Abstract
Dynamic connectivity networks identify directed interregional interactions between modeled brain regions in neuroimaging. However, problems arise when the regions involved in a task and their interconnections are not fully known a priori. Objective measures of model adequacy are necessary to validate such models. We present a connectivity formalism, the Switching Linear Dynamic System (SLDS), that is capable of identifying both Granger-Geweke and instantaneous connectivity that vary according to experimental conditions. SLDS explicitly models the task condition as a Markov random variable. The series of task conditions can be estimated from new data given an identified model providing a means to validate connectivity patterns. We use SLDS to model functional magnetic resonance imaging data from five regions during a finger alternation task. Using interregional connectivity alone, the identified model predicted the task condition vector from a different subject with a different task ordering with high accuracy. In addition, important regions excluded from a model can be identified by augmenting the model state space. A motor task model excluding primary motor cortices was augmented with a new neural state constrained by its connectivity with the included regions. The augmented variable time series, convolved with a hemodynamic kernel, was compared to all brain voxels. The right primary motor cortex was identified as the best region to add to the model. Our results suggest that the SLDS model framework is an effective means to address several problems with modeling connectivity including measuring overall model adequacy and identifying important regions missing from models.
Keywords: Dynamic systems, fMRI, Effective connectivity, Motor systems, Computational modeling
Introduction
In order for the field of neuroimaging to advance beyond phenomenology, computational models capable of generating behavior from neural level variables are needed. One approach for creating these models linking brain and behavior proceeds from first-principles toward observed data. Simple dynamic models mimicking quasi-neural level computational units (e.g., individual spiking neurons, cortical columns, etc.) are combined based on prior hypotheses of interregional connectivity or network patterns derived from anatomical data (Sporns et al., 2000; Breakspear et al., 2003; Honey et al., 2007; Izhikevich and Edelman, 2008). The resulting simulated behavior of the model is examined for its relation to real neuroimaging data or overt behavior (Arbib et al., 1995; Tagamets and Horwitz, 1998; Horwitz and Tagamets, 1999; Husain et al., 2004; Chadderdon and Sporns, 2006; Morgan and Soltesz, 2008; Honey et al., 2009).
A complementary method, which we pursue here, begins with experimental neuroimaging data and seeks to derive generative models of these data by describing the interactions among variables defined at a lower level of analysis. Ultimately, this lower level of analysis should approach a neural-like level and the models should describe how the imaging data map onto this level. Several modeling methods examining Granger-Geweke causality (i.e., temporally directed connectivity) have been successfully applied to neuroimaging data. Though often defined in hemodynamic space, pair-wise connectivity measures have been derived for full-brain exploratory analyses that could in principle be mapped to a lower, neural-like level via simple deconvolution methods (Goebel et al., 2003; Valdés-Sosa, 2004; Roebroeck et al., 2005; Abler et al., 2006; Gao et al., 2008). More hypothesis driven connectivity models containing a small number of regions of interest (ROI) using multivariate autoregressive (MAR) models have also been examined for neuroimaging data (Harrison et al., 2003; Bressler et al., 2007; David et al., 2008; Seth, 2008). Using a separately estimated hemodynamic response function as the forward model mapping lower-level representations to functional magnetic resonance imaging (fMRI) data, interregional Granger-Geweke causality has been examined at a quasi-neural level with close agreement between the identified Granger-Geweke connectivity and intercranial recordings (David et al., 2008).
Such models have been proven useful, can be relatively computationally simple, and provide a powerful method for exploratory functional connectivity. However, for complex hemodynamic response functions, separating the hemodynamic estimation from the connectivity estimation may induce additional error. For full-brain analyses where detailed hemodynamic models cannot be tractably applied to each voxel, the variability in the hemodynamic response across subjects and regions (Aguirre et al., 1998; Handwerker et al., 2004) presents a potential limitation for accurate directional estimates in time based analyses (David et al., 2008). In addition, known experimental variables such as sensory inputs and task conditions have not always been included in these models potentially reducing their representational power for typical fMRI experiments (though see Riera et al., 2004).
Recently, Friston and colleagues have developed a method, Dynamic Causal Modeling (DCM), partially as a means of identifying ROI based directional connectivity models at a quasi-neural level while addressing these two issues of hemodynamic variability and representational power (Friston et al., 2003; Kiebel et al., 2007; Stephan et al., 2007, 2008). For fMRI data, they proposed a bilinear dynamic system model with a continuous time, deterministic state space, and a biologically based, stochastic, nonlinear forward model of the hemodynamic response. These models have achieved widespread use in neuroimaging to confirm hypotheses regarding the interactions among a small number of ROIs (e.g., Bitan et al., 2005; Mechelli et al., 2005; Fairhall and Ishai, 2007; Leff et al., 2008; Seghier and Price, 2009).
With all modeling efforts, it is important to evaluate the accuracy of any identified model before it can be analyzed and interpreted. Model validation in DCM is typically performed using Bayesian Model Selection (BMS) methods to select the best model among a set of specified candidate models (Penny et al., 2004; Stephan et al., 2009). Although a full treatment of BMS is beyond the scope of this investigation, a few points are relevant to the current discussion. BMS, as it has been applied to DCM for fMRI, searches among a pre-specified set of models for the model with the highest “model evidence” which is the one that best approximates the true conditional distribution of the observed fMRI data given the model integrated over possible parameter values which amounts to a penalty for the number of parameters required to model that distribution. If model complexity is equated and maximum likelihood estimates of the parameters in each model are used, the best model will have the highest likelihood of observing the fMRI data given the model parameters. It can be shown that for finite dimensional data, BMS will, on average, select the model representing the true posterior distribution from a set of competing false models (Bishop, 2006). Obviously however, this is the case only if the true model does indeed exist within the test set (Bishop, 2006). When all examined models are poor approximations of the true posterior distribution, BMS will still identify a “best” model. While BMS may essentially equate models with no relation to the true posterior, a real danger is the inclusion of a model in the test set with a minimal relation to the true posterior among many other models with no relation at all. The poor model may be a clear winner in terms of its Relative Bayes Factor or likelihood ratio, but still be a very poor generative model. Relative Bayes Factors or other measures based on relative likelihoods among known models say little about the sufficiency of any single model, they only indicate sufficiency relative to the others tested.
This issue is unlikely to be just a hypothetical problem given the large number of cortical and subcortical regions involved in all but the most simplistic of tasks, the massive interregional connectivity in the brain, and the limited dimensionality of DCM and similar ROI based models. Therefore model validation using the conditional probability of the fMRI data and BMS alone can only reasonably be applied in cases where preexisting data or theory clearly identifies the complete candidate connectivity model or the goal is simply adjudication between a small number of theoretically well defined models. BMS cannot be used for validation of any of the models themselves. Data likelihood based BMS as the sole validation tool is inadequate for exploratory analyses, analyses where the tested theory does not specify all relevant regions and the full interregional connectivity, or analyses of complex tasks where the number of regions known to participate in the task exceeds the number of regions that can be tractably included in a model. What is desired then is a validation method for the model itself that can be combined with previously described BMS procedures to evaluate the sufficiency of the identified best model.
Here, we present a method for connectivity estimation, the Switching Linear Dynamic System (SLDS), which has been extensively studied in several fields such as econometrics, data visualization, speech recognition, target tracking in aeronautics, and communications (Hamilton, 1989; Li and Bar-Shalom, 1993; Moore and Krishnamurthy, 1994; Zoeter and Heskes, 2003; Mesot and Barber, 2007). SLDS models are strongly related to DCM and can exhibit a number of its advantages with respect to hemodynamic variability and task representation while additionally providing an objective measure of the overall quality and sufficiency of an identified model. The SLDS formalism can also be used to identify candidate brain regions missing from poorly performing models. Here, we first present the SLDS as both an extension to and simplification of DCM. We describe a simple process for fitting SLDS models to fMRI data and describe an objective validation measure for the SLDS based on prediction of the experimental task condition. We next demonstrate the feasibility of SLDS for fMRI using data from a simple motor task. We describe simple methods for examining the estimated dynamics within these models. Finally, we describe how the SLDS method can be used to identify potentially important brain regions that should be included to improve the sufficiency of poor models.
Switching linear dynamic systems
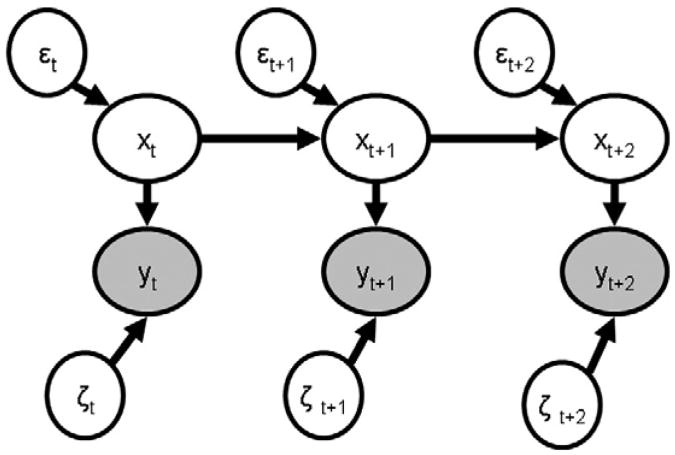
We begin by examining a simpler model, the linear dynamic system upon which SLDS is based. Discrete, linear, time-invariant dynamic systems subject to Gaussian noise have been well studied in signal processing and engineering (Bar-Shalom et al., 2001; Haykin, 2002; Simon, 2006). Linear dynamic systems are directly related to auto-regressive models such as MAR such that the autoregressive model can be described as in a formally equivalent linear dynamic system. They can be depicted as in Eq. (1) and graphically as in Fig. 1. In this formulation, the continuous-valued, n dimensional vector yt
Fig. 1.
A schematic representation of a linear dynamic system. In this diagram, filled ovals represent observed continuous-valued variables while unfilled ovals represent continuous-valued variables that must be estimated from the data. Arrows indicate directional dependency.
| (1) |
contains the n observations of the system at single time t and is an instantaneous linear function of an m dimensional vector xt and additive noise ζt∼N(0,R)1. The continuous-valued, normally distributed variable x represents the stochastic, possibly high-dimensional, unknown true state of the dynamic system to be estimated from the observed data. The n by m matrix C defines the linear relation between x and y. The unobserved (hidden) state xt+1 is itself a linear function of the previous hidden state xt with additive noise εt+1∼N(0,Q). It is assumed that the errors are uncorrelated; E[εζ]= 0. The m by m matrix A describes the discrete time dynamics of the linear system. It contains the directed interactions among the hidden states and can be thought of as an effective connectivity matrix. External inputs to the system have been left out for simplicity. Within this formalism, the goal is to infer the state of the dynamic system, xt, given the measurement observations of the system y1:τ (i.e., maximizing p(xt|y1:τ), approximated by a Gaussian distribution). This inference can be done efficiently using the Kalman filter for τ≤t or smoothing algorithms such as the Rauch-Tung-Stribel or Mayne-Fraiser smoothers for τ>t (Bar-Shalom et al., 2001; Haykin 2002; Simon 2006). What separates this model from other hidden variable models such as Principle Components Analysis or Independent Components Analysis is the inclusion of a direct causal relation (in an information sense) through the A matrix between successive hidden states (Roweis and Ghahramani 1999). Thus, if the current state of the system xt is estimated, it can be used to predict future states of the system xt+τ which in turn can generate predictions of future measurements yt+τ.
While powerful, time-invariant systems with constant A and Q matrices may not fully capture the dynamics of neuroimaging data from most experiments since the experimental manipulation would likely have an impact on the state dynamics (connectivity) and potentially the noise variance. Indeed it is the impact of the experimental manipulation that is often of interest. Complicated nonlinear models of the regional interactions in the full experimental paradigm could be attempted, as well as simplifications of this nonlinear model such as the bilinear model used in DCM. However, stationary models such as DCM would not capture differences in noise covariance between experimental conditions which may be useful if this noise is related to the influences of unmodeled regions on the modeled regions (Penny et al., 2005).
Here we use a piece-wise linear approximation to the full non-stationary nonlinear dynamic system using multiple linear models each with their own noise covariance. One linear sub-model is identified for each cognitive regime and the full model switches between these different sub-models. Different cognitive regimes may be equated with different conditions of an experiment. Switching the sub-model with each cognitive regime allows not only the system dynamics matrix to vary as a function of the experimental condition as in DCM, but also the state and observation noise covariances may vary if necessary. The cognitive regime itself is incorporated into the model by introducing a new nominal variable, ut, to index the experimental condition or regime within which the model is currently operating. This cognitive regime variable can be assumed to follow Markov dynamics, making the current cognitive regime dependent upon the previous cognitive regime.
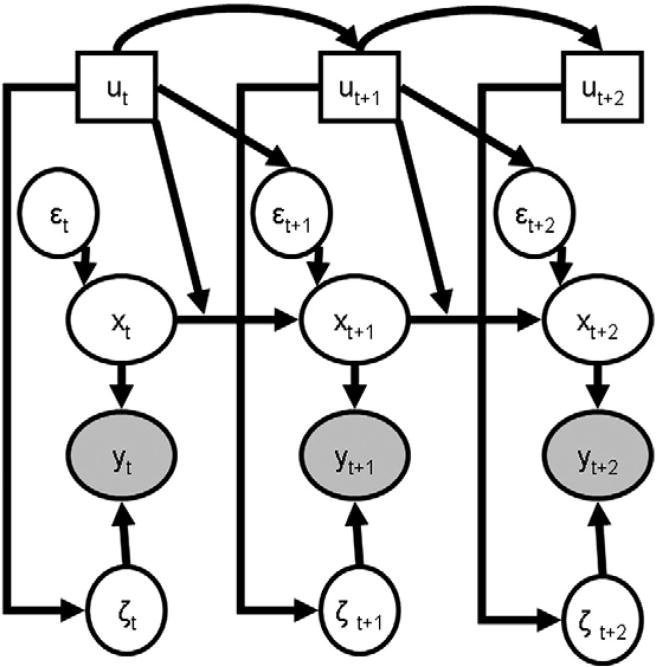
The form of an SLDS model for p>1 experimental conditions is shown in Eq. (2) and graphically in Fig. 2. Here, Π is a matrix
Fig. 2.
A schematic representation of the SLDS. In this diagram, ovals represent continuous-valued variables while boxes represent discrete valued variables. Filled nodes are observed while unfilled nodes are estimated. Arrows indicate directional dependency. In contrast to standard Bayesian network depictions, here we have drawn the arrow from ut to the connection between xt and xt+1 to emphasize the dependence of the connectivity on the cognitive state.
| (2) |
describing the transition probabilities of the cognitive regime u (i.e., P (ut+1=i|ut=j) with a prior probability on u=π. The diagonal elements of Π govern the expected time the system remains in each cognitive regime as given by (1/(1–Π(i,i))). The u superscript indicates a matrix that changes with the change of the state of the experimental condition variable u. Thus there are p A matrices each describing the dynamics for a single experimental condition. External input has been introduced to the system via the vector vt which is the known experimental input at time t and the matrix Dut which is the cognitive regime-dependent impact of vt on the state space variables at time t+1. In addition we change the distribution of the noise variables with regime such that ζt∼N(0,Ru) and εt∼N(0,Qu). Note that by switching between multiple noise covariance matrices, the noise distribution of the full model takes a non-Gaussian form approximated by a mixture of Gaussians.
To show the connection between SLDS and DCM, compare the hidden state dynamics of Eq. (2) of the SLDS with hidden state dynamics of a discrete bilinear system similar to DCM as shown in
| (3) |
Eq. (3)2. Consider the case where there are p=2 non-overlapping experimental conditions: rest and task (i.e., u={0,1}). In the bilinear model, during rest when ut =0, the bilinear dynamics reduce to xt+1=Adxt+Ddvt since Bdut=0. The discrete time SLDS representation of this system would simply set Au=0 to equal Ad from the bilinear model. During the active task condition (ut =1) the bilinear state space dynamics are given as xt+1=(Ad+Bd)xt+Ddvt. Here the SLDS representation would set Au=1 to equal (Ad+Bd) from the bilinear model. Thus, the SLDS model can describe similar dynamics as a bilinear model such as DCM, although defined in discrete as opposed to continuous time. However, unlike SLDS, in the original formulation of DCM the state transitions are deterministic (i.e., εt =0) which is unlikely when other unmodeled regions are interacting with the ROIs in the model (Penny et al., 2005). In addition, the D and R matrices are typically fixed in DCM but can vary as a function of the cognitive regime in the SLDS formulation.
It is important to note that there is no a priori reason that the SLDS model must operate in only a single cognitive regime at a time. In fact, SLDS estimation methods often explicitly assume that the models operate in a mixture of regimes and allow the mixing proportions to be determined (cf., Li and Bar-Shalom, 1993; Murphy, 1998; Barber, 2006). For simplicity, in the analyses presented here we assume that a single cognitive regime is active at a given time when fitting the parameters of a model but allow probabilistic regime mixing when estimating the states for new data (see below).
For fMRI data, the dynamics described in Eq. (2) would occur in a hemodynamic space. Thus the connections would describe interactions among hemodynamic entities, not among more neural level variables where the interactions should occur (Gitelman et al., 2003). To form a quasi-neural hidden state space for the SLDS we follow Penny et al. (2005) and add an additional variable Zt between x and y.
| (4) |
The matrix Zt is (h+1) by m containing h lags of orders 0 though h of each xt. The full SLDS for fMRI model incorporating this lag variable is shown in Eq. (4). The matrix Φ contains a basis set capable of spanning most of the variability in observed hemodynamic responses (e.g., a canonical hemodynamic response function and its derivatives with respect to time and dispersion), while the matrix β provides ROI specific weights for these bases (see Penny et al., 2005 for implementation details for a single ROI). The matrix R contains the hemodynamic space noise covariance. The matrices Z and β are formulated such that each hidden state outputs to one and only one observed variable as in DCM, keeping the regional specificity of the model. The resulting estimated output is equivalent to the convolution of each quasi-neural state space with an ROI specific hemodynamic response function. As in Penny et al. (2005), we instantiate the Z variable by augmenting the state space with lagged versions of the quasi-neural variables and increase the dimension of the A matrices with appropriate sub-diagonal entries to create the lags. The details of this operation are given in Appendix A. Below, we do not allow the β and R matrices to vary with the cognitive state though this could be done if evidence existed that the hemodynamic response function itself varied as a function of task condition or differences in measurement noise were expected due to for example differences in subject motion. In addition, instantaneous (i.e., non-lagged) interactions between the quasi-neural state space variables can be modeled by off-diagonal entries in the Qu matrices. These off-diagonal entries represent coherent relations between the quasi-neural variables that are not captured by the dynamics in the A matrices and are a function of the instantaneous non-directional correlation between the variables. It can be shown that correlational models can be captured in the linear dynamic systems framework by setting the A matrix to a zero matrix allowing the Q matrix to capture all interregional relations (cf., Roweis and Ghahramani, 1999). Such instantaneous interactions may be useful for fMRI data at longer TR where the autoregressive model may be less appropriate3 but will not be used here.
Parameter estimation in an SLDS for fMRI model
All parameters in SLDS models can be estimated from a given dataset, including entries in the involved matrices as well as an estimate of the u vector if the experimental ordering is unknown. Exact inference for the true posterior distribution of u is computationally intractable for all but the most trivial cases, though several approximate solutions exist (Shumway and Stoffer, 1991; Kim, 1994; Kim and Nelson, 1999; Logothetis and Krishnamurthy, 1999; Murphy, 1998; Doucet and Andrieu, 2001; Barber, 2006). However, in typical fMRI experiments, the experimental condition ordering and thus the likely cognitive regime is known by the experimenter. This information can be exploited to decrease the computational burden, reducing the problem to one of fitting several LDS models. Parameter estimation for LDS can be accomplished via several methods (Shum-way and Stoffer, 1991; Ghahramani and Hinton, 1998; Logothetis and Krishnamurthy, 1999; Murphy, 1998; Oh et al., 2005). Here we use the Expectation Maximization (EM) algorithm as described by Murphy (1998). The maximum likelihood parameters of the model, denoted
| (5) |
by the vector θ, are identified by inferring the posteriors of the quasi-neural variables p(xt|y1:T, θold) with the Rauch-Tung-Strieber smoother and maximizing the expected complete data likelihood shown in Eq. (5) with respect to θ. This is accomplished in an iterative fashion as follows. The complete data likelihood is calculated holding the parameters fixed at some value. The partial derivative of the log likelihood with respect to each of the parameters is calculated and new parameters are identified where this derivative is zero. The data likelihood is then recalculated with these new parameters and the process iterated until the likelihood ceases to change. Derivatives of the data likelihood with respect to each of the model parameters are given in the study of Ghahramani and Hinton (1996) for the case where the cognitive regimes are known and in the study of Murphy (1998) for the case of unknown and mixing regimes. Derivatives for the hemodynamic parameters are described in Penny et al. (2005). For completeness, the full EM algorithm is given in Appendices B and C. A priori hypotheses regarding entries in any of these matrices, including equivalence of values between conditions, if desired can be included in EM estimation. More complex methods than EM such as Variational Bayes may provide more accurate parameter estimates and more elegantly incorporate prior information though at increased computational cost (Ghahramani and Hinton, 1998).
Validating an SLDS for fMRI model
Several tests exist for the consistency of a single (S)LDS model for a single dataset (Bar-Shalom et al., 2001; Lütkepohl, 2007). Here we focus on cross-validation of a model; using a single model to predict data from multiple subjects with multiple experimental condition orderings. While agreement between the hemodynamic data estimated by the model and the actual measured fMRI signal could be used as the cross-validation measure, it is unclear how well the fMRI data from a single subject should match that of another. However, given a reasonable experimental design and a valid SLDS model of the different interregional connectivity patterns in that experiment, what should be in good agreement are estimates of the cognitive regime based on the model with the actual experimental condition. As noted previously all parameters of the SLDS, including u, can be estimated from a given dataset. It is also possible to take the parameters from a previously identified model and use them to estimate x1:T and u1:T for fMRI data from a similar experiment by computing p(xt|y1:τ) and p(ut| xt, y1:τ). Agreement between the estimated and actual u has both a theoretical lower and upper limit (chance and 100% respectively) allowing for meaningful objective comparisons between multiple SDLS models as well as between SLDS and different model formalisms which cannot be directly compared using Relative Bayes factors. It is important to note here that the distinction between the models of different cognitive regimes is solely a distinction between different patterns of interregional connectivity. Thus estimating the most likely cognitive regime at a given time is essentially estimating the most likely pattern of connectivity at that time. Expected regional activation in a cognitive regime is not included in the model. We further note that this is in essence using BMS at each time point to identify the most likely cognitive regime.
Here we apply the Generalized Pseudo-Bayes Two (GPB2) algorithm to compute smoothed estimates of u via p(ut|ut+ 1,y1:T). Details of the algorithm as applied to SLDS are in the study of Murphy (1998). In essence, the initial distribution of the hidden state p(x1) is a mixture of p Gaussians, one for each of the p different cognitive regimes. This state is propagated through p equations to time 2 making p(x2) a mixture of p2 Gaussians. The posterior of the hidden state at a generic time t, given by p(xt|y1:t) is thus a mixture of pt Gaussians. The GPB2 algorithm will approximate this intractably large distribution by maintaining only p2 Gaussians. Gaussians representing states that differ in their history of more than two time steps are collapsed together via moment matching. The details of the algorithm as applied here are given in Appendix B. Since we do not yet update the model parameters when estimating the cognitive regimes, the complex GPB2 algorithm is not strictly necessary and simpler algorithms could be used instead including algorithms that do not allow mixtures of regimes. We use GPB2 in anticipation of learning all model parameters including regime simultaneously in future studies. In the remainder of the document, we demonstrate the feasibility and versatility of SLDS for fMRI by applying the model to a simple motor task.
Methods
Participants
Two healthy male participants, both aged 35 years with normal vision, participated in the study. Both were identified as strongly right-handed using a standard handedness inventory (Oldfield, 1971). Both gave written informed consent in accordance with the National Institutes of Health Institutional Review Board. One participant was compensated for his time.
Experimental task
Participants performed a blocked visually cued finger opposition task with three conditions (left hand finger tapping, right hand finger tapping and rest). Both active conditions involved sequentially tapping each finger to the opposing thumb of the indicated hand. Timing of the tapping during scanning was self paced. Prior to scanning, the tapping sequences were briefly practiced at approximately 5Hz. In the control (rest) condition, participants were instructed to rest both their hands at their sides. The identity of each condition was indicated to the participant by presenting the words “Left”, “Right” or “Rest” in 48 point white Ariel font on a black back ground at the beginning of each block for1 s using a projector and semi-transparent screen. During the remaining time in each block, a white fixation cross was presented in the center of the screen. Participants performed two scanning runs. In each run participants performed seven 22-s blocks of each condition in a different pseudorandom order. An additional rest block ended each scanning run and 8 s of fixation began each run. Participants were observed during each scanning session and task compliance was visually verified.
MR image acquisition
Functional imaging data were collected with a 3 T GE whole body system (General Electric, Milwaukee, WI) equipped with an 8-channel head coil. Twenty-two interleaved T2*-weighted axial slices (64×64 matrix, 4.3 mm slice thickness) covering the top of the brain approximately down to the bottom of the thalamus were acquired using an EPI BOLD sequence with ASSET acceleration (TE=20 ms, TR=1.0 s, FOV=240 mm, flip angle=90°, acceleration factor=2). Four-hundred ninety-six volumes were obtained for each of the two runs for each participant and the first four volumes of each run were discarded from subsequent analysis to allow for signal intensity saturation.
In addition, a T1-weighted anatomical image was collected at the same slice locations as the functional data (TE=2.4 ms, TR=300 ms, flip angle=70°, slice thickness=2.6 mm, matrix=256×256, FOV=240 mm) and a T1-weighted FSPGR image (TE=2.4 ms, prep time=450 ms, flip angle=12° FOV=240 mm, slice thickness=1.3 mm matrix=256×256, phase field of view=80%) covering the entire brain was collected for spatial normalization.
MR image preprocessing
Image quality was reviewed for each participant. The functional image sequences for each participant were preprocessed using the Statistical Parametric Mapping software package (SPM2, Wellcome Department of Cognitive Neurology, London). For each participant, functional images were corrected for differences in slice timing, realigned to the first image of the first run then to the mean of both runs, corrected for interactions between motion and susceptibility distortion and resliced using 4th degree beta splines. The partial brain T1 image was coregistered to the mean functional image and then the full-brain T1 image was coregistered to the partial. The full-brain image was segmented into gray matter, white matter and CSF and the gray matter image was normalized to the gray matter template provided with the SPM2 package. Functional images were normalized using the parameters from the segmented T1 image, resliced to 2 mm isotropic voxels and spatially smoothed with a 6 mm FWHM Gaussian kernel.
ROI selection
Regions exhibiting task-related BOLD signal increases relative to rest were identified for each task condition using the general linear model as implemented in SPM2. Because of the well-studied nature of the motor task (cf., Smith et al., 2006), we chose five (n=5) anatomical regions of interest a priori: left and right primary sensory-motor cortex, left and right premotor cortex and medial supplementary motor cortex. Coordinates of peak activation were identified in each of the preselected anatomical regions. Different coordinates were selected for each subject. Each of the five regions selected was identified as active above the resting baseline (p<0.05 svc) for both tapping conditions. For each run, the first eigenvector of a 216 mm3 volume (27 voxels) ROI centered on each of the coordinates was calculated. The eigenvector was oriented such that its sum was positive and the ROI was projected onto this vector to create a single time series per ROI. Each time series was detrended using polynomials up to 3rd order and normalized to zero mean and unit variance.
SLDS modeling
All SLDS model estimation and validation procedures were implemented in Matlab (Mathworks Inc., Natick, MA) using custom software. Subject one run one was selected as the training dataset. Parameters for an SLDS model were estimated with this dataset using the EM algorithm. The lag for hemodynamic response estimation was set to 15 time points making the hemodynamic responses 16 s in length. The input vector v was set to zeros except for the first TR of each block where it was set to one to model the visual input and the cognitive regime vector u was set to index the task condition ordering of run one. The input vector did not vary as a function of task condition. The A, D, and Q matrices were switched based on the task condition u. The Q matrices were forced to be diagonal (i.e., no instantaneous connectivity) as was the R matrix (i.e., uncorrelated error at each ROI). In keeping with the exploratory nature of the model fitting, the A matrices were not restricted and all-to-all connectivity, including self connections, was allowed. The model parameters (Au, Du, Qu, R, β) were updated until the relative change in log likelihood dropped below 1e−7. Since the experimental conditions were known, Π and π were known and thus not updated. To help avoid over-fitting of the model, the observation error variances in R were not allowed to go below 1e−3. The model identification procedure was repeated ten times with different random starts in the Au matrices and the model with the highest log likelihood was selected. The parameters of this best model were then fixed and the model used to predict the fMRI data and task condition vector for each of the four datasets using the GPB2 algorithm.
Results
The SLDS model was able to fit the fMRI data from the first run of one participant quite well with correlations between the estimated fMRI data and the actual fMRI data above .925 for all five regions (mean±SD r=.945±0.018). This accuracy compares favorably with predictions of each ROI based on linear regression using the known task conditions convolved with a canonical hemodynamic response (mean±SD r= .744±0.091). The SLDS accuracy also compares well with predictions based on linear regression using the other four ROIs to predict each ROI (mean±SD r= .851±0.002). While the SLDS and regression models are not directly comparable given the differences in complexity, the comparison demonstrates that the SLDS model was able to predict the fMRI data with considerable accuracy.
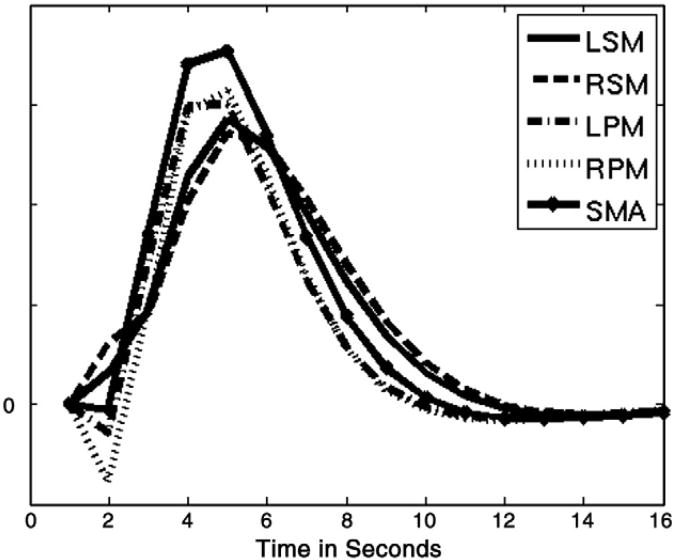
The hemodynamic response estimates were all reasonable and within the range of normal variability. The identified regional hemodynamic responses are shown in Fig. 3. All responses were qualitatively very similar. The hemodynamic response functions in the primary sensory-motor cortices had no initial negativity and showed a slightly longer time to peak and slower decay than the other regions. Given the block design of the motor experiment regional or subject variability in the hemodynamic response may have a limited impact on the model. Hemodynamic variability would likely have a larger effect in event related designs. It remains to be seen how well SLDS performs in this case.
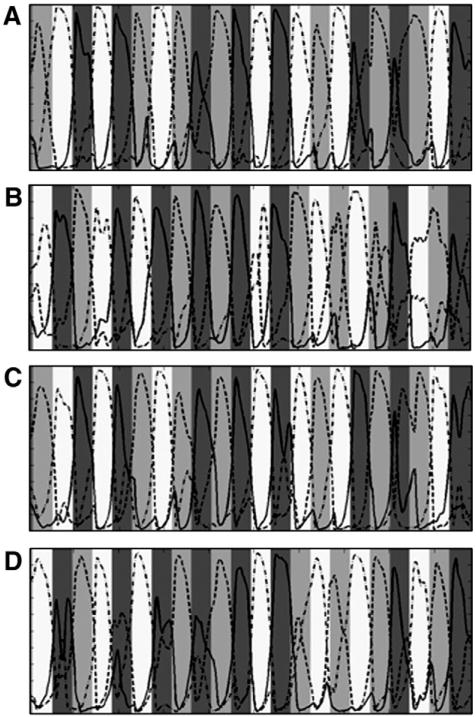
Fig. 3.
Graphical depiction of the estimated cognitive state probabilities in the motor task. In each graph, shading of the background indicates the correct cognitive regime, dark gray indicates the rest condition time points, light gray indicates the left hand tapping condition time points and white indicates the right hand tapping time points. Lines indicate the estimated probability [0:1] from the model that a cognitive regime is active at that time point. The solid line indicates the rest connectivity pattern probabilities, the dark dashed line indicates the left hand connectivity pattern probabilities, and the light dashed line indicates the right hand connectivity pattern probabilities. A. Estimated cognitive regime probabilities for subject one run one. This is the data set used to estimate the model parameters. B. Estimated cognitive regime probabilities for subject one run two. C. Estimated cognitive regime probabilities for subject two run one. D. Estimated cognitive regime probabilities for subject two run two.
All parameters in this SLDS model were fixed and then used to estimate the most likely u vector from both runs from both subjects separately using GPB2 as described above. Thus, p(ut=i|y1:492) was estimated and the most likely condition for each time point was identified. The predictions were compared to the actual experimental design. The predictions were accurate, exceeding 82% in all cases with chance level being approximately 33% (mean±SD, percent correct=85.47%±3.47%; see Table 1). The estimated state probabilities p(ut=i|y1:492), are shown graphically for each run in Fig. 4. Again, these predictions are based solely on the ability of the different patterns of interregional connectivity contained in the Au matrices to predict the observed fMRI data; task condition differences in mean level of regional activity play no direct role.
Table 1.
| Percent correct labeling (n/492) | Run one | Run two |
|---|---|---|
| Subject one | 86.99% | 82.92% |
| Subject two | 89.63% | 82.32% |
Fig. 4.
Estimated hemodynamic response functions for each region. Abbreviations: LSM, left sensory-motor cortex; RSM, right sensory-motor cortex; LPM, left premotor cortex; RPM, right premotor cortex; SMA, supplementary motor area.
One goal of dynamic models such as SLDS is to gain insight into the patterns of interregional connectivity that generate the observed imaging data and the relation of these patterns to the cognitive regime. Here we separate two analysis issues; identifying changes in individual parameters and identifying changes in overall dynamics. These two analysis issues are not equivalent. Comparisons of individual connection magnitudes from one connectivity matrix to another ignore changes in the relative contribution of connections and the overall pattern of connectivity. The value of individual connections can only be fully understood when examined relative to all others. Here we provide a means of computing both the significance of individual connections and their changes between regimes as well as a simple means of examining changes in dynamics.
It can be shown that under a set of non-restrictive regularity assumptions the difference between the estimated maximum likelihood model parameters and the true parameters of a state space model is asymptotically multivariate normal with mean 0 and covariance given by the inverse of the expected Fisher Information matrix (Cavanaugh and Shumway, 1996). The observed Fisher Information matrix of the parameters can be computed or estimated for a specified model given a specific dataset as the hessian (second derivative) matrix of the likelihood function maximized by the EM algorithm above (Cavanaugh and Shumway, 1996; Klein and Neudecker, 2000; Klein et al., 2000). The inverse of the Fisher Information matrix can then be used to calculate 100*(1–α)% confidence intervals for each parameter in a model (Neumaier and Schneider, 2001). When the model observations are not a simple function of the state space as is the case here, calculation of the exact hessian can be cumbersome (cf., Klein et al., 2000). Bootstrap estimates of the inverse Fisher Information matrix can also be calculated that are asymptotically correct (Stoffer and Wall, 1991) but rely on replicable accuracy of the model estimation method. Here we build an approximation of the hessian matrix of the likelihood function of the SLDS model for the Au matrices only using the method of finite differences using direct evaluations of the likelihood function of the SLDS model. Note that estimating the hessian this way assumes
| (6) |
that the variances of the Au parameters are independent from the other parameters of the model. We then construct confidence intervals for each parameter using Eq. (6) where H is the estimate of the hessian (see Eq. (41) of Neumaier and Schneider (2001)). Parameters where the confidence interval does not cross zero are considered statistically significant, and two parameters whose confidence intervals do not intersect are considered significantly different. Because the estimated hessian can potentially underestimate parameter variance (Stoffer and Wall, 1991) we use an α=0.01 confidence interval Bonferoni corrected for multiple comparisons.
For the rest condition, all connections were identified as non-zero except the connection from the right premotor cortex to the right primary sensory-motor cortex. In both the left and right hand tapping conditions, all connections were identified as non-zero except the connection from the right primary sensory-motor cortex to itself. In the left hand tapping condition, connections from the left premotor cortex to all regions but itself as well as the connection from the right premotor cortex to itself did not differ from the rest condition. All other connections differed significantly between the left and rest conditions. In the right hand tapping condition, the connection from the right premotor cortex to the left primary sensory-motor cortex, left premotor cortex, and supplementary motor cortex did not differ from their values in the rest condition. All other connections differed significantly between the right and rest conditions.
Connections that are non-zero may still play a minimal role in the dynamics of the system. To determine the prominent modes of interaction within the systems different methods are needed. A full discussion of linear dynamics analysis is far beyond the scope of this article and several approaches exist. One approach to understanding the connectivity in dynamic networks is to reduce their dimensionality and examine the prominent or dominating patterns of interaction of the network. There are several techniques of varying degrees of sophistication in the literature that can provide such pattern based analysis of the network dynamics (Hasselmann, 1988; Huang et al., 1998; Farrell and Ioannou, 2001; Liao, 2003). Here we use
| (7) |
a simple analysis based on the singular value decomposition (SVD) to illustrate some of the information one can obtain from complex connectivity matrices. The SVD of a matrix A is given in Eq. (7) (cf., Strang, 1988). The columns of U and V form orthonormal bases of the column space and the row space of the matrix A respectively. The columns of U and V are called left and right singular vectors or modes of the matrix A (Strang, 1988). The diagonal matrix S contains the singular values of A. From S we obtain the fractional singular values by squaring each diagonal element and dividing each by their sum of squares which gives the relative scaling factor of the corresponding singular vector. That is, the diagonal elements of S can be used to give the percentage of variance within the matrix in the direction of the corresponding singular vector. Prominent directions in an Au matrix extracted via SVD can be interpreted as indicating prominent interactions among the variables during a cognitive regime. If a single direction dominates the variance, the dynamics described by the system can be well expressed in a lower dimensional space and the pattern analysis is informative. If all directions contain an approximately equal percentage of the variance, the dimension of the dynamics cannot be reduced via linear methods and the SVD pattern analysis is not appropriate.
Here we do SVD on each of the Au matrices from the motor model estimated from the first run of the first subject that generalized well to the other runs. The first mode of each of these matrices accounts for a large proportion of the matrix variance in each case (56%, 47%, and 41% for rest, left tapping, and right tapping respectively) while subsequent modes accounted for far less of the variance. Thus the dominant mode of each Au matrix alone summarizes much of dynamic interactions in their respective regime. The Au matrices are then reconstructed using these dominant modes and thresholded to see the locus of these major interactions in the mode.
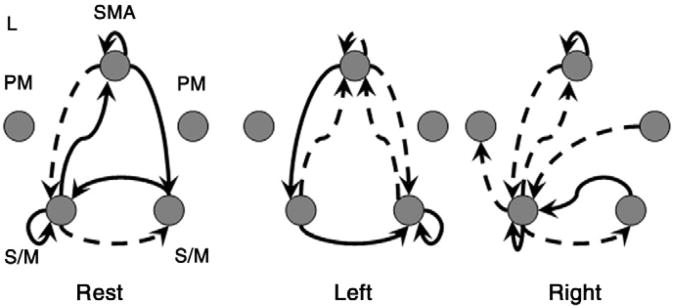
Fig. 5 shows the dominant patterns of the connectivity matrices for the different regimes. The values of the reconstructed matrices are thresholded at an arbitrary value based on their empirical distribution. Positively valued connections between regions exceeding the threshold are shown with solid lines while negatively valued connections exceeding the threshold are shown with dashed lines. The observed patterns of connectivity are generally consistent with previous studies such as the bilateral representation of the task, particularly with the non-dominant hand (Rao et al., 1993; Singh et al., 1998; Cramer et al., 1999; Smith et al., 2006). Direct interactions between lateralized primary motor areas as seen in all conditions have been highlighted in several studies using transcortical magnetic stimulation as well as other methods (Kapur, 1996; Allison et al., 2000; Kobayashi et al., 2000; Zhuang et al., 2005). The lack of supra-threshold connectivity to or from the premotor cortices in the dominant modes of the rest and left hand tapping regimes suggests that these regions play less of a role in this task and could potentially be excluded from the model.
Fig. 5.
Dominant modes of connectivity in the three cognitive regimes. Depicted are the effective connectivity patterns reconstructed from the dominant singular vectors and thresholded within a matrix at an arbitrary level based on the empirical distribution. Solid lines indicate positively valued connections while dashed lines indicate negatively valued connections. Abbreviations: S/M, primary sensory-motor cortex; PM, premotor cortex; SMA, supplementary motor area; L, left hemisphere.
The patterns in Fig. 5 show the major source of interactions between the regions in each regime but cannot be used to directly compare and contrast conditions. Here we extend the logic of the above SVD analysis and use generalized eigenvectors to directly identify prominent directions of variance in one matrix with minimal projection in another. The most prominent general eigenvector of one Au matrix relative to another describes a pattern of interactions between brain regions that simultaneously has maximum energy in the first matrix and minimal energy in the second. Thus, the general eigenvector represents a connectivity dynamic strongly operational in one cognitive regime but not in another (see Diamantaras and Kung, 1996 for a similar use of general eigenvectors for signal enhancement against known noise sources). Note that this is not the subtractive difference between condition dynamics which says little about the actual dynamics in a condition. If the two matrices are unrelated or the second matrix has uniform variance in all directions, the general eigenvector will be in the same direction as the eigenvector. As in the SVD analysis, if all general eigenvectors describe an equal amount of variance, the dynamics cannot be reduced to a lower dimension via linear methods. As before, the dynamics of an Au matrix not contained in another can be reconstructed from its dominant general eigenvector, z-score thresholded at |z|>2, and examined for large connections.
We first examine each of the active tapping dynamics relative to the resting dynamics via the generalized eigenvector method. For left hand tapping, 86.08% of the relative dynamics was contained in the first general eigenvector. The prominent dynamic in left hand tapping not in rest is primarily a positive effective connectivity from right sensory-motor cortex to itself (z=2.51) as well as a negative effective connectivity from the supplementary motor area to right sensory-motor cortex (z= −3.1). For right hand tapping, 89.51% of the relative dynamics was contained in the first general eigenvector. The prominent dynamic in right hand tapping not in rest is primarily a negative effective connectivity from the left to right sensory-motor cortices (z= −3.45).
Next we examined the general eigenvectors contrasting the two active task conditions. For left hand tapping relative to right, 58.92% of the relative dynamics was contained in the first general eigenvector. The prominent dynamic in left hand tapping not in right hand tapping was a negative effective connection from the left sensory-motor cortex to the left premotor cortex(z=− 2.18). For right hand tapping relative to left, 59.50% of the relative dynamics was contained in the first general eigenvector. The prominent dynamic in right hand tapping not in left hand tapping was a positive effective connectivity from left sensory-motor cortex to itself (z= 2.55) and a positive effective connection from the left sensory-motor cortex to the supplementary motor area (z= 2.17).
To further examine the utility of the SLDS model we created two more models of this simple motor task. The first model was a reduced motor model where we purposefully ignored important motor regions, thus simulating in complete knowledge of the appropriate task network. We included three regions in the model, left and right premotor cortex and medial supplementary motor cortex; primary sensory-motor cortices were left out of the model. Because the most task relevant regions were removed from the model, SLDS should perform worse in this case. The same SLDS fitting procedures were used as in the full model above. The reduced model was estimated using one run of one participant, fixed to the best fitting parameters, then used to predict the task condition vector of both runs from both participants. The reduced model was still able to predict the experimental task vector above chance levels though less convincingly (mean±SD percent correct=55.28%±16.77%; see Table 2).
Table 2.
| Percent correct labeling (n/492) | Run one | Run two |
|---|---|---|
| Subject one Subject two | 71.14% 42.68% | 68.29% 39.02% |
The next model was also based on the same reduced set of three motor regions. However, in this model we augmented the quasi-neural state space variable dimension by one. This additional state space variable was allowed to interact with the other state space variables through the Au matrices, but could not directly impact the estimated fMRI data (i.e., entries in β for this additional state space variable were set to 0). Note that the SLDS is itself estimating a time series for the augmented quasi-neural variable. Rather than using the fMRI data directly to estimate this augmented variable, its estimation is based on its interactions with the other quasi-neural variables and the effect this has on their predictions of their respective ROIs. This can be considered as estimating functionally relevant regions not included in the model that have high effective connectivity with the included regions.
This augmented model was estimated using the same procedures as above on one run from one subject with the following exception. All identified models had roughly equivalent likelihood values. To identify a best model, each was fixed, and used to predict the task condition vector of the second run from the training participant. The model with the best cross-validation accuracy on this second run was selected as best and used to predict the task condition vector from the other participant. The predictions were more accurate than the reduced model in all cases though not as accurate as the full model (mean percent correct±SD, 66.77%±12.47%; see Table 3).
Table 3.
| Percent correct labeling (n/492) | Run one | Run two |
|---|---|---|
| Subject one | 82.52% | 65.65% |
| Subject two | 66.87% | 52.03% |
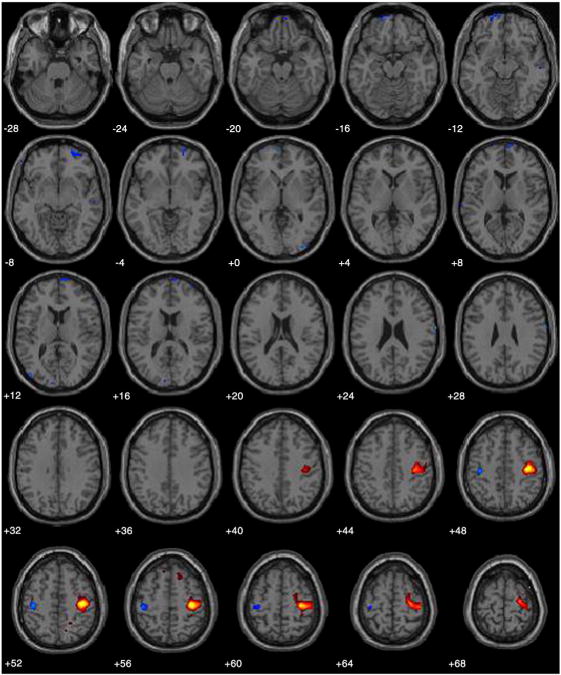
To identify the brain region most related to the quasi-neural augmented variable we convolved the time series of this variable estimated from the training run with the canonical hemodynamic response from SPM2 and used the resulting hemodynamic time series to predict each voxel in the brain for that same run via linear regression. The augmented variable was maximally related to the right primary motor cortex (t490 =14.22, p< 0.001 FWE corrected; see Fig. 6).
Fig. 6.
Location of the augmented variable. Depicted are t values thresholded at pfwe<0.05, from the regression analysis using the augmented variable time series convolved with a canonical hemodynamic response as the predictor variable plotted on a standard atlas included in the SPM2 software package in neurological convention. The maximum t value occurs in the right primary sensory motor cortex (t490=14.22, p< 0.001 FWE corrected).
Discussion
We presented switching linear dynamic systems for fMRIasa means to identify task dependent interregional effective connectivity at a quasi-neural level and to objectively validate these models using their ability to predict the cognitive regime in new data. This objective criterion assesses the ability of a model to predict what is often the most critical element of a functional imaging study, the cognitive states induced by the study design. This criterion should help identify poorly performing models in more exploratory analyses. Here, identification of task condition or cognitive state is accomplished by identifying the most probable connectivity pattern operational at each given time point. While task condition specific mean activation levels could also be included in the model to improve accuracy and better reflect the activity and connectivity dependence of cognition, for this study we focused on differential connectivity only. Using simple point-estimation procedures, an SLDS model was identified for fMRI data from a simple motor task. The model was able to predict the fMRI data from the training subject better than regression models without a temporal dependence. The model generalized well to a second run from the same subject with a different task ordering as well as two runs from a different subject. A reduced model without the primary sensory-motor cortices did not generalize as well, adding face validity to the model validation procedure. Finally, the reduced model was augmented, allowing the SLDS estimation procedure to identify a quasi-neural time series that was best able to assist in understanding the connectivity of the included regions. This time series was identified as being strongly localized to the right primary sensory-motor cortex.
Two methods were described for examining the interregional connectivity identified by the SLDS model. Simple confidence intervals based on the Fisher Information matrix were used to identify changes in individual parameters and to compare them against zero. Singular value decomposition was used to identify prominent patterns of connectivity within the connectivity matrices and generalized eigenvalues were used to identify prominent patterns of connectivity within an Au matrix that were not present in another Au matrix. The confidence interval tests suggested that essentially all connectivity parameters were non-zero and that the majority changed from one condition to another. Whether this reflects the true nature of the neural implementation of the task is unknown and requires further research. However these individual parameter tests are related to the curvature of the local maximum identified by the EM algorithm in the direction of the parameter. They do not indicate how important a connection is, only how confident the estimation scheme is of its value. A connection may be non-zero yet play a minimal role in governing the interregional dynamics. Importance of the connections or of the change in connections is seen in the (general) eigen-analysis of the connectivity matrices. These analyses indicate if a connection has a prominent impact on the quasi-neural state dynamics. Unfortunately, such analyses are fairly qualitative as “importance” is ill defined. While statistical tests can be derived for the participation of a region in a prominent eigenvector (cf., Neumaier and Schneider, 2001) more research is required to derive similar tests for individual connections. It may be possible to use the predictions of the cognitive regime as a measure of connection importance by for example removing a connection and testing the specificity of the resulting model. Development of such tests is an important area for further research for SLDS and similar modeling efforts.
The ability of SLDS to estimate the task condition vector on novel datasets is an important advantage over DCM, MAR, and similar models where the task conditions must always be pre-specified. Given that the number of regions included in these models must be limited for computational considerations, some regions important for the task may be excluded. This may result in models with greater than chance connectivity that nonetheless fail to capture the true nature of the task. The reduced motor model identified above without primary motor cortices was able to predict both the fMRI data and the cognitive state at a greater than chance level. It is likely that BMS methods using the data likelihood alone would identify some connectivity model among these three regions as better than a chance or null model. However, the reduced motor model excluding the primary motor cortices hardly constitutes a valid model of the finger opposition task. By cross-validating an SLDS model on multiple datasets and using agreement between the predicted and actual task condition vectors as a validation measure, a better sense of the external validity of a model can be gained. Such a measure should help to avoid simply identifying the best among many poor models.
The ability to predict the cognitive regime is not simply useful for model validation. There are many interesting experimental situations where the cognitive regime either is unknown or else an unknown mixture of multiple conditions. For such experiments, DCM and MAR cannot be used because the correct condition segmentation cannot be provided to the model. However, methods exist for simultaneously fitting all parameters of an SLDS model including the task conditions (Ghahramani and Hinton, 1998; Murphy, 1998). Or, as was shown here, given an SLDS model with parameters identified when the segmentation was known, the cognitive regime vector can be estimated for additional data sets where the regime is unknown (cf., Li and Bar-Shalom, 1993; Murphy, 1998; Barber, 2006). For example, one might wish to identify an SLDS model with differential connectivity during correct versus incorrect trials. For some experiments, trial correctness may not be known if no overt response is measured. SLDS modeling may be used in such instances to identify correct trials based on interregional connectivity. If Kalman filtering rather than a smoothing algorithm is used for likelihood estimation, this identification can potentially be accomplished in close to real time. Other examples where SLDS cognitive regime estimation would be useful are recognition memory experiments examining old-new judgments. These experiments often seek to distinguish item recall processes from item familiarity though both processes may be at work to some degree in any given trial. Because the GPB2 algorithm used here generates a probability weighted mixture of the connectivity models, the SLDS formulism may be used to identify the mixture of recall and familiarity for a given trial. SLDS modeling of a probabilistically mixed cognitive regime may also have application as a clinical measure; identifying for example the probability that a given trial was performed using a pre-identified healthy or diseased network. One could also use the SLDS formulism to examine theoretical relations between low-level tasks and higher cognition. For example, possible relations between procedural memory and syntactic processing have been discussed (Ullman, 2004). Using an SLDS model of a procedural memory task to predict the cognitive regime vector in a syntactic processing task may help elucidate the connection between these processes.
The utility of the augmented SLDS method to identify additional task-connectivity relevant brain regions for poorly performing models represents a significant advance over current effective connectivity methods such as structural equation modeling and DCM. We showed here how poorly performing models such as the reduced motor model can be augmented to identify additional regions based on connectivity patterns that may be useful to include in the model. Here, augmented SLDS clearly identified the right primary motor cortex as the optimal region to include in the model. While there is no guarantee that this augmentation procedure will always clearly identify a single region, it may still help identify a candidate set of potentially useful regions whose connectivity with the pre-specified regions should be further examined. It may eventually be extendable to an iterative procedure where a small set of ROIs are augmented and modeled, the best matching region to the augmented series is identified and added to the model and the procedure repeated until the cognitive regime prediction accuracy asymptotes. Further study is necessary to determine the utility of this iterative procedure.
We see the SLDS modeling formalism as complimentary to existing methods of modeling temporally dependent connectivity such as DCM and pair-wise Granger-Geweke causality. For testing well specified a priori hypotheses regarding the interactions between a small number of regions where all relevant regions and connections are identified, DCM with its nonlinear and biologically meaningful hemodynamic model is likely superior. For more exploratory analyses, where most task relevant regions are unknown and connectivity models cannot be pre-specified, pair-wise Granger-Geweke causality with a seed ROI and simple deconvolution methods are likely more informative. SLDS will likely be optimal in the cases between these extremes where some regions and some connections are theoretically specified while others are unknown. SLDS may be particularly useful for patient studies where disease and/or compensatory mechanisms cause the effective connectivity network used by patients to diverge substantially in an unknown manner from the network used in the normal population. Here, since only a small subset of regions may be known, the “best poor model” problem is even more pertinent and validation methods selecting among a set of pre-specified candidate models are inadequate.
A major issue with SLDS as presented here as well as with DCM and other Granger-Geweke causal methods applied to fMRI, is the validity of high-frequency signals in the quasi-neural or fMRI time series (Goebel et al., 2004). The hemodynamic response is essentially a low-pass filter with a cutoff frequency near 0.2 Hz (Henson, 2003). Signals much above this frequency are essentially removed from the data. To the extent that a given cognitive task is dominated by fast, transient neural activity (on the order of milliseconds), temporally dependent causal measures based on fMRI data are inappropriate. In this case SLDS has an advantage over DCM since the Qu matrices can be used to model instantaneous connectivity. However the Au matrix in this case may be dominated by noise possibly inducing meaningless high-frequency signals into the quasi-neural time series and leading to spurious conclusions regarding directional connectivity. We acknowledge that this is indeed a problem. This issue again points to a need for a validation measure such as the one presented here that is based on something other than the likelihood of the observed fMRI signal. If the effective connectivity matrices are indeed able to distinguish different cognitive regimes in a cross-validation test set then they must contain at least some portion of the meaningful signal. Using SVD to analyze the Au matrices should also identify cases where the Granger dynamics are essentially noise. Another intriguing possible solution to the low-pass filter problem is to incorporate data from simultaneous fMRI and EEG recordings into a single model (cf., Martínez-Montes et al., 2004). Though the direct connection between fMRI and EEG signals remains largely unknown, the higher sampling rate of the EEG could help avoid fitting noise in the quasi-neural variables.
Conclusions
The SLDS model framework is an effective means to address several problems with Granger-Geweke connectivity and DCM including measuring overall model adequacy by estimating the cognitive regime and identifying important regions left out of incomplete models. Extensions of the model can handle unknown cognitive regimes, mixtures of multiple cognitive regimes and instantaneous connectivity. SLDS provides additional useful tools for identifying cognitive regime-dependent effective connectivity at a quasi-neural level.
Acknowledgments
This work was supported by the intramural program of the National Institute on Deafness and Other Communication Disorders, the National Institutes of Health, and a National Institutes of Health Intramural Research Training Award to JFS. The authors would also like to acknowledge support from the National Institute on Aging (AG19610 AG025526 [GEA]), the Evelyn F. McKnight Brain Institute [GEA], and the State of Arizona [GEA, KC].
Appendix A.
We first describe the embedding of the fMRI model into a conventional state space representation. The SLDS model can be rewritten as
| (A.1) |
Here we absorb Z from equation 4 into x by adding a lagging matrix to A
| (A.2) |
where I is an appropriately sized identity matrix and 0 is an appropriately sized matrix of zeros and
| (A.3) |
The covariance of the quasi-neural state error Qu is similarly padded with zeros.
| (A.4) |
When analyzing the resulting conventional system, only the Au, Du, and Qu submatrices are relevant. The observation matrix C is formed piecemeal for each output region from β and Φ. Using the Matlab colon operator for notational purposes,
| (A.5) |
represents the hemodynamic response with Φ a hemodynamic basis set and βi the weights for the ith region. All elements of C not explicitly indexed in Eq. (A.5) are zero to maintain a one-to-one correspondence between the quasi-neural state and an observed region.
Appendix B.
The E-step.
Here, we describe a solution to the cognitive regime inference problem (i.e., maximizing p(ut=i|y1:t)) using the parameters of a specified SLDS model. The solution is due to Murphy (1998) and is represented here for completeness. If the regime is known the computations can be simplified as described below and used as the E-step of the EM algorithm for learning the model parameters. Using the GPB2 algorithm, the statistics of two consecutive time steps need to be saved. First, we define the necessary notation. Following Murphy (1998) we define the following expectations on the quasi-neural state means:
| (B.1) |
Here, the superscript refers to the cognitive state. The value of the cognitive state at time t is identified by the superscript within the parenthesis. The value of the cognitive state at time t–1 is identified by the superscript to the left of the parenthesis and the value of the cognitive state at time t+1 is identified by the superscript to the right of the parenthesis. We also require terms for the covariance of the quasi-neural state. Again using the superscript notation to identify regime define:
| (B.2) |
Finally we require terms for probabilities on the cognitive regime itself and the observed data given a regime:
| (B.3) |
We can now compute the expectation in two passes. The Kalman filter recursions are used to compute the filtered (using data up to time t) statistics followed by the RTS smoother to compute smoothed (using full data) statistics. First, filtered estimates of ut are computed using Filter() to indicate a single forward step of a standard Kalman filter using the indicated conditional moments indicated. The likelihood of the update is returned in L. MomentMatch() indicates computation of the optimum unconditional moments given conditional moments and mixing coefficients.
| (B.4) |
Here the effect of Π is evident. The values in Π provide priors for the regime transitions such that the likelihood of a regime transition from i to j is the probability that the regime was i during the previous time multiplied by Π(i,j) (the prior probability of the transition) multiplied by the likelihood of the data given the new regime. Next smoothed estimates of ut are computed using Smooth() to indicate a single backwards step of the RTS smoother using the conditional moments indicated:
| (B.5) |
Note that if the correct cognitive regime is known, the probabilities of each state Mt|t and Mt|T are either 1 or 0 making the mixing proportion in the moment matching procedures Wi|k and Wk|j also reduce to {0,1}. Thus for calculation of the E-step for EM when ut is known filtering and smoothing only need to be performed once at each time step with the parameters of the correct regime. In addition, for parameter learning, the smoothed cross-variance term is required which if the regime is known can be computed as
| (B.6) |
where Jt is the RST smoother gain at time t.
Appendix C.
M-step.
To compute new parameters we seek to maximize the full data log likelihood:
| (C.1) |
Assuming the regime is known, additional terms for π and π are ignored. To compute the parameter updates from this equation, we identify the partial derivative of this function with respect to each of the parameters, set to zero and solve. The following smoothed terms from the E-step are required for this operation:
| (C.2) |
Note that these terms are no longer conditioned on the cognitive regime. We compute each of the following terms in order. Any known parameters can be set to their known value (e.g., a 0 connection in an Au matrix) rather than using the computed value. First, the weights for the hemodynamic bases are computed for each region independently and assuming no distinction between regimes.
| (C.3) |
Other parameters can also be computed ignoring regime distinctions to equate parameters across regimes. Then the new estimate of the observation matrix C is formed piecemeal using the new βi as in Eq. (A.5). The observation noise covariance is computed again assuming no distinction between regimes:
| (C.4) |
The system dynamics matrices, the quasi-neural state noise, and the initial statistics of the system are all updated using weighted versions of the equations in Ghahramani and Hinton (1996).
| (C.5) |
Again, since the correct regime at each time is known the weights Wut reduce to {0,1} such that the sums for regime specific matrices only need computed for times when that regime is active. Note that for Au and Qu, only the first p dimensions of each matrix are used where p is the dimension of the un-lagged quasi-neural state space. These matrices are then placed within the larger matrices as in Appendix A.
Footnotes
Notation: We denote matrices with capital letters, vectors as lower case letters and scalars as lower case letters in italics. The subscript t on a variable denotes the value of the variable at some time while a superscript T indicates the vector or matrix transpose. The tilde denotes “distributed as” and N(g,G) indicates a Gaussian distribution with mean g and covariance G. E[x] denotes the expected value of x, and p(x|y) denotes a probability density of x conditional on y while P(x) denotes a specific probability.
Because of the discretization, the A matrix from a continuous time DCM is not equal to the discrete version Ad but they can be related for example by Ad = eAΔT where ΔT is the discrete sampling rate.
Consider the extreme case where the TR equals the experimental block length. Here the autoregressive portion of the model, A, would identify the between block relations while the within block relations (task effects) are “instantaneous” to the model and thus contained in Q.
References
- Abler B, Roebroeck A, Goebel R, Höse A, Schönfeldt-Lecuona C, Hole G, Walter H. Investigating directed influences between activated brain areas in a motor-response task using fMRI. Magnetic Resonance Imaging. 2006;24(2):181–185. doi: 10.1016/j.mri.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Aguirre GK, Zarahn E, D'Esposito M. The variability of human, BOLD hemodynamic responses. NeuroImage. 1998;8(4):360–369. doi: 10.1006/nimg.1998.0369. [DOI] [PubMed] [Google Scholar]
- Allison JD, Meador KJ, Loring DW, Figueroa RE, Wright JC. Functional MRI cerebral activation and deactivation during finger movement. Neurology. 2000;54:135–142. doi: 10.1212/wnl.54.1.135. [DOI] [PubMed] [Google Scholar]
- Arbib MA, Bishoff A, Fagg AH, Grafton ST. Synthetic PET: analyzing large-scale properties of neural networks. Human Brain Mapping. 1995;2:225–233. [Google Scholar]
- Barber D. Expectation correction for smoothed inference in switching linear dynamical systems. Journal of Machine Learning Research. 2006;7:2515–2540. [Google Scholar]
- Bar-Shalom Y, Li XR, Kirubarajan T. Estimation with Applications to Tracking and Navigation. John Wiley and Sons; Hoboken NJ: 2001. [Google Scholar]
- Bishop CM. Pattern Recognition and Machine Learning. Springer; New York, NY: 2006. [Google Scholar]
- Bitan T, Booth JR, Choy J, Burman DD, Gitelman DR, Mesulam MM. Shifts of effective connectivity within a language network during rhyming and spelling. Journal of Neuroscience. 2005;25(22):5397–5403. doi: 10.1523/JNEUROSCI.0864-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear M, Terry J, Friston K. Synchronization and complex dynamics in neuronal dynamics. Neurocomputing. 2003;52:151–158. [PubMed] [Google Scholar]
- Bressler SL, Richter CG, Chen Y, Ding M. Cortical functional network organization from autoregressive modeling of local field potential oscillations. Statistics in Medicine. 2007;26:3875–3885. doi: 10.1002/sim.2935. [DOI] [PubMed] [Google Scholar]
- Cavanaugh JE, Shumway RH. On computing the expected Fisher information matrix for state-space model parameters. Statistics and Probability Letters. 1996;26:347–355. [Google Scholar]
- Chadderdon GL, Sporns O. A large-scale neurocomputational model of task-oriented behavior selection and working memory in prefrontal cortex. Journal of Cognitive Neuroscience. 2006;18(2):242–257. doi: 10.1162/089892906775783624. [DOI] [PubMed] [Google Scholar]
- Cramer SC, Finklestein SP, Schaechter JD, Bush G, Rosen BR. Activation of distinct motor cortex regions during ipsilateral and contralateral finger movements. Journal of Neurophysiology. 1999;81:383–387. doi: 10.1152/jn.1999.81.1.383. [DOI] [PubMed] [Google Scholar]
- David O, Guillemain I, Saillet S, Reyt S, Deransart C, Segebarth C, Depaulis A. Identifying neural drivers with functional MRI: an electrophysiological validation. PLOS Biology. 2008;6(12):2683–2697. doi: 10.1371/journal.pbio.0060315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamantaras KI, Kung SY. Principal Component Neural Networks: Theory and Applications. John Wiley and Sons; New York, NY: 1996. [Google Scholar]
- Doucet A, Andrieu C. Iterative algorithms for state estimation of jump Markov linear systems. Institute of Electrical and Electronics Engineers Transactions on Signal Processing. 2001;49(6):1216–1227. [Google Scholar]
- Fairhall SL, Ishai A. Effective connectivity within the distributed cortical network for face perception. Cerebral Cortex. 2007;17(10):2400–2406. doi: 10.1093/cercor/bhl148. [DOI] [PubMed] [Google Scholar]
- Farrell BF, Ioannou PJ. Accurate low-dimensional approximation of the linear dynamics of fluid flow. Journal of the Atmospheric Sciences. 2001;58(18):2771–2789. [Google Scholar]
- Friston KJ, Harrison L, Penny WD. Dynamic causal modeling. Neuroimage. 2003;19(4):1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Gao Q, Chen H, Gong Q. Evaluation of the effective connectivity of the dominant primary motor cortex during bimanual movement using Granger causality. Neuroscience Letters. 2008;443(1):1–6. doi: 10.1016/j.neulet.2008.07.036. [DOI] [PubMed] [Google Scholar]
- Ghahramani Z, Hinton GE. Technical Report CRG-TR-96-2. Department of Computer Science, University of Toronto; 1996. Parameter estimation for linear dynamic systems. [Google Scholar]
- Ghahramani Z, Hinton GE. Variational learning for switching state space models. Neural Computation. 1998;12(4):963–996. doi: 10.1162/089976600300015619. [DOI] [PubMed] [Google Scholar]
- Gitelman DR, Penny WD, Ashburner J, Friston KJ. Modeling regional and psychophysiologic interactions in fMRI: the importance of hemodynamic deconvolution. NeuroImage. 2003;19:200–207. doi: 10.1016/s1053-8119(03)00058-2. [DOI] [PubMed] [Google Scholar]
- Goebel R, Roebroeck A, Kim DS, Formisano E. Investigating directed cortical interactions in time-resolved fMRI data using vector autoregressive modeling and Granger causality mapping. Magnetic Resonance Imaging. 2003;21:1251–1261. doi: 10.1016/j.mri.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Hamilton J. A new approach to the economic analysis of nonstationary time series and the business cycle. Econometrica. 1989;57(2):357–384. [Google Scholar]
- Handwerker DA, Ollinger JM, D'Esposito M. Variation of BOLD hemodynamic responses across subjects and brain regions and their effect of statistical analyses. NeuroImage. 2004;21(4):1639–1651. doi: 10.1016/j.neuroimage.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Harrison L, Penny WD, Friston KJ. Multivariate autoregressive modeling of fMRI time series. Neuroimage. 2003;19(4):1477–1491. doi: 10.1016/s1053-8119(03)00160-5. [DOI] [PubMed] [Google Scholar]
- Hasselmann K. PIPs and POPs: the reduction of complex dynamical systems using principal interaction and oscillation patterns. J Geophys Res. 1988;93:11015–11021. [Google Scholar]
- Haykin S. Adaptive Filter Theory. Upper Saddle River, NJ: Prentice-Hall; 2002. [Google Scholar]
- Henson RNA. Analysis of fMRI time series. In: Frackowiak RSJ, Friston KJ, Frith C, Dolan R, Friston KJ, Price CJ, Zeki S, Ashburner J, Penny WD, editors. Human Brain Function. Academic Press; 2003. [Google Scholar]
- Honey CJ, Kotter R, Breakspear M, Sporns O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proceedings of the National Academy of Science USA. 2007;104(24):10240–10245. doi: 10.1073/pnas.0701519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proceedings of the National Academy of Sciences USA. 2009;106(6):2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz B, Tagamets MA. Predicting human functional maps with neural net modeling. Human Brain Mapping. 1999;8:137–142. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<137::AID-HBM11>3.0.CO;2-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NE, Shen Z, Long SR, Wu MC, Shih HH, Zheng Q, et al. The empirical mode decomposition and the Hilbert spectrum for nonlinear and non-stationary time series analysis. Proceedings: Mathematical, Physical and Engineering Sciences. 1998:903–995. [Google Scholar]
- Husain FT, Tagamets MA, Fromm SJ, Braun AR, Horwitz B. Relating neuronal dynamics for auditory object processing to neuroimaging activity: a computational modeling and an fMRI study. NeuroImage. 2004;21:1701–1720. doi: 10.1016/j.neuroimage.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Izhikevich EM, Edelman GM. Large-scale model of mammalian thalamocor-tical systems. Proceedings of the National Academy of Sciences USA. 2008;105(9):3593–3598. doi: 10.1073/pnas.0712231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur N. Paradoxical functional facilitation in brain–behavior research. A critical review. Brain. 1996;46:184–189. doi: 10.1093/brain/119.5.1775. [DOI] [PubMed] [Google Scholar]
- Kiebel SJ, Klöppel S, Weiskopf N, Friston KJ. Dynamic causal modeling: a generative model of slice timing in fMRI. NeuroImage. 2007;34(4):1487–1496. doi: 10.1016/j.neuroimage.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Kim CJ. Dynamic linear models with Markov-switching. Journal of Econometrics. 1994;60:1–22. [Google Scholar]
- Kim CJ, Nelson CR. State-Space Models With Regime Switching. MIT Press; Cambridge, MA: 1999. [Google Scholar]
- Klein A, Mélard G, Zahaf T. Construction of the exact Fisher information matrix of Gaussian time series models by means of matrix differential rules. Linear Algebra and its Applications. 2000;321:209–232. [Google Scholar]
- Klein A, Neudecker H. A direct derivation of the exact Fisher information matrix of Gaussian vector state space models. Linear Algebra and its Applications. 2000;321:233–238. [Google Scholar]
- Kobayashi M, Hutchinson S, Schlaug G, Pascual-Leone A. Ipsilateral motor cortex activation on functional magnetic resonance imaging during unilateral hand movements is related to interhemispheric interactions. NeuroImage. 2000;20:2259–2270. doi: 10.1016/s1053-8119(03)00220-9. [DOI] [PubMed] [Google Scholar]
- Leff AP, Schofielf TM, Stephan KE, Crinion JT, Friston KJ, Price CJ. The cortical dynamics of intelligible speech. Journal of Neuroscience. 2008;28(49):13209–13215. doi: 10.1523/JNEUROSCI.2903-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XR, Bar-Shalom Y. Performance prediction of the interacting multiple model algorithm. IEEE Transactions on Aerospace and Electronic Systems. 1993;29(3):1015–1022. [Google Scholar]
- Liao S. Beyond Perturbation: Introduction to the Homotopy Analysis Method. 1st. Chapman & Hall/CRC; 2003. [Google Scholar]
- Logothetis A, Krishnamurthy V. Expectation maximization algorithms for MAP estimation of jump Markov linear systems. IEEE Transactions of Signal Processing. 1999;47(8):2139–2156. [Google Scholar]
- Lütkepohl H. New Introduction to Multiple Time Series Analysis. Springer-Verlag; Berlin: 2007. [Google Scholar]
- Martínez-Montes E, Valdés-Sosa PA, Miwakeichi F, Goldman RI, Cohen MS. Concurrent EEG/FMRI analysis by multiway partial least squares. NeuroImage. 2004;22:1023–1034. doi: 10.1016/j.neuroimage.2004.03.038. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Crinion JT, Long S, Friston KJ, Lambon Ralph MA, Patterson K, McClellanf JL, Price CJ. Dissociating reading processes on the basis of neuronal interactions. Journal of Cognitive Neuroscience. 2005;17(11):1753–1765. doi: 10.1162/089892905774589190. [DOI] [PubMed] [Google Scholar]
- Mesot B, Barber D. Switching linear dynamical systems for noise robust speech recognition. IEEE Transactions on Audio, Speech and Language Processing. 2007;15(6):1850–1858. [Google Scholar]
- Moore JB, Krishnamurthy V. De-interleaving pulse trains using discrete-time stochastic dynamic-linear models. IEEE Transactions on Signal Processing. 1994;42(11):3092–3103. [Google Scholar]
- Morgan RJ, Soltesz Nonramdom connectivity of the epileptic dentate gyrus predicts a major role for neuronal hubs in seizures. Proceedings of the National Academy of Sciences USA. 2008;105(16):6179–6184. doi: 10.1073/pnas.0801372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KP. Switching Kalman filters. Technical report, DEC/Compaq Cambridge Research Labs; 1998. [Google Scholar]
- Neumaier A, Schneider T. Estimation of parameters and eigenmodes of multivariate autoregressive models. ACM Transactions on Mathematical Software. 2001;27(1):27–57. [Google Scholar]
- Oh SM, Rehg JM, Balch T, Dellaert F. Data-driven MCMC for learning and inference in switching linear dynamic systems. Proceedings of the 22nd AAAI national conference on AI; Pittsburgh, PA. 2005. pp. 944–949. [Google Scholar]
- Oldfield RC. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9:97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- Penny WD, Ghahramani Z, Friston KJ. Bilinear dynamical systems. Philosophical Transactions of the Royal Society B. 2005;360(1457):983–993. doi: 10.1098/rstb.2005.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny WD, Stephan KE, Mechelli A, Friston KJ. Comparing dynamic causal models. NeuroImage. 2004;22(3):1157–1172. doi: 10.1016/j.neuroimage.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Rao SM, Binder JR, Bandettini PA, Hammeke TA, Yetkin FZ, Jesmanowicz A, Lisk LM, Morris GL, Mueller WM, Estkowski LD, Wong EC, Haughton VM, Hyde JS. Functional magnetic resonance imaging of complex human movements. Neurology. 1993;43:2311–2318. doi: 10.1212/wnl.43.11.2311. [DOI] [PubMed] [Google Scholar]
- Riera J, Bosch J, Yamashita O, Kawashima R, Sadato N, Okada T, Ozaki T. fMRI activation maps based on the NN-ARx model. NeuroImage. 2004;23(2):690–697. doi: 10.1016/j.neuroimage.2004.06.039. [DOI] [PubMed] [Google Scholar]
- Roebroeck A, Formisano E, Goebel R. Mapping directed influence over the brain using Granger causality and fMRI. NeuroImage. 2005;25:230–242. doi: 10.1016/j.neuroimage.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Roweis S, Ghahramani Z. A unifying review of linear Gaussian models. Neural Computation. 1999;11(2):305–345. doi: 10.1162/089976699300016674. [DOI] [PubMed] [Google Scholar]
- Seghier ML, Price CJ. Reading aloud boosts connectivity through the putamen. Cerebral Cortex Electronic publication ahead of print. 2009 doi: 10.1093/cercor/bhp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A. Causal networks in simulated neural systems. Cognitive Neurodynamics. 2008;2(1):49–64. doi: 10.1007/s11571-007-9031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumway RH, Stoffer DS. Dynamic linear models with switching. Journal of the American Statistical Association. 1991;86(414):763–769. doi: 10.1080/01621459.1991.10475086. [DOI] [PubMed] [Google Scholar]
- Simon D. Optimal state estimation: Kalman, H infinity, and nonlinear approaches. John Wiley and Sons; Hoboken NJ: 2006. [Google Scholar]
- Singh LN, Higano S, Takahashi S, Abe Y, Sakamoto M, Kurihara N, Furuta S, Tamura H, Yanagawa I, Fujii T, Ishibashi T, Maruoka S, Yamada S. Functional MR imaging of cortical activation of the cerebral hemispheres during motor tasks. American Journal of Neuroradiology. 1998;19:275–280. [PMC free article] [PubMed] [Google Scholar]
- Smith JF, Chen K, Johnson S, Morrone-Strupinsky J, Johnson SC, Reiman EM, Nelson A, Moeller JR, Alexander GE. Network analysis of single-subject fMRI during a finger opposition task. NeuroImage. 2006;32(1):325–332. doi: 10.1016/j.neuroimage.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Edelman GM. Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cerebral Cortex. 2000;10:127–141. doi: 10.1093/cercor/10.2.127. [DOI] [PubMed] [Google Scholar]
- Strang G. Linear Algebra and Its Applications. 3rd. Brooks Cole; 1988. [Google Scholar]
- Stephan KE, Harrison L, Keibel SJ, David O, Penny WD, Friston KJ. Dynamic causal models of neural system dynamics: current state and future extensions. Journal of Biosciences. 2007;32(1):129–144. doi: 10.1007/s12038-007-0012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Kasper L, Harrison L, Daunizeau J, den Ouden H, Breakspear M, Friston KJ. Nonlinear dynamic causal models for fMRI. Neuroimage. 2008;42(1):1–9. doi: 10.1016/j.neuroimage.2008.04.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Penny WD, Daunizeau J, Moran R, Friston KJ. Bayesian model selection for group studies. Neuroimage. 2009;46(3):1004–1014. doi: 10.1016/j.neuroimage.2009.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffer DS, Wall KD. Bootstrapping state-space models: Gaussian maximum likelihood estimation and the Kalman filter. Journal of the American Statistical Association. 1991;86(416):1024–1033. [Google Scholar]
- Tagamets MA, Horwitz B. Integrating electrophysiological and anatomical experimental data to create a large-scale model that simulates a delayed match-to-sample human brain imaging study. Cerebral Cortex. 1998;8:310–320. doi: 10.1093/cercor/8.4.310. [DOI] [PubMed] [Google Scholar]
- Ullman MT. Contributions of neural memory circuits to language: the declarative/procedural model. Cognition. 2004;92(1-2):231–270. doi: 10.1016/j.cognition.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Valdés-Sosa PA. Spatio-temporal autoregressive models defined over brain manifolds. Neuroinformatics. 2004;2(2):239–250. doi: 10.1385/NI:2:2:239. [DOI] [PubMed] [Google Scholar]
- Zhuang J, LaConte S, Peltier S, Zhang K, Hu X. Connectivity exploration with structural equation modeling: an fMRI study of bimanual coordination. NeuroImage. 2005;25:462–470. doi: 10.1016/j.neuroimage.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Zoeter O, Heskes T. Hierarchical visualization of time-series data using switching linear dynamical systems. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2003;25(10) [Google Scholar]