface has identified a group of amino acids that are required for binding but do not have direct contact with the carbohydrate antigens, implying that these residues may be involved in the structural integrity of the receptor binding interface. Taken together, our study provides new insights into the carbohydrate/capsid interactions which are a valuable complement to the atomic structures in understanding the virus/host interaction and in the future design of antiviral agents.
Introduction
Noroviruses, a group of single-stranded, positive sense RNA viruses within the family Caliciviridae, are the major viral pathogens of epidemics of acute gastroenteritis worldwide. The norovirus capsid is composed of 180 monomers of the capsid protein that organizes into 90 dimers (Prasad et al., 1999). The capsid protein can be divided into two principle domains, the S domain constitutes the interior icosahedral shell, while the P domain forms an arch-shaped protrusion emanating from the shell. The P domain can be further divided into P1 and P2 subdomains, with the P2 subdomain located at the outer surface of the viral capsid. The primary sequence of the P2 subdomain is highly variable among norovirus strains, suggesting its involvement in host interaction. Expression of the P domain in vitro results in dimerization (P dimer) and the formation of P particles that retain HBGA-binding function (Seto et al., 2005; Tan, Hegde, and Jiang, 2004; Tan, Meller, and Jiang, 2006; Tan et al., 2004). The involvement of the P2 subdomain domain in receptor binding has been confirmed by mutagenesis and crystallographic studies (Bu et al., 2008; Cao et al., 2007; Tan et al., 2003).
Noroviruses recognize human histo-blood group antigens (HBGAs) as receptors, in a strain-specific manner [Reviewed in (Tan and Jiang, 2005; Tan and Jiang, 2007)]. Eight distinct HBGA-binding patterns of noroviruses have been described (Huang et al., 2003; Huang et al., 2005). Strain VA387 belongs to GII-4 genotype that is predominant in many countries (Ramirez et al., 2008; Siebenga et al., 2008; Tu et al., 2008; Verhoef et al., 2008). VA387 represents one of the eight receptor binding patterns and binds to A, B and H antigens. Other HBGA-binding patterns include the H and A binders of Norwalk virus and C59, the A binder of BUDS, the B binder of Snow Mountain virus, the A and B binder of MOH and etc. Human HBGAs are complex carbohydrates that are composed of 3-8 monosaccharides linked to proteins or lipids on the surface of red blood cells and mucosal epithelia of the respiratory, genitourinary, and digestive tracts, or as free oligosaccharide in biological fluids such as saliva. HBGAs are highly polymorphic and three major HBGA families, namely the ABO, Lewis, and secretor families, are involved in norovirus recognition (Tan and Jiang, 2005; Tan and Jiang, 2007). Direct evidence linking HBGAs to norovirus infections and illness has been obtained in volunteer studies by challenging with Norwalk virus (Hutson et al., 2005; Hutson et al., 2002; Lindesmith et al., 2003). Outbreak studies of GII-4 viruses also support this linkage (Tan et al., 2008; Thorven et al., 2005). However, such evidence for other strains representing other receptor binding patterns remains lacking.
Recently, the 2.0 Å crystal structures of the P dimer of VA387 complexed with the type A-{α-L-fucose-(1,2)-[α-D-acetylgalactosamine-(1,3)]-D-galactose} and type B-{α-L-fucose-(1,2)-[ α-D-galactose-(1,3)]-D-galactose} trisaccharides, respectively, have been determined (Cao et al., 2007) (Fig 1). This has led to the identification of the HBGA-binding interface on the outermost P2 surface and the recognition of the importance of dimerization of the P domain in the receptor binding. The crystal structure revealed an extensive hydrogen-bonding network between the trisaccharides and the nearby amino acid residues of the receptor-binding interface. Three groups of amino acids, each interacting with different sugars of the trisaccharide, have been described (Cao et al., 2007). Although both the A- and the B-trisaccharides were observed to bind to the same open pocket, the crystal structures did not provide detail information on the binding specificity of noroviruses to HBGAs. In fact, no direct interaction was observed between P domain and the acetamide of the A-trisaccharide, the only structural difference between the A and the B antigens (Bu et al., 2008; Cao et al., 2007).
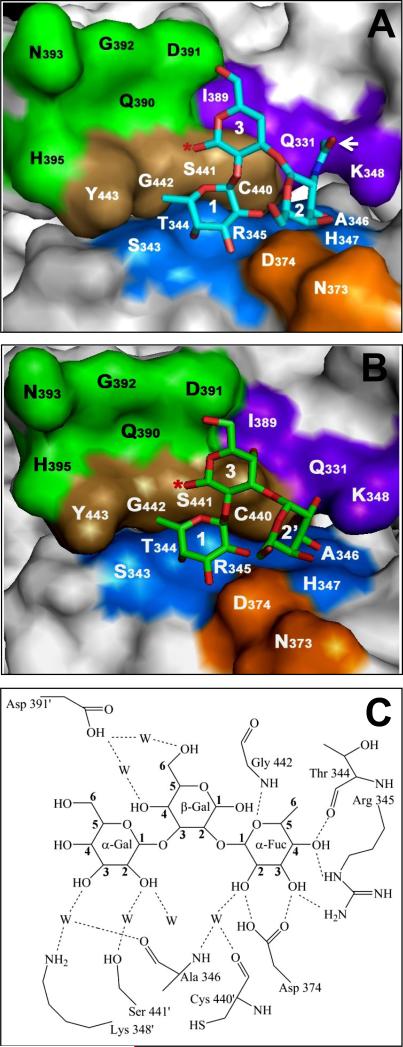
Fig 1.
The HBGA-binding interface of the norovirus capsid of VA387 (GII-4) interacting with the type A- and B-trisaccharides. The binding interfaces in (A) and (B) are in surface representation. The A- and B-trisaccharides are shown in stick representation, with nitrogen, oxygen, and carbon atoms colored blue, red, and cyan (A-antigen) or green (B-antigen), respectively. The saccharides are indicated as 1=α-1,2-fucose, 2=α-1,3-N-acetylgalactoseamine, 2’=α-1,3-galactose, 3=β-1,3-galactose. The star symbol indicates the junction linking to the remaining portion of the antigen. The white arrow shows acetamide group in (A). The nearby open pocket is shown in green at the top-left corner. (C) schematically shows hydrogen bonds (doted lines) between the individual saccharides of the B-trisaccharide and the nearby amino acids of the capsid. The water-bridged hydrogen bonds are indicated by W. (A) and (B) were prepared by software PyMOL version 1.0 (Delano Scientific), while (C) by software ChemDraw Pro version 11.0 (Adept Scientific).
In this study, we performed systematic mutagenesis to explore the binding specificity of the HBGA-binding interface of VA387 and identified a site defined by three amino acids (Q331, K348, and I389) that are sterically close to the acetamide of the A antigen and that is required for binding to A- but not to the B-antigens, suggesting the involvement of the site generated by these residues in interactions with the acetamide of the A antigen. Similar effects were also observed in a nearby open cavity. In addition, a group of other residues that do not have direct contact with HBGAs but are required for binding has been identified. These new data provide important insights into the carbohydrate/capsid interactions and would be useful for future antiviral design against noroviruses.
Materials and methods
Construction of mutant P particles and P dimers of VA387
The wild type P particle and the P dimer of VA387 were constructed and expressed in bacteria as described previously (Seto et al., 2005; Tan, Hegde, and Jiang, 2004; Tan, Meller, and Jiang, 2006). The mutant P particles and P dimers of VA387 with single amino acid substitutions were constructed by site-directed mutagenesis using the expression constructs of P particle (P-RGD4C in pGEX-4T-1) (Seto et al., 2005; Tan, Meller, and Jiang, 2006) or P dimer (HP in pGEX-4T-1) (Seto et al., 2005; Tan, Hegde, and Jiang, 2004), respectively, as templates. Site-directed mutagenesis was performed using the QuickChange Site-Directed Mutagenesis Kit (Stratagen, La Jolla, CA) and the corresponding primer pairs (Table 1) as described elsewhere (Seto et al., 2005; Tan, Hegde, and Jiang, 2004; Tan, Meller, and Jiang, 2006). The wild type and mutant P particles, as well as P dimers, were expressed and purified as described previously (Seto et al., 2005; Tan, Hegde, and Jiang, 2004; Tan, Meller, and Jiang, 2006). Briefly, after sequence confirmation through DNA sequencing, the mutant constructs were expressed in E. coli strain BL21 with IPTG (0.5 mM) induction at room temperature (~25°C) overnight. The P protein-GST fusion proteins were purified using the Glutathione Sepharose 4 flow (GE Health Care Bio-Sciences, Piscataway, NJ) according to the manufacturer's protocol. P proteins were released from GST by thrombin (GE Health Care Bio-Sciences, Piscataway, NJ) digestion. The formation of P particle and P dimer was determined by gel filtration using a size-exclusion column Superdex 200 (GE Health Care Bio-Sciences, Piscataway, NJ) powered by an AKTA-FPLC system (model 920, GE Health Care Bio-Sciences, Piscataway, NJ) , in which the P particles form a peak at ~830 kDa and the P dimer at ~69 kDa, respectively (Seto et al., 2005; Tan, Hegde, and Jiang, 2004; Tan, Meller, and Jiang, 2006).
Table 1.
Primers used for site-directed mutagenesis to generate single amino acid mutation.
| # | Name | Primer sequence (5’ to 3’) | Sense | Mutation |
|---|---|---|---|---|
| 1 | P981 | CGTGGGAAAGATCGCAGGCATGCTCACCCAAACC | + | Q331A |
| 2 | P982 | GGTTTGGGTGAGCATGCCTGCGATCTTTCCCCAG | – | Q331A |
| 3 | P983 | GGGAAAGATC CAAGGCGCGCTCACCCAAACCAC | + | M333A |
| 4 | P984 | GTGGTTTGGGTGAGCGCGCCTTGGATCTTTCCC | – | M333A |
| 5 | P1102 | CAGTGAGCACTGGGACTGTCCACTTCACTCC | + | S335A |
| 6 | P1103 | GGAGTGAAGTGGACAGTCCCAGTGCTCACTG | – | S335A |
| 7 | P985 | CCAAGGCATGCTCACCGCAACCACAAGAGAGGATGGC | + | Q336A |
| 8 | P986 | GCCATCCTCTCTTGTGGTTGCGGTGAGCATGCCTTGG | – | Q336A |
| 9 | P929 | GCATGCTCACCCAAACCGCAAGAGAGGATGGCTCG | + | T338A |
| 10 | P930 | CGAGCCATCCTCTCTTGCGGTTTGGGTGAGCATGC | – | T338A |
| 11 | P931 | GCTCACCCAAACCACAGCAGAGGATGGCTCGACC | + | R339A |
| 12 | P932 | GGTCGAGCCATCCTCTGCTGTGGTTTGGGTGAGC | – | R339A |
| 13 | P943 | CACAAGAGAGGATGGCGCGACCCGCGCCCAC | + | S343A |
| 14 | P944 | GTGGGCGCGGGTCGCGCCATCCTCTCTTGTG | – | S343A |
| 15 | P925 | GAGAGGATGGCTCGGCCCGCGCCCACAAAGC | + | T344A |
| 16 | P926 | GCTTTGTGGGCGCGGGCCGAGCCATCCTCTC | – | T344A |
| 17 | P927 | GAGGATGGCTCGACCGCCGCCCACAAAGTTAC | + | R345A |
| 18 | P928 | GTAGCTTTGTGGGCGGCGGTCGAGCCATCCTC | – | R345A |
| 19 | P991 | GGATGGCTCGACCCGCTCCCACAAAGCTACAGTG | + | A346S |
| 20 | P992 | CACTGTAGCTTTGTGGGAGCGGGTCGAGCCATCC | – | A346S |
| 21 | P1285 | GGCTCGACCCGCGGCCACAAAGCTACAG | + | A346G |
| 22 | P1286 | CTGTAGCTTTGTGGCCGCGGGTCGAGCC | – | A346G |
| 23 | P1100 | GCTCGACCCGCGCCGCCAAAGCTACAGTGAGC | + | H347A |
| 24 | P1101 | GCTCACTGTAGCTTTGGCGGCGCGGGTCGAGC | – | H347A |
| 25 | P1001 | CGACCCGCGCCCACGCAGCTACAGTGAGCAC | + | K348A |
| 26 | P1002 | GTGCTCACTGTAGCTGCGTGGGCGCGGGTCG | – | K348A |
| 27 | P993 | CCACTGACACAAACGCTGATTTTCAAACTGGC | + | N373A |
| 28 | P994 | GCCAGTTTGAAAATCAGCGTTTGTGTCAGTGG | – | N373A |
| 29 | P933 | CACTGACACAAACAATGCTTTTCAAACTGGCC | + | D374A |
| 30 | P934 | GGCCAGTTTGAAAAGCATTGTTTGTGTCAGTG | – | D374A |
| 31 | P995 | GACACAAACAATGATGCTCAAACTGGCCAAAACACG | + | F375A |
| 32 | P996 | CGTGTTTTGGCCAGTTTGAGCATCATTGTTTGTGTC | – | F375A |
| 33 | P1259 | CACCCCAGTCGGCGTCGCCCAGGATGGTAATAACC | + | I389A |
| 34 | P1260 | GGTTATTACCATCCTGGGCGACGCCGACTGGGGTG | – | I389A |
| 35 | P1287 | CCCAGTCGGCGTCGTCCAGGATGGTAATAACC | + | I389V |
| 36 | P1288 | GGTTATTACCATCCTGGACGACGCCGACTGGG | – | I389V |
| 37 | P1261 | CCAGTCGGCGTCATCGCGGATGGTAATAACCACC | + | Q390A |
| 38 | P1262 | GGTGGTTATTACCATCCGCGATGACGCCGACTGG | – | Q390A |
| 39 | P1106 | CGGCGTCATCCAGGCTGGTAATAACCACC | + | D391A |
| 40 | P1107 | GGTGGTTATTACCAGCCTGGATGACGCCG | – | D391A |
| 41 | P1263 | GCGTCATCCAGGATGCTAATAACCACCAAAATG | + | G392A |
| 42 | P1264 | CATTTTGGTGGTTATTAGCATCCTGGATGACGC | – | G392A |
| 43 | P1265 | CGTCATCCAGGATGGTGCTAACCACCAAAATGAACC | + | N393A |
| 44 | P1266 | GGTTCATTTTGGTGGTTAGCACCATCCTGGATGACG | – | N393A |
| 45 | P1267 | CCAGGATGGTAATAACGCCCAAAATGAACCCCAGC | + | H395A |
| 46 | P1268 | GCTGGGGTTCATTTTGGGCGTTATTACCATCCTGG | – | H395A |
| 47 | P1108 | CCACTATGCCCGGGGCTAGCGGGTATCCC | + | C440A |
| 48 | P1109 | GGGATACCCGCTAGCCCCGGGCATAGTCC | – | C440A |
| 49 | P1110 | CTATGCCCGGGTGTGCCGGGTATCCCAAC | + | S441A |
| 50 | P1111 | GTTGGGATACCCGGCACACCCGGGCATAG | – | S441A |
| 51 | P997 | GCCCGGGTGTAGCGCGTATCCCAACATG | + | G442A |
| 52 | P998 | CATGTTGGGATACGCGCTACACCCGGGC | – | G442A |
| 53 | P935 | CCCGGGTGTAGCGGGGCTCCCAACATGAATCTGG | + | Y443A |
| 54 | P936 | CCAGATTCATGTTGGGAGCCCCGCTACACCCGGG | – | Y443A |
| 55 | P999 | GGGTGTAGCGGGTATGCCAACARGAATCTGG | + | P444A |
| 56 | P1000 | CCAGATTCATGTTGGCATACCCGCTACACCC | – | P444A |
| 57 | P937 | CCAATGGTTATTTTAGATTTGCTTCCTGGGTCAACCAG | + | D517A |
| 58 | P938 | CTGGTTGACCCAGGAAGCAAATCTAAAATAACCATTGG | – | D517A |
| 59 | P939 | GGTTATTTTAGATTTGATGCCTGGGTCAACCAGTTCTAC | + | S518A |
| 60 | P940 | GTAGAACTGGTTGACCCAGGCATCAAATCTAAAATAACC | – | S518A |
HBGA binding assay
The saliva- and synthetic oligosaccharide-based binding assays were performed basically as described elsewhere (Huang et al., 2003; Huang et al., 2005). The affinity-column purified P particles and P dimers were first diluted to 1 mg/ml as starting solutions, then they were diluted further in a 3-fold-series to 33.3 μg/ml (30 x), 11.1 μg/ml (90 x), 3.70 μg/ml (270 x), 1.23 μg/ml (810 x), 0.41 μg/ml (2,430 x), 0.14 μg/ml (7290 x), 0.05 μg/ml (21,870 x), 0.02 μg/ml (65,610 x) directly on the Elisa plates, on which saliva- or synthetic oligosaccharide have been coated. Alternatively, the proteins were diluted to 10 μg/ml (100 x), 3.3 μg/ml (300 x), 1.1 μg/ml (900 x), 0.37 μg/ml (2,700 x), 0.12 μg/ml (8100 x), 0.04 μg/ml (24,300 x), 0.01 μg/ml (72,900 x). The three saliva samples used in this study were well-defined in our laboratory (Huang et al., 2003; Huang et al., 2005), each containing high level of A-, B-, and H-antigen as determined by monoclonal antibodies against A, B, H1, H2, H3 antigens (Signet Laboratories Inc, Dedham/MA). The synthetic oligosaccharide-based binding assays were performed using a panel of oligosaccharides representing 9 different HBGAs (A, B, H1, H2, H3, Lea, Leb, Lex, and Ley) as reported previously (Huang et al., 2003; Huang et al., 2005).
Crystal structure visualization and analysis
The crystal structures of the P dimer of VA387 complexed with type A- and B-trisaccharides were analyzed using the PyMOL software (DeLano Scientific LLC, Palo Alto, CA) and the Polyview-3D server (http://polyview.cchmc.org). The PDB files of the P protein in complex with A-trisaccharide (2OBS) and with B-trisaccharide (2OBT) were downloaded from the Protein Data Bank at the Rutgers University, New Brunswick, NJ (http://www.rcsb.org).
Results
Evidence for the role of the acetamide of the A antigen in norovirus-HBGA binding specificity
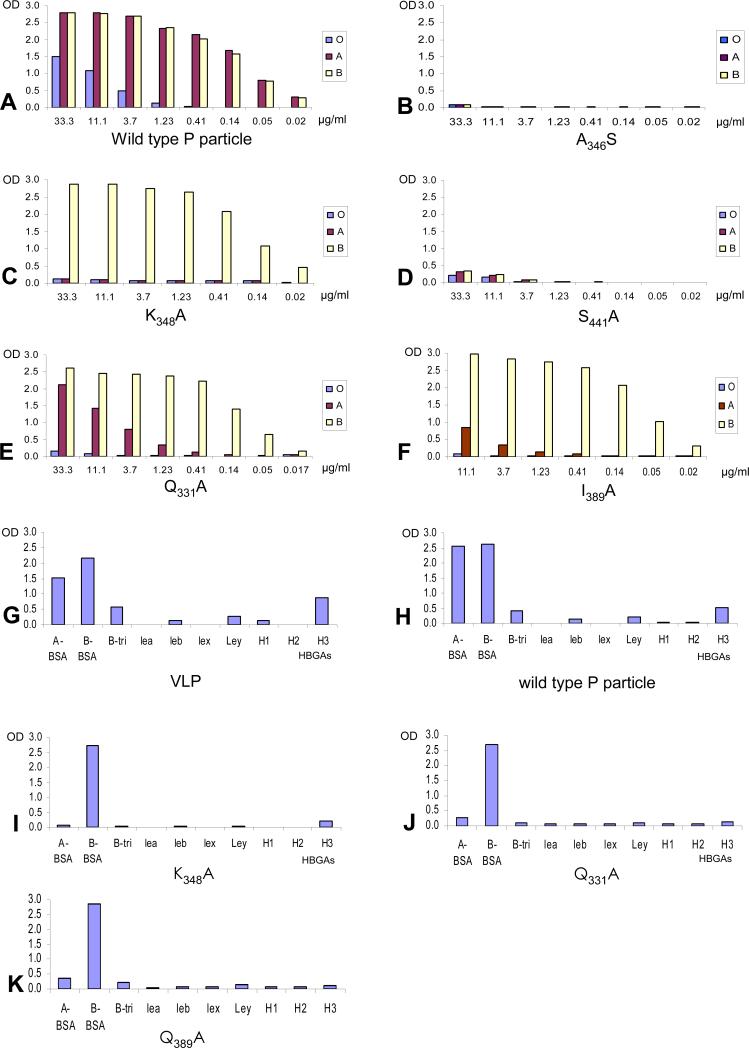
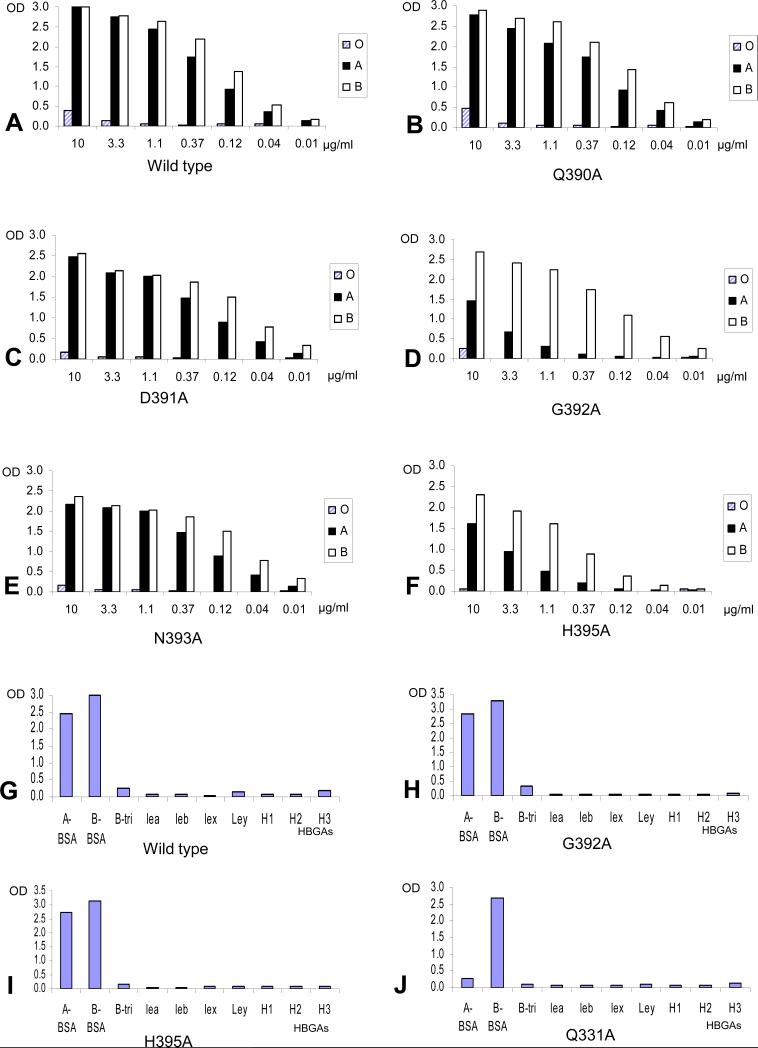
Our study began with the characterization of the site on the capsid interacting with the α-1,3-glactose of the B-trisaccharide as suggested by the crystal structure (Cao et al., 2007). The A- and B-antigens are structurally identical except for the presence of an acetamide group at the terminal sugar α-1,3-N-acetylgalactosamine of the A-antigen, but not the α-1,3-glactose of the B-antigen (Fig 1). Three amino acids (A346, S441, and K348) were predicted to interact via solvent molecules with the α-1,3-glactose of the B-antigen based on the crystal structure (Cao et al., 2007) (Fig 1). Mutations A346S and S441A completely prevented binding of the mutant P particles to all A-, B-, and H-antigens (Fig 2B and D, Table 2), indicating that these two residues are critical for binding to both A and B antigens. However, the third mutation (K348A) resulted in complete loss of binding to the A- but did not affect binding to the B-antigen in both saliva- and synthetic oligosaccharide-based binding assays (Fig 2C and 2I, Table 2), suggesting that this residue is involved in the binding specificity to the A antigen. The crystal structure of the HBGA-binding interface (Fig 1) indicates that residue K348 is positioned away from the center of the binding interface, close to the acetamide of the A-antigen, although no direct interaction between the acetamide and K348 is seen (Cao et al., 2007). Interestingly, replacement of two other residues (Q331 and I389) in the vicinity of K348 (Fig 1) with alanines also resulted in los of binding to the A- but not B- antigen (Fig 2E-F, J-K; Table 2). These data suggested that the three clustered residues Q331, K348, and I389 might form an interaction interface specifically needed by the A antigen.
Fig 2.
Binding of mutant P particles with amino acid substitutions in or around the α-galactose-binding site to the saliva samples (A-F) and synthetic oligosaccharide representing different HBGAs (G-K). The X-axes of (A) to (F) show the protein concentrations of the P particles and the Y-axes indicate the optical densities at 450 nm (OD450) that were the average value of triplicate experiments. “O”, “A”, and “B” represent the type O (containing H antigen), A, and B saliva, respectively. The X-axes of (G) to (K) shows a panel of synthetic oligosaccharides representing 9 different HBGAs (A, B, Lea, Leb, Lex, Ley, H1, H2, and H3). BSA indicates that those oligosaccharides were linked to bovine serum albumin as backbones (A-BSA and B-BSA), while the remaining oligosaccharides were conjugated to polyacrylamide (PAA) as backbones. The protein concentrations of VLP (G) and P particle (H-K) were 1 μg/ml and 2 μg/ml, respectively.
Table 2.
Summary of the systematic mutagenesis analysis on residues in and around the HBGA-binding interface of VA387 norovirus
| Mutants3 | Relative binding affinity to HBGAs1 |
|||||
|---|---|---|---|---|---|---|
| P particles2 | P dimers | |||||
| O | A | B | O | A | B | |
| Wild type | + | ++++ | ++++ | + | ++ | ++ |
|
The α-fucose-binding site: | ||||||
| T344A* | + | ++++ | ++++ | + | ++ | ++ |
| R345A* | - | - | - | - | - | - |
| A346S* | - | - | - | / | / | / |
| N373A | - | - | - | / | / | / |
| D374A* | - | - | - | - | - | - |
| C440A* | + | ++++ | ++++ | + | ++ | ++ |
| G442A* | - | - | - | - | - | - |
| Y443A | - | - | - | - | - | - |
| P444A | - | - | - | / | / | / |
|
Residues that support the conformation of the α-fucose-binding site: | ||||||
| Q336A | - | - | - | / | / | / |
| T338A | - | - | - | - | - | - |
| S343A | +/- | ++ | ++ | +/- | + | + |
| H347A | - | - | - | / | / | / |
|
The α-glactose/α-N-acetylglalactoseamine-binding site: | ||||||
| Q331A | - | + | ++++ | / | / | / |
| A346S* | - | - | - | / | / | / |
| A346G* | +++ | ++++ | ++++ | / | / | / |
| K348A* | - | - | ++++ | / | / | / |
| I389A | - | - | ++++ | / | / | / |
| I389V | + | +++ | ++++ | / | / | / |
| S441A* | - | - | - | / | / | / |
|
β-Galactose-binding site: | ||||||
| D391A* | + | +++ | +++ | / | / | / |
|
2nd open cavity: | ||||||
| Q390A | +/- | +++ | +++ | / | / | / |
| G392A | - | + | +++ | / | / | / |
| N393A | +/- | +++ | +++ | / | / | / |
| H395A | +/- | ++ | +++ | |||
|
Negative control: | ||||||
| T335S | + | ++++ | ++++ | / | / | / |
| M333A | + | ++++ | ++++ | / | / | / |
| R339A | + | ++++ | ++++ | + | ++ | ++ |
| D517A | + | ++++ | ++++ | + | ++ | ++ |
| S518A | + | ++++ | ++++ | + | ++ | ++ |
Number of “+” indicated the relative binding affinity of the mutant P particles and P dimers to HBGAs. “-” indicated a complete loss of binding. / = not determined.
The P particle formation of wild type and all mutants were confirmed by gel filtration.
Amino acids with a star symbol are predicted to interact with type A- and/or B-trisaccharides by crystallographic study.
Systematic analysis of residues in and around the binding interface
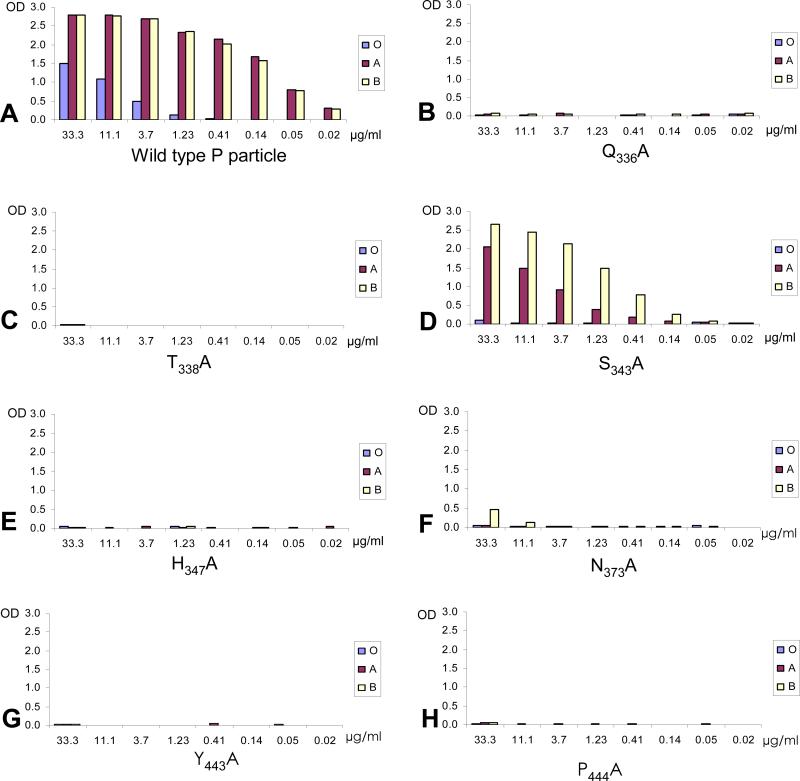
The findings of the two residues (Q331 and I389) that have no direct contact with the trisaccharides but affect the receptor binding suggested that a systematic analysis of individual residues in and surrounding the HBGA-binding interface might be informative. The crystal structure of the VA387 P dimer bound to the A- and B-trisaccharides revealed eleven amino acids in the receptor-binding interface that interact with the trisaccharide via direct or water-mediated hydrogen bonds (Cao et al., 2007) (Fig 1). We replaced all of them with either alanines or serines, including the three residues interacting with the α-1,3-glactose described above, and found that the mutations affected HBGA-binding as predicted by the crystal structures (see below).
In addition, 12 amino acids that have no direct contact with the trisaccharides but surround the binding interface as shown by the crystal structures (Fig 1) were also subject to mutagenesis. Many of them also affected HBGA-binding (see below), indicating that these residues are also required for the structural integrity of the receptor binding interface. Finally, five other mutations (T335S, M333A, R339A, D517A, and S518A) that are far away from the binding interface were introduced into the P particles and none of them affected the HBGA-binding function. None of the single amino-acid mutations described in this study affected P particle or P dimer formation as shown by gel filtration analysis (Seto et al., 2005; Tan, Hegde, and Jiang, 2004; Tan, Meller, and Jiang, 2006).
The α-fucose plays a central role in norovirus/receptor interactions
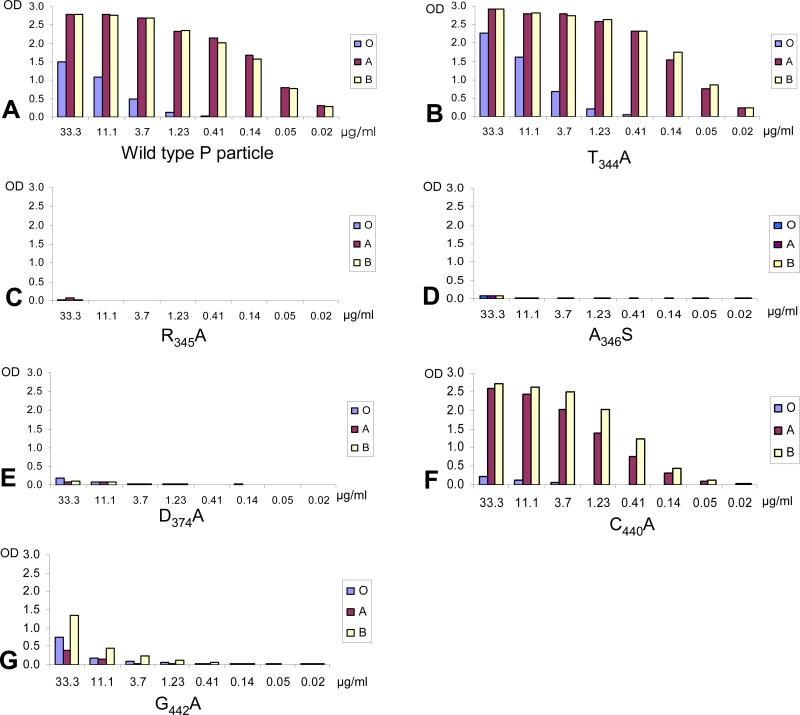
The crystal structures suggested six amino acids (T344, R345, A346, D374, C440, and G442) interacting with the α-1,2-fucose of the B-trisaccharide (Cao et al., 2007). These residues were replaced by alanine or serine and 4 of the mutants lost their binding to all A-, B-, and H- antigens completely or nearly completely (G442A) (Fig 3 and Table 2). The binding to HBGAs of the remaining 2 mutants (T344A and C440A) did not change, although the crystal structures showed hydrogen bonds between the two amino acids and the fucose (Fig 1). This discrepancy could be explained by the fact that both residues interact with the α-1,2-fucose through their backbone oxygen atoms (Cao et al., 2007) (Fig 1), which would remain the same in the substituted alanines. Thus all six fucose-interacting amino acids predicted by the crystal structures are most likely required for binding to the carbohydrate receptors.
Fig 3.
Binding of mutant P particles with amino acid changes in or around the α-fucose-binding site to the saliva samples. The X-axes show the protein concentrations of the P particles, while the Y-axes indicate the optical densities at 450 nm (OD450) that were the average value of triplicate experiments. “O”, “A”, and “B” represent the type O (containing H antigen), A, and B saliva samples, respectively.
In addition, seven other amino acids, none of which have contact with the A- and B-trisaccharides, were also examined. Four (S343, Q336, T338, and H347) of them were suggested to support the conformation of the α-fucose-binding site (Cao et al., 2007), while the other three (N373, Y443, and P444) were nearby. Replacement of each of these seven amino acids with alanines decreased binding to all A-, B- and H-antigens significantly (Fig 4 and Table 2). Taken together, except the six amino acids that have direct contact with the α-fucose, at least seven additional residues are likely to be required for the structural integrity of the binding site, further highlighting the importance of this sugar in the norovirus/HBGA interaction as suggested by previous studies (Cao et al., 2007; Huang et al., 2005; Tan and Jiang, 2005; Tan and Jiang, 2007).
Fig 4.
Binding of mutant P particles with single mutations at the 4 amino acids that were predicted to support the structural integrity of the α-fucose-binding site. The X-axes show the protein concentrations of the P particles, while the Y-axes indicate the optical densities at 450 nm (OD450) that were the average value of triplicate experiments. “O”, “A”, and “B” represent the type O (containing H antigen), A, and B saliva samples, respectively.
The β-galactose may not be critical for HBGA-binding
The crystal structures indicated that the β-1,3-glalactose of both type A- and B-trisaccharide is relatively distant from the P dimer surface (Cao et al., 2007) (Fig 1). Only D391 was seen interaction with the β-galactose via two water-bridged hydrogen bonds (Fig 1C). However, replacement of D391 with an alanines only slightly reduced binding to HBGAs (Fig 5C and Table 2), suggesting that the interaction between D391 and the β-1,3-glalactose may not be critical for binding to HBGAs.
Fig 5.
Binding of mutant P particles with single amino acid change at the second open pocket to the saliva samples (A-F) and synthetic oligosaccharide representing different HBGAs (G-J). The X-axes of (A) to (F) show the protein concentrations of the P particles and the Y-axes indicate the optical densities at 450 nm (OD450) that were the average value of triplicate experiments. “O”, “A”, and “B” represent the type O (containing H antigen), A, and B saliva, respectively. The X-axes of (G) to (J) shows a panel of synthetic oligosaccharides representing 9 different HBGAs (A, B, Lea, Leb, Lex, Ley, H1, H2, and H3). BSA indicates that those oligosaccharides were linked to bovine serum albumin as backbones (A-BSA and B-BSA), while the remaining oligosaccharides were conjugated to polyacrylamide (PAA) as backbones. The protein concentrations of the P particles in G to J were 2 μg/ml. Mutant Q331A (J) served as control of selective inhibition of binding to A-antigen.
A nearby pocket affects binding to the A-antigen
Another open pocket near the binding interface that is made up of residues Q390 and H395 in the bottom and D391, G392, N393 and Y443 as the walls of the pocket was suggested to have additional interaction with the HBGAs (Cao et al., 2007) (Fig 1). When these residues were replaced by alanines, except Y443 that has been shown to be important for binding to A-, B-, and H-antigens probably by interacting with the α-1,2-fucose (Fig 3H), only G392 and H395 showed reduction in binding to A- but not B-antigens (Fig 5A to F). Interestingly, the reduction in binding to A-antigen by G392A and H395A mutations occurred only in the saliva- but not the synthetic oligosaccharide-based binding assays (Fig 5H to I). These data suggest that some unknown features of the native HBGAs (saliva) that are missing on the synthetic oligosaccharides may be responsible for the interaction with this nearby pocket.
The receptor binding interface is highly conserved among known GII-4 viruses
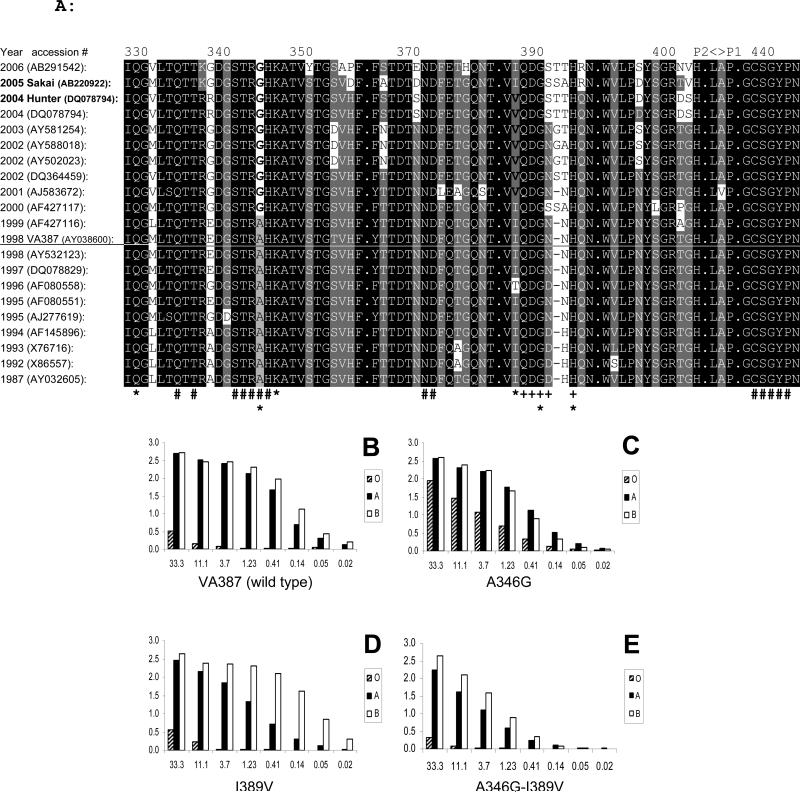
Following the identification of the receptor binding interface and the functional involvement of the amino acids described above, we performed a sequence alignment (Fig 6A) of the major GII-4 variants, including several strains with known binding patterns to all A-, B- and H-antigens and two strains that lost binding to all A-, B- and H-antigens (Huang et al., 2003; Huang et al., 2005; Lindesmith et al., 2008), in an attempt to elucidate the mechanism and residues that are responsible for the receptor binding changes. Among the 19 amino acids (Fig 6A and Table 2) important for HBGA-binding identified in this study, only two amino acids have changed since 1987: A346 to a glycine (G) in 2000 and I389 to a valine (V) in 2001 (Fig. 6A). These are very similar amino acids with only a methyl group (CH3) difference in their side chains between A and G or between I and V. These changes appear not to correlate with the phenotypic changes of the binding patterns (Huang et al., 2003; Huang et al., 2005; Lindesmith et al., 2008). Mutant A346G developed for VA387 did not affect the binding to types A and B saliva but increased binding to type O saliva (Fig 6C), mutant I389V showed a small decrease in binding to type A but not B saliva (Fig 6D), while mutant with double mutations of A346G and I389V resulted in stronger reduction in binding to both A- and B-antigens but the bindings to all A-, B- and H-antigens clearly remain (Fig 6E). These data suggested that subtle changes of the binding interface can vary receptor binding, but neither A346 nor I389 is responsible for the complete loss of HBGA-binding of the two variants, Hunter (2004) and Sakai (2005) viruses.
Fig 6.
Sequence alignment (A) of the HBGA-binding interfaces and the surrounding regions in the P domain of GII-4 viruses selected from different years since 1987. Each virus is indicated by the year of isolation and accession number in the GenBank. Dark background indicates highly conserved residues and gray background represents less conserved amino acids. Dots in the dark background indicate omission of conserved residues. The numbers on the top indicate the positions of the residues according to VA387 (underlined). The Hunter virus (2004, DQ078794) and Sakai virus (2005, AB220922) that loss their binding activities (Lindesmith et al., 2008) are highlighted by bold letters. “#” indicates amino acids that are required for HBGA-binding, “*” indicates residues that affect the binding specificity, “+” indicates amino acids that form the nearby open pocket and “< >” indicates the boundary of the P1 and P2 subdomains. B to E, binding of mutant P particles of VA387 with single mutations [A346G (C) or I389V (D)], or double mutation [A346G and I389V (E)] to saliva samples. The X-axes show the protein concentrations of the P particles and the Y-axes indicate the optical densities at 450 nm (OD450) that were the average value of triplicate experiments. “O”, “A”, and “B” represent the type O (containing H antigen), A, and B saliva, respectively.
Discussion
In this study we performed systematic mutagenesis on 27 amino acids in and around the HBGA-binding interface of a GII-4 virus (VA387) to examine their roles in norovirus-receptor binding affinity and specificity. Among 11 amino acid residues that have direct contact with the carbohydrate receptors based on the crystal structures (Cao et al., 2007), 7 residues are absolutely required for receptor-binding as they knock out binding to both A- and B-antigens when they are replaced by alanines. One residue (K348) is required for binding to A- but not to B-antigens and the remaining three residues (T344, C440 and D391) are also likely involved in the binding although the possibility exists that we might not have selected proper replacement of amino acids in our mutagenesis. In conclusion, except T344, C440, and D391, whose roles for HBGA-binding remain to be confirmed, all remaining 8 amino acids constituting the binding interface observed in the crystal structure (Cao et al., 2007) participate in norovirus receptor binding.
In addition, we have identified another 11 amino acids that are important for HBGA-binding, although they do not make direct contact with the carbohydrate receptors according to the crystal structures. These residues are likely responsible for the conformational integrity of the receptor binding interface. Finally, we have demonstrated that the HBGA-binding interface is highly conserved among the known GII-4 viruses, which is consistent with our observation that the majority of GII-4 viruses bind to all A, B and H antigens (unpublished data) and appear to explain their predominance in the epidemiology. However, we still cannot exclude the possibility of additional residues other than those described in this study also being involved in norovirus-HBGA binding because some GII-4 viruses lose complete or partial receptor-binding but retain the conserved binding interface. Further investigations of this issue are necessary.
Here we have provided direct evidence suggesting that the acetamide group of the A antigen is responsible for the binding specificity differences between the A and the B antigens. The identification of three amino acids (H348, Q331, and I389) responsible for the binding specificity may help future genetic analysis and classification of noroviruses. For example, noroviruses differ in recognizing the A and B antigens. VA387 (GII-4) described in this study and a few other strains (MOH, MxV, and Paris Island virus) recognize both A and B antigens; the prototype Norwalk virus and BUDS recognize A but not B; while the Snow Mountain virus recognizes the B but not A antigen (Huang et al., 2005; Tan and Jiang, 2005; Tan and Jiang, 2007; Tan et al., 2004). The identification of the site responsible for acetamide recognition on the capsid would allow correlation between sequence and phenotype and consequently allow a more detailed classification of norovirus strains.
The demonstration of the involvement of the open pocket near the binding interface in the HBGA interaction is also significant since it indicates that additional features of the HBGA molecules may also participate in norovirus recognition. The involvement of this site in binding to HBGAs was also observed previously through a mutant VLP (D393G) of another GII-4 strain (Lindesmith et al., 2008). In addition, the binding difference of the two mutants (G393A and H395A) to the native HBGAs in saliva and synthetic carbohydrate receptors (Fig 5) further supports our hypothesis. The native HBGAs are much more complicated containing 3 to 8 saccharides either free or linked to proteins or lipids as backbones, while the synthetic carbohydrates used in the crystallographic and the mutagenesis studies contain only 2 to 4 sugars either free or linked to bovine serum albumin (BSA) or polyacrylamide (PAA) as backbones. Thus either additional sugars or the backbones of the native receptors could be involved in the interaction with the nearby pocket. The impact of the backbones in norovirus-receptor interaction has also been previously suggested (Huang et al., 2005) but the site on the viral capsid for this interaction remains unknown. The potential interacting site of additional sugars could be investigated by co-crystallization of the viruses or P dimers with more complicated carbohydrate molecules.
Our study also provides new evidence supporting the central role of the α-1,2-fucose- (H-epitope) in norovirus-host interaction which was first suggested by our previous characterization of receptor binding variations of variable noroviruses (Huang et al., 2003; Huang et al., 2005) and then by the crystal structure of a GII-4 virus (Cao et al., 2007). Structurally the α-1,2-fucose is the terminal saccharide of the H-related antigens, including A, B, Leb, Ley, and H types 1, 2, 3 antigens, and thus is the key determinant for binding to noroviruses. We now show functional evidence for the requirement of the 6 residues interacting with the α-1,2-fucose in GII-4 norovirus binding to all A-, B-, and H-antigens. In addition, we demonstrated the involvement of a few other residues interacting with the α-1,3-N-acetylglalactoseamine- (A-epitope), which may further strengthen the binding affinity and determine the binding specificity. The complete loss of binding activity to all A-, B- and H-antigens by single mutations in these binding sites imply that they may be important targets for broadly effective antivirals against different noroviruses with different binding patterns.
The observed involvement of the acetamide of the A antigen in the binding specificity of VA387 provides an example of how a minor structural modification of a HBGA can affect the receptor binding pattern and potentially change the host range of a norovirus. Human HBGA system is highly polymorphic under the control of multiple gene families. In addition, modifications by other chemical groups such as the sialic epitopes and subtypes of the ABO, secretor and Lewis families of HBGAs make the human HBGA system and its recognition with noroviruses even more complicated. The demonstration of the relationship between the three amino acids (Q331, K348, and I389) and the acetamide helps explain the mechanism of the strain-specific norovirus-HBGA interaction. Another example are the residues in the nearby open cavity that could interact with additional structures of HBGAs and reduce binding to the A- more than to the B-antigen [Fig 5, (Lindesmith et al., 2008)], although the detailed mechanism still needs to be confirmed.
In an attempt to explore the mechanism of the HBGA-binding variations, evolution, and epidemiology of different GII-4 viruses from different countries, we have shown that the majority of GII-4 strains isolated in the past 20 years retain a broad spectrum of host specificity to all A, B and H antigens, although binding to each of the antigen may be variable [(Huang et al., 2005), unpublished data]. In this study we performed a sequence alignment of different GII-4 strains and found that 17 of the 19 amino acids that are critical for HBGA-binding are highly conserved among all GII-4 viruses, while the remaining two residues are replaced by very similar amino acids. These data suggest that the human HBGAs might be a selection marker in the evolution of noroviruses and our observation that the majority of GII-4 viruses bind to all A, B and H antigens (unpublished data) appears to explain the predominance of GII-4 viruses over other types that recognize narrower spectrum of human HBGA receptors. Two mutations (A345G and I389V) affected the binding specificity in a different way suggesting that a subtle conformational change in the binding interface could result in changes of the binding pattern. Unfortunately, we were unable to identify any genetic markers responsible for the phenotypic change of the GII-4 variants and the reason for the complete loss of binding to the A, B and/or H antigens of Hunter and Sakai viruses (Lindesmith et al., 2008) remains unknown. One possibility is that residues other than those described in this study are also involved in HBGA-binding of noroviruses. Those residues could directly interact with the carbohydrate receptors by as yet unidentified structures or be involved in defining the local or global conformation that is required for the receptor binding function. One example is the arginine cluster at the C-terminus of the P domain that is far from the HBGA-binding interface but is required for receptor binding (Tan, Meller, and Jiang, 2006). These issues will require further structural and solution studies.
Acknowledgement
We thank Sahle Amsalu, Weiming Zhong, and Xiaoyun Deng for technical support in protein expression. In addition, we thank Dr Weiming Bu for his help in preparing Fig. 1 C. The research described in this article was supported by the National Institute of Health, the National Institute of Allergy and Infectious diseases (R01 AI37093 and R01 AI55649) and National Institute of Child Health (PO1 HD13021), the Department of Defense (PR033018) to X.J., and “863” Project (China, 2006AA02A322) to X.L. This work was also supported by a grant from the Translational Research Initiative of Cincinnati Children's Hospital Medical Center (SPR102032) to M.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bu W, Mamedova A, Tan M, Xia M, Jiang X, Hegde RS. Structural basis for the receptor binding specificity of Norwalk virus. J Virol. 2008;82(11):5340–7. doi: 10.1128/JVI.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol. 2007;81(11):5949–57. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Farkas T, Marionneau S, Zhong W, Ruvoen-Clouet N, Morrow AL, Altaye M, Pickering LK, Newburg DS, LePendu J, Jiang X. Noroviruses Bind to Human ABO, Lewis, and Secretor Histo-Blood Group Antigens: Identification of 4 Distinct Strain-Specific Patterns. J Infect Dis. 2003;188(1):19–31. doi: 10.1086/375742. [DOI] [PubMed] [Google Scholar]
- Huang P, Farkas T, Zhong W, Tan M, Thornton S, Morrow AL, Jiang X. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J Virol. 2005;79(11):6714–22. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson AM, Airaud F, LePendu J, Estes MK, Atmar RL. Norwalk virus infection associates with secretor status genotyped from sera. J Med Virol. 2005;77(1):116–20. doi: 10.1002/jmv.20423. [DOI] [PubMed] [Google Scholar]
- Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis. 2002;185(9):1335–7. doi: 10.1086/339883. [DOI] [PubMed] [Google Scholar]
- Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, Stewart P, LePendu J, Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9:548–53. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- Lindesmith LC, Donaldson EF, Lobue AD, Cannon JL, Zheng DP, Vinje J, Baric RS. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008;5(2):e31. doi: 10.1371/journal.pmed.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286(5438):287–90. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- Ramirez S, Giammanco GM, De Grazia S, Colomba C, Martella V, Arista S. Genotyping of GII.4 and GIIb norovirus RT-PCR amplicons by RFLP analysis. J Virol Methods. 2008;147(2):250–6. doi: 10.1016/j.jviromet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Seto Y, Iritani N, Kubo H, Kaida A, Murakami T, Haruki K, Nishio O, Ayata M, Ogura H. Genotyping of Norovirus Strains Detected in Outbreaks between April 2002 and March 2003 in Osaka City, Japan. Microbiol Immunol. 2005;49(3):275–83. doi: 10.1111/j.1348-0421.2005.tb03718.x. [DOI] [PubMed] [Google Scholar]
- Siebenga J, Kroneman A, Vennema H, Duizer E, Koopmans M. Food-borne viruses in Europe network report: the norovirus GII.4 2006b (for US named Minerva-like, for Japan Kobe034-like, for UK V6) variant now dominant in early seasonal surveillance. Euro Surveill. 2008;13(2) [PubMed] [Google Scholar]
- Tan M, Hegde RS, Jiang X. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J Virol. 2004;78(12):6233–42. doi: 10.1128/JVI.78.12.6233-6242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Huang P, Meller J, Zhong W, Farkas T, Jiang X. Mutations within the P2 Domain of Norovirus Capsid Affect Binding to Human Histo-Blood Group Antigens: Evidence for a Binding Pocket. J Virol. 2003;77(23):12562–71. doi: 10.1128/JVI.77.23.12562-12571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Jiang X. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 2005;13(6):285–93. doi: 10.1016/j.tim.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Tan M, Jiang X. Norovirus-host interaction: implications for disease control and prevention. Expert Rev Mol Med. 2007;9(19):1–22. doi: 10.1017/S1462399407000348. [DOI] [PubMed] [Google Scholar]
- Tan M, Jin M, Xie H, Duan Z, Jiang X, Fang Z. Outbreak studies of a GII-3 and a GII-4 norovirus revealed an association between HBGA phenotypes and viral infection. J Med Virol. 2008;80(7):1296–1301. doi: 10.1002/jmv.21200. [DOI] [PubMed] [Google Scholar]
- Tan M, Meller J, Jiang X. C-terminal arginine cluster is essential for receptor binding of norovirus capsid protein. J Virol. 2006;80(15):7322–31. doi: 10.1128/JVI.00233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Zhong W, Song D, Thornton S, Jiang X. E. coli-expressed recombinant norovirus capsid proteins maintain authentic antigenicity and receptor binding capability. J Med Virol. 2004;74(4):641–9. doi: 10.1002/jmv.20228. [DOI] [PubMed] [Google Scholar]
- Thorven M, Grahn A, Hedlund KO, Johansson H, Wahlfrid C, Larson G, Svensson L. A homozygous nonsense mutation (428G-->A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J Virol. 2005;79(24):15351–5. doi: 10.1128/JVI.79.24.15351-15355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu ET, Bull RA, Greening GE, Hewitt J, Lyon MJ, Marshall JA, McIver CJ, Rawlinson WD, White PA. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin Infect Dis. 2008;46(3):413–20. doi: 10.1086/525259. [DOI] [PubMed] [Google Scholar]
- Verhoef L, Depoortere E, Boxman I, Duizer E, van Duynhoven Y, Harris J, Johnsen C, Kroneman A, Le Guyader S, Lim W, Maunula L, Meldal H, Ratcliff R, Reuter G, Schreier E, Siebenga J, Vainio K, Varela C, Vennema H, Koopmans M. Emergence of New Norovirus Variants on Spring Cruise Ships and Prediction of Winter Epidemics. Emerg Infect Dis. 2008;14(2):238–243. doi: 10.3201/eid1402.061567. [DOI] [PMC free article] [PubMed] [Google Scholar]