Abstract
Pinellia pedatisecta agglutinin (PPA) is a specific mannose-binding plant lectin accumulated in the tuber of P. pedatisecta. In the work presented, the cytotoxicity of PPA to cancer cells was investigated through exogenous expression. A PPA gene was transduced into normal and cancer cell lines through plasmid vectors, and the effect of PPA expression was examined. Results showed that PPA translocated into the nucleus, colocalized with DNA and induced cell death. A mannose-binding motif and a V103-W130 region directed the nuclear translocation of PPA. Coprecipitation, mass spectrometry and western blotting analysis further indentified that PPA was associated with the methylosome, which contains methylosome protein 50 and protein arginine methyltransferase 5 (PRMT5). Knockdown of PRMT5 significantly inhibited the PPA-induced cell death, suggesting that PPA used the methylosome as a target. Furthermore, Ad.surp-PPA, an adenovirus vector in which the PPA gene was controlled by a survivin promoter (surp), selectively inhibited the proliferation of cancer cell lines. Taken together, the expression of PPA gene elicited significant cytotoxicity to cancer cells through targeting the methylosome and might be developed into a novel agent in cancer gene therapy.
Keywords: methylosome, Pinellia pedatisecta agglutinin, MEP50, PRMT5, nuclear translocation
Introduction
Lectins are carbohydrate-binding proteins containing at least one non-catalytic domain that binds reversibly with mono- or oligosaccharide with high specificity and affinity.1, 2 An increasing number of three-dimensional structures of lectins has been revealed, which has led to better understanding of the interaction between sugars and carbohydrate recognition domains in lectins.3, 4 Carbohydrates are a highly diverse group because of their variability in saccharide monomers and complicating structures. Interestingly, the diversity of carbohydrate decoration in mammalian cell membranes and viral particles can be recognized by specific plant lectins, which are originally involved in plant defense and root symbiosis.3, 4, 5 At present, plant lectins have been used in studies of mammalian cells and viruses, such as peanut agglutinin in identifying hematopoietic cell sub-populations,6, 7 soybean agglutinin in preparation of proper cell fraction for bone marrow transplantation,8 Galanthus nivalis agglutinin, hippeastrum hybrid agglutinin, cymbidium agglutinin and Urtica dioica agglutinin in targeting HIV,9, 10 and a variety of plant lectins in providing markers and therapeutic agents for cancer cells.11
Monocot mannose-binding lectins (MBLs) constitute a superfamily of mannose-specific lectins, which have mainly been isolated and cloned from families of Alliaceae, Amarylliadaceae, Orchidaceae, Liliaceae, Iridaceae and Araceae.12, 13 Pinellia pedatisecta agglutinin (PPA) is a mannose-binding lectin accumulated in the tuber of P. pedatisecta, an Araceae species. Based on the genomic and complementary DNA sequences of PPA reported previously,14, 15 a recombinant PPA has been used in labeling fractions of myeloid leukemia cells in our laboratory.16
In this work, a PPA gene was transfected into a variety of human cells including normal and cancer cell lines through plasmid vectors. The exogenous PPA expression and the underlying mechanism of PPA-induced cell death were investigated. To further evaluate the antiproliferative effect of the PPA gene, a replication-defective adenovirus harboring the PPA gene under the control of a survivin promoter (surp), Ad.surp-PPA, was constructed. The selective cytotoxicity of this adenovirus to cancer cells was analyzed as well.
Results
Exogenous expression of PPA in normal and cancer cells
The amino-acid sequence of PPA was shown in Figure 1a. To determine the exogenous expression of PPA in human normal and cancer cells, pcDNA3.1/His-PPA or pcDNA3.1/His plasmids were transfected into a panel of cells and the expression of PPA was examined by western blotting analysis with an anti-6his antibody. Results showed that 6his-PPA with a molecular weight as expected was detected in normal lung cell line WI38, lung cancer cell lines A549, H460 and H1299 (Figure 1b), as well as hepatocellular carcinoma cell lines PLC and Hep3B (Figure 1c). To further determine the exogenous expression of PPA, lung cancer cell line H1299 was transfected with pcDNA3/FLAG-PPA or pcDNA3/FLAG plasmids followed by western blotting analysis with an anti-FLAG antibody. As compared with pcDNA3/FLAG-transfected cells, FLAG-PPA was detected in pcDNA3/FLAG-PPA-transfected cells (Figure 1d). Our results indicate that PPA gene can be expressed in a variety of human cells including both normal and malignant cell lines.
Figure 1.
The amino acid sequence and exogenous expression of PPA in human cells. (a) The amino acid sequence of PPA. Underlined amino acid sequences show three MBMs. (b) The exogenous expression of his-PPA in a normal lung cell line WI38 and lung cancer cell lines A549, H460 and H1299. Cells were transfected with pcDNA3.1/His or pcDNA3.1/His-PPA. Cell lysates were analyzed by western blot with antibodies against 6His or Actin. (c) The exogenous expression of his-PPA in liver cancer cell lines PLC or Hep3B. Cells were transfected with pcDNA3.1/His or pcDNA3.1/His-PPA. Cell lysates were analyzed by western blot with antibodies against 6His or Actin. (d) The exogenous expression of FLAG-PPA in H1299 cells. Cells were transfected with pcDNA3/FLAG or pcDNA3/FLAG-PPA. Cell lysates were analyzed by western blot with antibodies against FLAG or Actin.
PPA translocated into the nucleus and induced cell death
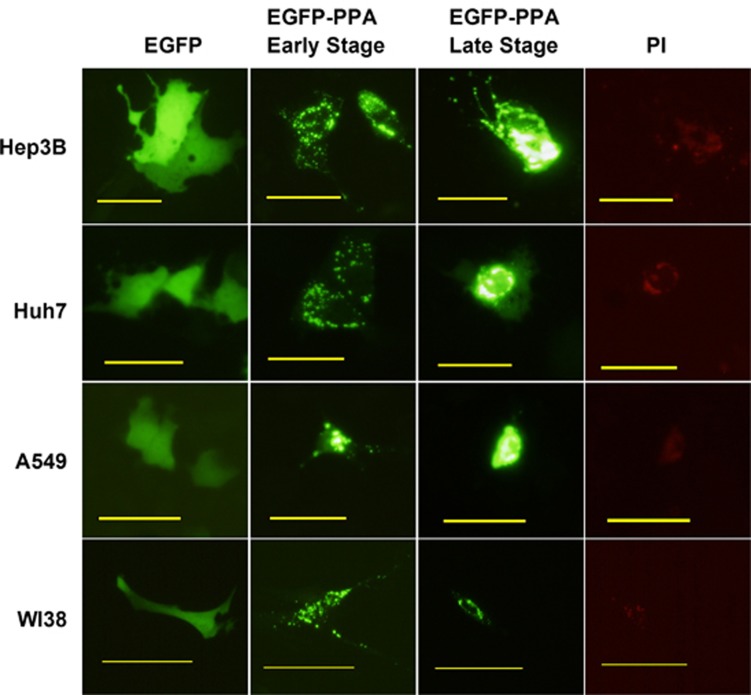
We then examined the effect of PPA expression in both normal and cancer cells. Plasmids pEGFP-C1 or pEGFP-PPA-C1 were transfected into hepatocellular carcinoma cell lines Hep3B and Huh7, lung cancer cell line A549 and normal lung cell line WI38 followed by fluorescent microscope observation. As compared with pEGFP-C1 transfected cells, most of cells transfected with pEGFP-PPA-C1 exhibited a number of green fluorescence dots at the early stage (before 24 h). At the late stage (after 48 h), dots in majority of cells were finally accumulated in a central area, which can be stained by propidium iodide, a DNA staining agent for damaged cells. A condensation of DNA was also observed (Figure 2). Results suggested that PPA entered the nucleus and colocalized with DNA, which might subsequently alter the chromatin structure and induce cell death.
Figure 2.
PPA translocated into the nucleus and induced cell death. Cells were transfected with pEGFP-C1 or pEGFP-PPA-C1 followed by fluorescent microscope observation after 24 h (early stage) or 48 h (late stage). Cells transfected with pEGFP-PPA-C1 were further stained with propidium iodide after 48 h. Bars show 100 μm.
A mannose-binding motif and a V103-W130 region directed the nuclear translocation of PPA
The entrance of PPA into the nucleus suggested that there might exist a nuclear localization signal in PPA. As PPA is a specific mannose-binding lectin, we proposed that the mannose-binding motifs (MBMs) in PPA might have a role in nuclear translocation. To test this hypothesis, a sequence of MBM in PPA (QDNGFGVVY) indicated in Figure 1a was cloned into pEGFP-C1 plasmids to form pEGFP-MBM-C1. Plasmids pEGFP-MBM-C1 and pEGFP-C1 were then transfected into hepatocellular carcinoma cell lines PLC and Huh7, as well as lung-cancer cell line H1299. As observed under a fluorescence microscope, MBM obviously directed enhanced green fluorescent protein (EGFP) to translocate into the nucleus (Figure 3a). The nuclear localization of EGFP was further verified by a Hoechst33342 staining in H1299 cells (Figure 3b). The intact morphology of the nucleus stained by Hoechst33342 shown in Figure 3b also suggested that MBM alone did not induce obvious cell damage. Our results indicated that the MBM was responsible for directing PPA to translocate into the nucleus.
Figure 3.
Regions in directing the nuclear translocation of PPA. (a) Cells were transfected with pEGFP-MBM-C1 and pEGFP-C1 followed by fluorescent microscope observation after 48 h. (b) H1299 cells transfected with pEGFP-MBM-C1 were further stained with Hoechst33342 after 48 h followed by fluorescent microscope observation. (c) H1299 cells were transfected with pEGFP-C1 or pEGFP-(V103-W130)-C1 followed by fluorescent microscope observation after 48 h. Bars show 100 μm.
To further investigate the localization pattern of PPA, a sequence encoding the V103-W130 region without a predicted MBM was cloned into pEGFP-C1 to generate pEGFP-( V103-W130)-C1. As compared with H1299 cells transfected with pEGFP-C1, nuclear localization of EGFP was observed in cells transfected with pEGFP-( V103-W130)-C1 (Figure 3c). Therefore, our results suggest that the localization pattern of PPA is complicated and the nuclear translocation of PPA may be achieved by a cooperation of several regions including the mannose-binding region and V103-W130 region.
The PPA expression did not alter the caspase signaling
To analyze the mechanism of PPA-induced cell death, apoptotic signal elements including caspase3, caspase8 and poly (ADP-ribose) polymerase (PARP) in H1299 and H460 cells, transfected with pcDNA3.1/His-PPA or pcDNA3.1/His-EGFP were examined. As shown in Figure 4, PPA did not significantly induced activation of caspase3 and caspase8, as compared with EGFP expression. Interestingly, an elevated total level of PARP was observed in cells with PPA expression. The functions of PARP previously have been linked to DNA repair and caspase-independent cell death.31, 32 The elevated level of PARP in response to PPA expression may be induced by the coagulation of DNA as observed in Figure 2.
Figure 4.
PPA-induced PARP level. H1299 and H460 cells were transfected with pcDNA3.1/His-PPA or pcDNA3.1/His-EGFP. After 48 h, cells lysates were analyzed by western blot with antibodies against caspase3, caspase8, PARP and Actin.
PPA-induced cell death through targeting the methylosome
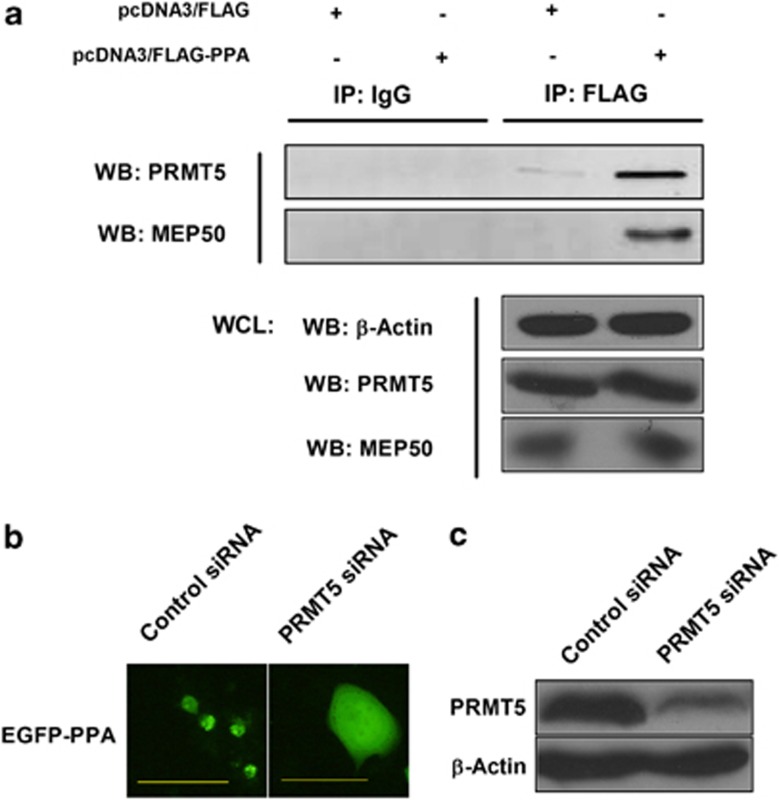
We then further investigated the underlying mechanism of the PPA-induced cell death through examining intracellular proteins interacting with PPA. H1299 cells were transfected with pcDNA3/FLAG-PPA or pcDNA3/FLAG plasmids followed by immunoprecipitation with a FLAG antibody conjugated gel. A specific band in SDS–polyacrylamide gel electrophoresis (SDS–PAGE) was subjected to mass spectrometry analysis, and the protein was identified as methylosome protein 50 (MEP50) (Table 1), a component of the methylosome.24 To verify the association of PPA with the methylosome, anti-FLAG precipitated complexes were subjected to western blotting analysis. As shown in Figure 5a, both MEP50 and protein arginine methyltransferase 5 (PRMT5) were found to be associated with PPA, indicating the interaction of PPA with the methylosome.
Table 1. MEP50 specific peptides identified by mass spectrometry.
| Peptides | Calulated mass | Observed mass | ±p.p.m. | Start | End | Sequence |
|---|---|---|---|---|---|---|
| 1 | 916.525 | 916.5231 | −2 | 192 | 198 | ILLWDTR |
| 2 | 1115.5704 | 1115.5734 | 3 | 293 | 301 | SQAHRDFVR |
| 3 | 1244.6997 | 1244.7007 | 1 | 4 | 15 | ETPPPLVPPAAR |
| 4 | 1372.7947 | 1372.7911 | −3 | 3 | 15 | KETPPPLVPPAAR |
| 5 | 1627.7356 | 1627.7494 | 8 | 16 | 29 | EWNLPPNAPACMER |
| 6 | 1722.9133 | 1722.9197 | 4 | 36 | 52 | YRSDGALLLGASSLSGR |
Abbreviation: MEP50, methylosome protein 50.
A 48 kDa protein was subjected to trypsin digestion and mass spectrometry. Six peptides were identified and matched to MEP50.
Figure 5.
PPA-induced cell death through targeting the methylosome. (a) H1299 cells were transfected with pcDNA3/FLAG-PPA or pcDNA3/FLAG. Cell lysates were immunoprecipitated with a FLAG antibody conjugated gel. Precipitation with a normal mouse IgG plus a protein G conjugated agarose served as a control. The precipitated complexes were examined by western blotting analysis with antibodies against MEP50 and PRMT5. Actin, MEP50 and PRMT5 in whole-cell lysates was monitored for expression levels. (b) H1299 cells were transfected with a control siRNA or PRMT5 siRNA followed by a second transfection with pEGFP-PPA-C1 after 48 h. Bars show 100 μm. (c) H1299 cells were transfected with a control siRNA or PRMT5 siRNA. After 48 h, the whole-cell lysates were examined by western blot with antibodies against PRMT5 and actin.
To further elucidate the role of the methylosome in the PPA-induced cell death, H1299 cells were transfected with a control small interfering RNA (siRNA) or PRMT5 siRNA followed by a second transfection with pEGFP-PPA-C1. Results showed that the knockdown of PRMT5 expression significantly inhibited the accumulation and nuclear translocation of PPA in some cells, which was totally not observed in cells transfected with the control siRNA (Figure 5b). The knockdown efficiency of PRMT5 was shown in Figure 5c. Taken together, our data indicated that PPA-induced cell death through using the methylosome as a target.
Controlled expression of PPA elicited selective toxicity to cancer cells
To evaluate the antiproliferative effect of PPA gene against cancer cells, Ad.surp-PPA, a replication-defective adenovirus harboring the PPA gene under the control of a survivin promoter, was constructed. Previous studies have demonstrated that survivin is an apoptosis inhibitor highly expressed in most cancers and low expressed in terminally differentiated cells,29 and the survivin promoter has been used as a cancer-specific promoter to control gene expression in generating oncolytic viruses.30 Ad.surp-PPA was infected into normal lung cell line WI38, lung-cancer cell line H1299 and hepatocellular carcinoma cell lines Huh7 and PLC at multiplicity of infections indicated. Figure 6 showed that Ad.surp-PPA elicited a minimal toxicity to WI38. However, cell proliferation was significantly inhibited in H1299, Huh7 and PLC cells by Ad.surp-PPA at a dosage-dependent manner, and the maximum inhibition was achieved in PLC cells. Our data indicated that the cancer-specifically controlled expression of PPA elicited selective toxicity to cancer cells.
Figure 6.
Ad.surp-PPA elicited a selective toxicity to cancer cells. Normal lung cell line WI38, lung-cancer cell line H1299 and liver cancer cell lines PLC and Huh7 were treated with Ad or Ad.surp-PPA at multiplicity of infections indicated. After 48 h, cell viability was analyzed by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Values are shown as mean±s.d.
Discussion
PPA is a mannose-binding lectin accumulated in the tuber of P. pedatisecta. In the work presented, the PPA gene was exogenously expressed in a variety of human normal and cancer cell lines. Results showed that PPA translocated into the nucleus, colocalized with DNA, and induced cell death through targeting the MEP50/PRMT5 methylosome. A MBM in PPA and the V103-W130 region directed the nuclear translocation of PPA. Furthermore, Ad.surp-PPA, a replication-defective adenovirus carrying a survivin promoter controlled PPA gene elicited a selective cytotoxicity to cancer cells, suggesting an antiproliferative effect of the PPA gene against cancer cells.
PRMT5 is a type II protein arginine methyltransferase, which catalyzes monomethylation and symmetric dimethylation of arginine residues.17 PRMT5 methylates histones H2A, H3 and H4,18, 19, 20 as well as non-histone proteins such as Sm proteins, which are essential for the formation of spliceosomal small nuclear ribonucleoproteins.21 The diverse functions of PRMT5 are achieved through forming multiple complexes with a variety of proteins. For example, PRMT5 associated with SWI/SNF chromatin remodeling complexes methylates histone H3 arginine 8 and suppresses the expression of tumor repressor genes ST7 and NM23.19 A complex containing PRMT5 and Blimp1 methylates histone H2A and H4R3, and regulates germ cell development.22 Kruppel-like zinc finger protein ZNF224 forms a transcriptional repressor complex with PRMT5 and suppresses aldolase A gene expression.23 The methylosome containing PRMT5 and MEP50 has been shown in methylating Sm proteins and histones.24, 25 Recently, MEP50/PRMT5 complex was shown in mediating the nuclear cyclin D1/CDK4 kinase triggered neoplastic growth, suggesting an oncogenic role of PRMT5.26 In our results, PPA was determined to be associated with both the MEP50/PRMT5 methylosome and DNA, as well as resulted in DNA coagulation, suggesting a role of PPA in connecting the methylosome to chromatin structure alteration. Therefore, investigations into chromatin modifications may help to further elucidate the PPA-induced cell death.
We presented in this report that a MBM and the V103-W130 region directed the nuclear translocation of PPA. However, both the MBM and V103-W130 region were not sufficient to coagulate DNA and induce cell death. Our data imply that there might exist regions in PPA other than the MBM and V103-W130 region in interacting with DNA or chromatin structure. Therefore, dissecting the functional domains in PPA may provide further evidences for the PPA-induced chromatin structure alteration and cell death.
Cancer gene therapy represents a strategy that genetic materials are delivered to specific cells or tissues to cause cancer cell death, stimulate immune cells or restore normal cellular phenotypes through viral, non-viral or cell vectors.27, 28 In the work presented, a replication-defective adenovirus Ad.surp-PPA was constructed to deliver a survivin promoter controlled PPA gene into normal cells and cancer cells. As determined by others previously, survivin is an inhibitor of apoptosis overexpressed in a variety of cancer cells,29 and the survivin promoter has been used to target various cancer cells.30 As shown in our data, a selective cytotoxicity of Ad.surp-PPA to cancer cells was observed, suggesting that delivering PPA gene under cancer-specific control to malignant cells may be a useful strategy in cancer gene therapy. Thus, the PPA gene could be a novel anticancer gene in future cancer therapies.
In conclusion, our data indicate that exogenously expressed PPA in cancer cells translocated to the nucleus, colocalized with DNA, and induced cell death through acting on the methylosome as a target. A MBM and the V103-W130 region directed the translocation of PPA. Using adenoviral vectors harboring PPA gene under cancer-specific control could be a novel method in cancer gene therapy. However, the underlying mechanism of the PPA-induced cell death is still not completely revealed. Further investigations may help to further elucidate the PPA-induced cell death and provide insights into utilizing the PPA gene in cancer gene therapy.
Materials and methods
Plasmids
The plasmid pMD-18T-PPA containing full-length PPA genomic DNA sequence was constructed as described previously.16 A sequence encoding 234 amino-acids of PPA flanked by EcoRV and XhoI sites was amplified by PCR from pMD-18T-PPA. The sense primer was 5′-TATGATATCGTGGGAACCAACCACC TGCTGT-3′ and the antisense primer was 5′-TATCTCGAGCTACGCGGCAATTGG GCGCTTC-3′. The PCR product was inserted into the corresponding site of pcDNA3.1/His to generate the pcDNA3.1/His-PPA plasmid. The PPA sequence flanked by XhoI sites at both the ends was amplified from pMD-18T-PPA. The sense primer was 5′-TATCTCGAGTGGTGGGCACCAACTAC-3′ and the antisense primer was 5′-TATCTCGAGCTACGCGGCAATTGGGCGCTTC-3′. The PCR product was inserted into the corresponding site of pEGFP-C1 to generate pEGFP-PPA-C1 plasmid. The PPA sequence flanked by EcoRI and XhoI sites was cut from pEGFP-PPA-C1 and inserted into pcDNA3/FLAG to generate pcDNA3/FLAG-PPA plasmid. A pair of complementary oligonucleotides encoding a PPA MBM with an amino acid sequence LQDNGFGVVYG were synthesized and annealed to form a DNA duplex. The annealed oligonucleotides contained EcoRI and XhoI sites at both the ends. The sequences of the oligonucleotides were 5′-TCGAGACCTCCAGGACAACGGCTTCGGCGTCGTCTACGGCTAAG-3′ and 5′-AATTCTTAGCCGTAGACGACGCCGAAGCCGTTGTCCTGGAGGTC-3′. The annealed oligonucleotides were ligated into the EcoRI and XhoI sites of pEGFP-C1 to generate pEGFP-MBM-C1 plasmid. A sequence encoding the V103-W130 region flanked by EcoR I and XhoI was amplified by PCR and inserted into the corresponding sites of pEGFP-C1 to generate pEGFP-( V103-W130)-C1.
Adenoviral construction
Human CMV promoter was deleted from the pCA13 plasmid to generate pCA13-▵HCMV. A survivin promoter (surp) sequence flanked by SalI and EcoRI sites was inserted into pCA13-▵HCMV to form pCA13-surp plasmid. A sequence encoding 6xHis and 234 amino acids of PPA flanked by SalI and EcoRI sites was amplified by PCR from pMD-18T-PPA and inserted into pCA13-surp to generate pCA13-surp-PPA. Plasmids pCA13-surp-PPA and pBHGE3 were cotransfected into HEK293 cells, and adenovirus Ad.surp-PPA was subsequently produced through homologous recombination. Viral titration was determined through serial dilution and infection in HEK293 cells.
Cell culture and transfection
All cell lines were obtained from American Type Culture Collection (Rockville, MD, USA). Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin solution, and 1% ℒ-Glutamine. Appropriate amounts of plasmids were transfected into cells by Effectene Transfection Reagent (Qiagen Inc., Valenda, CA, USA) following the manufacturer's instruction.
Western blotting analysis
The cell extract or precipitated complexes were subjected to SDS–PAGE and electroblotted onto the nitrocellulose membrane. The membrane was then blocked with Tris-buffered saline and Tween 20 contaning 5% of bovine serum albumin at room temperature for 2 h and incubated with mouse anti-6 histidine (6 his) antibody, anti-caspase3 antibody, anti-caspase 8 antibody, rabbit anti-PARP antibody, goat anti-PRMT5 antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), mouse anti-caspase9, rabbit anti-MEP50 (Cell Signaling Technology, Inc., Danvers, MA, USA), mouse anti-FLAG antibody and rabbit anti-actin antibody (Beyotime Institute of Biotechnology, Shanghai, China) overnight at 4 °C. The membrane was washed and incubated with appropriate dilution of IRDye 800 donkey anti-mouse IgG, IRDye 800 donkey anti-goat IgG or IRDye 700 donkey anti-rabbit IgG (LI-COR, Inc., Lincoln, NA, USA) for 1 h at room temperature. After washing with Tris-buffered saline, the membrane was then analyzed by an Odyssey Infrared Imaging System (LI-COR, Inc.).
Immunoprecipitation and mass spectrometry
Cell lysates were prepared in a lysis buffer (Beyotime Institute of Biotechnology). The lysates were then centrifuged at 12 000 r.p.m. for 15 min. The supernatants were subjected to immunoprecipitation with an anti-6his antibody followed by protein A/G conjugated agarose when cells were transfected with pcDNA/His-PPA or pcDNA/His-EGFP plasmid. Precipitated immunocomplexes were washed three times in PBS and boiled in a loading buffer followed by SDS–PAGE and western blotting analysis. When cells were transfected with pcDNA3/FLAG-PPA or pcDNA3/FLAG plasmid, cell extracts were immunoprecipitated with an anti-FLAG M2 affinity gel (Sigma-Aldrich, St Louis, MO, USA). Precipitated complexes were washed three times with PBS and boiled in a loading buffer followed by SDS–PAGE and western blotting analysis. For mass spectrometry, precipitated complexes were subjected to SDS–PAGE followed by silver staining or coomassie brilliant blue staining. Specific bands were excised and digested with trypsin. Peptide mixtures were analyzed by an ABI 4700 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Framingham, MA, USA). A combined database search was performed using a Mascot software (Version 2.0; Matrix Science, London, UK).
RNA interference
Nonspecific RNA interference oligoribonucleotides and RNA interference oligoribonucleotides corresponding to PRMT5 were purchased from Santa Cruz Biotechnology. Cells were cultured in 24-well-plates at 1 × 105/well and transfected with 20 nmol of RNA interference for each well through a Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). After 2 days, cells were further transfected with plasmids pEGFP-PPA-C1 followed by observation under a fluorescent microscope (Olympus Corporation, Tokyo, Japan).
Cell viability assay
Cells were plated on 96-well plates at 1 × 104 per well one day before virus infection. Cells were then infected with viruses at indicated multiplicity of infections for 48 h. The cell viability was determined by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay.
Acknowledgments
This work was supported by National Natural Science Foundation of China Grants 30801379 and 30800093.
The authors declare no conflict of interest.
References
- Sharon N, Lis H. Lectins as cell recognition molecules. Science. 1989;246:227–234. doi: 10.1126/science.2552581. [DOI] [PubMed] [Google Scholar]
- Peumans WJ, van Damme EJM. Lectins as plant defense proteins. Plant Physiol. 1995;109:347–353. doi: 10.1104/pp.109.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N, Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiol. 2004;14:53R–62R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- Sharon N. Lectins: carbohydrate-specific reagents and biological recognition molecules. J Biol Chem. 2007;282:2753–2764. doi: 10.1074/jbc.X600004200. [DOI] [PubMed] [Google Scholar]
- De Hoff PL, Brill LM, Hirsch AM. Plant lectins: the ties that bind in root symbiosis and plant defense. Mol Genet Genomics. 2009;282:1–15. doi: 10.1007/s00438-009-0460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner Y, Linker-Israeli M, Sharon N. Separation of mouse thymocytes into two subpopulations by the use of peanut agglutinin. Cell Immunol. 1976;25:129–134. doi: 10.1016/0008-8749(76)90103-9. [DOI] [PubMed] [Google Scholar]
- London J, Berrih S, Bach JF. Peanut agglutinin. I. A new tool for studying T lymphocyte subpopulations. J Immunol. 1979;121:438–443. [PubMed] [Google Scholar]
- Reisner Y, Kapoor N, Kirkpatrick D, Pollack MS, Cunningham-Rundles S, Dupont B, et al. Transplantation for severe combined immunodeficiency with HLA-A,B,D,DR incompatible parental marrow cells fractionated by soybean agglutinin and sheep red blood cells. Blood. 1983;61:341–348. [PubMed] [Google Scholar]
- Balzarini J, Van Laethem K, Hatse S, Froeyen M, Peumans W, Van Damme E, et al. Carbohydrate-binding agents cause deletions of highly conserved glycosylation sites in HIV GP120: a new therapeutic concept to hit the achilles heel of HIV. J Biol Chem. 2005;280:41005–41014. doi: 10.1074/jbc.M508801200. [DOI] [PubMed] [Google Scholar]
- Balzarini J, Van Herrewege Y, Vermeire K, Vanham G, Schols D. Carbohydrate-binding agents efficiently prevent dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN)-directed HIV-1 transmission to T lymphocytes. Mol Pharmacol. 2007;71:3–11. doi: 10.1124/mol.106.030155. [DOI] [PubMed] [Google Scholar]
- De Mejía EG, Prisecaru VI. Lectins as bioactive plant proteins: a potential in cancer treatment. Crit Rev Food Sci Nutr. 2005;45:425–445. doi: 10.1080/10408390591034445. [DOI] [PubMed] [Google Scholar]
- Barre A, Van Damme EJ, Peumans WJ, Rougé P. Structure-function relationship of monocot mannose-binding lectins. Plant Physiol. 1996;112:1531–1540. doi: 10.1104/pp.112.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJ, Astoul CH, Barre A, Rougé P, Peumans WJ. Cloning and characterization of a monocot mannose-binding lectin from Crocus vernus (family Iridaceae) Eur J Biochem. 2000;267:5067–5077. doi: 10.1046/j.1432-1327.2000.01563.x. [DOI] [PubMed] [Google Scholar]
- Lin J, Zhou X, Fei J, Liao Z, Jin W, Sun X, et al. Genomic cloning and characterization of a PPA gene encoding a mannose-binding lectin from Pinellia pedatisecta. Biocell. 2006;30:15–25. [PubMed] [Google Scholar]
- Lin J, Zhou X, Gao S, Liu X, Wu W, Sun X, et al. cDNA cloning and expression analysis of a mannose-binding lectin from Pinellia pedatisecta. J Biosci. 2007;32:241–249. doi: 10.1007/s12038-007-0024-1. [DOI] [PubMed] [Google Scholar]
- Li GC, Li N, Zhang YH, Li X, Wang YG, Liu XY, et al. Mannose-exposing myeloid leukemia cells detected by the sCAR-PPA fusion protein. Int J Hematol. 2009;89:611–617. doi: 10.1007/s12185-009-0308-3. [DOI] [PubMed] [Google Scholar]
- Rho J, Choi S, Seong YR, Cho WK, Kim SH, Im DS. Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J Biol Chem. 2001;276:11393–11401. doi: 10.1074/jbc.M008660200. [DOI] [PubMed] [Google Scholar]
- Tee W-W, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Gene & Dev. 2010;24:2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the Rb family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol. 2008;28:6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Cesaro E, De Cegli R, Medugno L, Florio F, Grosso M, Lupo A, et al. The Kruppel-like zinc finger protein ZNF224 recruits the arginine methyltransferase PRMT5 on the transcriptional repressor complex of the aldolase A gene. J Biol Chem. 2009;284:32321–32330. doi: 10.1074/jbc.M109.043349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Wyce A, Paushkin S, Abel L, Rappsilber J, Mann M, et al. A novel WD repeat protein component of the methylosome binds Sm proteins. J Biol Chem. 2002;277:8243–8247. doi: 10.1074/jbc.M109984200. [DOI] [PubMed] [Google Scholar]
- Furuno K, Masatsugu T, Sonoda M, Sasazuki T, Yamamoto K. Association of polycomb group SUZ12 with WD-repeat protein MEP50 that binds to histone H2A selectively in vitro. Biochem Biophys Res Commun. 2006;345:1051–1058. doi: 10.1016/j.bbrc.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Aggarwal P, Vaites LP, Kim JK, Mellert H, Gurung B, Nakagawa H, et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18:329–340. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan RC. The basic science of gene therapy. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- Cross D, Burmester JK. Gene therapy for cancer treatment: past, present and future. Clin Med Res. 2006;4:218–227. doi: 10.3121/cmr.4.3.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ME, Howerth EW. Survivin: a bifunctional inhibitor of apoptosis protein. Vet Pathol. 2004;41:599–607. doi: 10.1354/vp.41-6-599. [DOI] [PubMed] [Google Scholar]
- Zhang KJ, Wang YG, Cao X, Zhong SY, Wei RC, Wu YM, et al. Potent antitumor effect of interleukin-24 gene in the survivin promoter and retinoblastoma double-regulated oncolytic adenovirus. Hum Gene Ther. 2009;20:818–830. doi: 10.1089/hum.2008.205. [DOI] [PubMed] [Google Scholar]
- Süsse S, Scholz C-J, Bürkle A, Wiesmüller L. Poly(ADP-ribose) polymerase (PARP-1) and p53 independently function in regulating double-strand break repair in primate cells. Nucl Acids Res. 2004;32:669–680. doi: 10.1093/nar/gkh227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai JB, Russell HM, Raman J, Jeevanandam V, Gupta MP. Increased expression of poly(ADP-ribose) polymerase-1 contributes to caspase-independent myocyte cell death during heart failure. Am J Physiol Heart Circ Physiol. 2005;288:H486–H496. doi: 10.1152/ajpheart.00437.2004. [DOI] [PubMed] [Google Scholar]