Abstract
The ga2 mutant of Arabidopsis thaliana is a gibberellin-deficient dwarf. Previous biochemical studies have suggested that the ga2 mutant is impaired in the conversion of copalyl diphosphate to ent-kaurene, which is catalyzed by ent-kaurene synthase (KS). Overexpression of the previously isolated KS cDNA from pumpkin (Cucurbita maxima) (CmKS) in the ga2 mutant was able to complement the mutant phenotype. A genomic clone coding for KS, AtKS, was isolated from A. thaliana using CmKS cDNA as a heterologous probe. The corresponding A. thaliana cDNA was isolated and expressed in Escherichia coli as a fusion protein. The fusion protein showed enzymatic activity that converted [3H]copalyl diphosphate to [3H]ent-kaurene. The recombinant AtKS protein derived from the ga2–1 mutant is truncated by 14 kD at the C-terminal end and does not contain significant KS activity in vitro. Sequence analysis revealed that a C-2099 to T base substitution, which converts Gln-678 codon to a stop codon, is present in the AtKS cDNA from the ga2–1 mutant. Taken together, our results show that the GA2 locus encodes KS.
GAs are a group of diterpene compounds, some of which are plant-growth regulators that control many aspects of plant development, such as seed germination, shoot elongation, and flower development. A number of GA-responsive dwarf mutants that are deficient in the biosynthesis of active GAs have been characterized in various plant species (for a recent review, see Hedden and Kamiya, 1997; Ross et al., 1997). Biochemical characterization of these mutants has contributed to the elucidation of GA biosynthetic pathways. ent-Kaurene is an early intermediate in GA biosynthesis and is synthesized by the cyclization of GGDP via CDP. In plants, this two-step cyclization involves two distinct enzymes: CPS catalyzes the cyclization of GGDP to CDP, which is then converted to ent-kaurene by KS. CPS and KS were formally called ent-kaurene synthases A and B, respectively (Hedden and Kamiya, 1997). Biochemical studies suggested that CPS and KS may interact with each other (Duncan and West, 1981) and that these enzymes are localized in plastids (Railton et al., 1984; Sun and Kamiya, 1994; Aach et al., 1995). There is some evidence that ent-kaurene biosynthesis is controlled by environmental conditions such as photoperiod (Zeevaart and Gage, 1993) and temperature (Moore and Moore, 1991), as well as during plant development (Chung and Coolbaugh, 1986; Silverstone et al., 1997).
From Arabidopsis thaliana six GA-responsive dwarf mutants (ga1 through ga6) have been isolated and characterized (Koornneef and van der Veen, 1980; Sponsel et al., 1997). Each mutant is blocked in a specific step in GA biosynthesis. The GA1 locus has been cloned (Sun et al., 1992) and shown to encode CPS (Sun and Kamiya, 1994). The GA4 and GA5 genes code for dioxygenases involved in later steps of the GA biosynthetic pathway (Chiang et al., 1995; Xu et al., 1995). The GA2, GA3, and GA6 loci may encode enzymes catalyzing steps in the GA biosynthetic pathway or regulators of these enzymes.
The ga2–1 mutant is a nongerminating, extreme dwarf, which is phenotypically similar to strong alleles of the ga1 and ga3 mutants. (Koornneef and van der Veen, 1980). The biochemical characterization of the ga2 mutant was described by Zeevaart and Talon (1992). The ga2 mutant is responsive to exogenous ent-kaurene and is lacking in the ability to accumulate ent-kaurene when treated with an inhibitor of ent-kaurene metabolism. A cell-free extract prepared from siliques of the ga2 mutant lacked KS activity. Therefore, the GA2 locus probably encodes either KS or a regulatory protein controlling the expression of the A. thaliana KS gene, AtKS. Several GA-responsive dwarf mutants from other plant species also contain reduced KS activity. Hedden and Phinney (1979) reported that the formation of ent-kaurene from mevalonic acid, GGDP, or CDP is reduced in cell-free preparations from the maize (Zea mays) dwarf-5 mutant. Studies using cell-free extracts suggested that the gib-3 mutant of tomato (Lycopersicon esculentum) is impaired in KS activity, leading to the GA-deficient mutant phenotype (Bensen and Zeevaart, 1990).
We previously purified KS from pumpkin (Cucurbita maxima; Saito et al., 1995) and isolated the corresponding cDNA (Yamaguchi et al., 1996). In this paper we report the isolation of the AtKS gene that we used to examine whether the GA2 locus encodes KS and to study the regulation of genes encoding the interacting enzymes CPS and KS in A. thaliana. Using the C. maxima KS (CmKS) cDNA, we cloned a homologous cDNA from A. thaliana and demonstrated that it encodes KS. Expression of the CmKS cDNA in ga2–1 complemented the mutant phenotype. We also demonstrated that the AtKS cDNA from the ga2–1 encodes a truncated protein that does not have any KS activity in an in vitro enzyme assay.
MATERIALS AND METHODS
Arabidopsis thaliana (L.) Heynh. (ecotype Lansberg erecta) plants were used in this study. The ga2–1 mutant seeds were obtained from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus). Before planting, the ga2–1 seeds were incubated in 100 μm GA3 solution at 4°C for 3 d. The plants were grown either on Murashige-Skoog agar medium (GIBCO-BRL) or in soil under 16-h light/8-h dark conditions at 22°C. The ga2–1 mutant was sprayed with 100 μm GA3 solution to produce seeds.
Complementation Test
To overexpress the CmKS cDNA, a gene fusion containing the cauliflower mosaic virus 35S-promoter with dual enhancers, the tobacco etch virus-nontranslated region, CmKS cDNA, and the nopaline synthase terminator was constructed in the binary vector pBI101.2 as follows. The nucleotide sequence around the first ATG codon of CmKS cDNA (Yamaguchi et al., 1996) was modified to contain an NcoI site, and an internal NcoI site near the 3′ end of the coding sequence was modified by PCR. Introduction of the NcoI site at the starting ATG converts the second codon TAT (Tyr) to GAT (Asp). Modification of the internal NcoI site does not alter the CmKS protein sequence. The forward (5′-ACCAGCCATGGATCTTTCCCGACCTACCGGCG; NcoI site is underlined) and reverse (5′-TTGAGCTCATTTGTTCAATAGTGCATCCAGATCCATAGGTT; SacI site and the altered NcoI site are underlined) primers were used to amplify a 2.4-kb DNA fragment from pCmKB-1 (Yamaguchi et al., 1996). The 2.4-kb DNA fragment was inserted into the NcoI-SacI site of pRTL2 (Restrepo et al., 1990). This plasmid was digested with HindIII and SacI, and the resulting 3.1-kb DNA fragment was ligated into the HindIII-SacI sites of the binary vector pBI101.2 (pBI/KSB101). Transgenic plants were generated by vacuum infiltration with Agrobacterium tumefaciens (Bechtold et al., 1993).
Isolation of Genomic and cDNA Clones
Plaque lifts from an A. thaliana (ecotype Columbia) genomic library (λDASHII; courtesy of Dr. M. Matsui, RIKEN Institute, Saitama, Japan) on nylon membranes (Hybond-N+, Amersham) were hybridized with a radiolabeled CmKS cDNA (2.7 kb; Yamaguchi et al., 1996) at 55°C in Rapid-hyb buffer (Amersham). The membranes were washed with 2× SSC (Sambrook et al., 1989) containing 0.1% SDS at 60°C. One of the clones isolated (λAtKS-42) was further characterized. A 3.5-kb DNA fragment, which was generated by EcoRI digestion from the λAtKS-42, was subcloned into the EcoRI site of pUC118 (Toyobo, Osaka, Japan) and was named pgAtKS1.
Isolation of the corresponding cDNA fragments was performed by using the Marathon cDNA amplification kit (Clontech, Palo Alto, CA), according to the manufacturer's protocol. Based on the DNA sequence of pgAtKS1, oligonucleotide primers AtKS1F (5′-TGAGCAAACAAAGGAGAAGATTAGG), AtKS1R (5′-CCTAATCTTCTCCTTTGTTTGCTCA), and AtKS2R (5′-CCCAACTAGTATCGTAGGCCG) were synthesized. Poly(A+) RNA was prepared from 14-d-old wild-type plants using a magnetic resin (PolyATtract mRNA isolation systems, Promega). For 5′ RACE, a reverse-transcription reaction was primed with the AtKS2R primer instead of the oligo(dT) primer. A cDNA template for 3′ RACE was prepared according to the Clontech manual. PCR was carried out using the gene-specific primers AtKS1R and AtKS1F (for 5′ and 3′ RACE, respectively) and the Expand High Fidelity PCR System (Boehringer Mannheim). The RACE products were subcloned into the pCRII vector (Invitrogen, San Diego, CA).
The DNA sequence of several independent clones (about 0.2 kb) from the 5′ RACE product was determined. Clones from the 3′ RACE (approximately 2.3 kb) were partially sequenced from both ends. The sequences from the RACE products were used to design a pair of primers 5′BamHI/AtKS (5′-TTGGATCCGTTGCTACGACGCCGTTTCGG) and 3′SalI/AtKS (5′-TGTCGACCTCTTCTTGTTCTGAAGCAAC) to amplify a 2.5-kb cDNA from the cDNA pool, which was prepared using 14-d-old A. thaliana mRNA. The PCR product was digested with BamHI and SalI and inserted into the BamHI-SalI site of pET28c vector (Novagen, Madison, WI) to generate pET/AtKS. The 2.5-kb cDNA from pET/AtKS was subcloned into pBluescript SK+ (pAtKS4) to determine the complete nucleotide sequence.
Heterologous Expression in Escherichia coli
A His-T7-tag-AtKS fusion protein was produced in E. coli BL21 (DE3) containing pET/AtKS. Expression of the fusion protein was induced by addition of 1 mm isopropyl β-d-thiogalactopyranoside when the E. coli cells reached an A600 of 1.0 at 37°C. After the addition of isopropyl β-d-thiogalactopyranoside, the bacteria were cultured at 20°C for 16 h. To detect the fusion protein by immunoblot analysis, total cell extracts were fractionated on an 8% SDS-polyacrylamide gel (Sambrook et al., 1989).
Isolation and overexpression of the AtKS cDNA from the ga2–1 mutant was performed as described above. The corresponding plasmid construct was named pET/ga2. To rule out the possibility that errors in the sequence were generated during PCR, we repeated these experiments using several pET/AtKS and pET/ga2 clones that were generated from independent PCRs.
Enzyme Assay of the Fusion Protein
KS activity in E. coli extracts was determined as described previously (Yamaguchi et al., 1996) using [3H]CDP and soluble crude extracts containing 40 μg of protein. Identification of ent-kaurene by full-scan GC-MS was performed using unlabeled GGDP as a substrate according to the methods of Ait-Ali et al. (1997).
DNA Sequencing
DNA sequences were determined using a DNA sequencer (model ABI373 or ABI377, Applied Biosystems). A series of deletion clones was generated from the genomic clone pgAtKS1 and the cDNA clone pAtKS4 using Exonuclease III and S1 nuclease (GIBCO-BRL). To sequence the AtKS cDNA from the ga2–1 mutant, cDNA fragments were amplified by two independent PCRs using double-stranded cDNA prepared from the ga2–1 mutant as the template. The PCR products were mixed and directly sequenced using internal primers.
Immunoblot Analysis
An alkaline phosphatase-conjugated T7-tag antibody (Novagen), which recognizes the sequence MASMTGGNN adjacent to His-tag, was used to detect the His-T7-tag fusion protein according to the manufacturer's protocol.
DNA and RNA Gel-Blot Analysis
DNA gel-blot analysis was performed using the 2.5-kb AtKS cDNA (from pAtKS4) as a probe. The membrane was hybridized at 55°C in Rapid-hyb buffer (Amersham) and washed with 2× SSC containing 0.1% SDS at 60°C (low stringency). The membrane was then washed with 0.1× SSC containing 0.1% SDS at 65°C (high stringency).
Poly(A+) RNAs were separated on a 1% agarose gel and blotted onto a nylon membrane. The 2.7-kb CmKS cDNA from pCmKB-1 (Yamaguchi et al., 1996) and the 2.5-kb AtKS cDNA were radiolabeled and used as probes to detect KS transcripts from pumpkin (Cucurbita maxima) and A. thaliana, respectively. For a loading control, the membranes were reprobed with radiolabeled cDNA for cytosolic cyclophilin (Lippuner et al., 1994). The membranes were exposed to a Storage Phosphor Screen and analyzed on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Quantitation of radioactivity in each band was performed using IMAGEQUANT software (Molecular Dynamics).
Physical Mapping of the AtKS Gene
Hybridization data presented at the web site (http://cbil.humgen.upenn.edu/~atgc/physical-mapping/physmaps.html) were used to estimate the map position of the BAC clone F12J12 (accession no. B08170), which contained an identical DNA sequence to the AtKS gene. The F12J12 hybridized to BAC F19N14, which hybridized to BACs F16N6 and F17C14. These BACs were identified by hybridization to the YAC CIC1E4, which spans the bottom of chromosome I (Creusot et al., 1995) and contains the restriction fragment-length polymorphism marker m132 (Chang et al., 1988). Because all of these BACs also hybridized to YAC yUP8H12, which has been mapped at the top of chromosome I, YAC yUP8H12 is likely to be chimeric.
Sequence Comparison and Alignments
The BLAST (Altschul et al., 1990) program was used to search for homologous sequences in the database. The GAP and PILEUP programs in the Genetics Computer Group programs (University of Wisconsin, Madison) were used for comparison of amino acid sequences and generation of sequence alignments.
RESULTS
Complementation of the ga2–1 Mutant
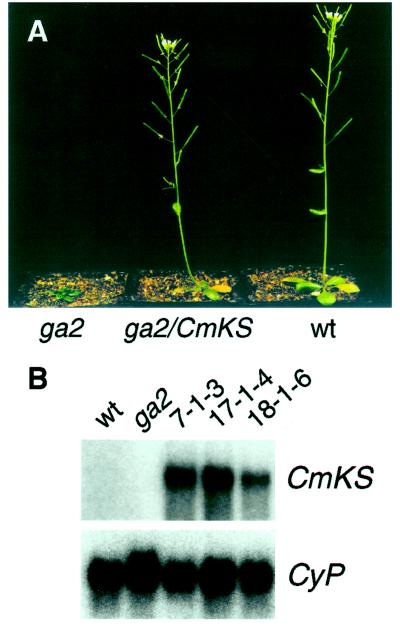
We used the CmKS cDNA to complement the mutant because the ga2 mutant is deficient in KS activity. The ga2–1 mutant was transformed with pBI/KSB101 via A. tumefaciens, and six independent kanamycin-resistant transgenic lines (T1) were isolated. In four lines, the T2 seeds segregated approximately 3:1 for kanamycin resistance versus sensitivity. The kanamycin-resistant seeds germinated in the absence of GA and grew identically to wild-type plants with normal fertility (Fig. 1A); conversely, the seeds that required GA for germination were kanamycin sensitive. These results demonstrated that expression of the CmKS cDNA was able to rescue the phenotype of the ga2 mutant. Three lines homozygous for the transgene (7–1–3, 17–1–4, and 18–1–6) were identified in the T3 generation. Expression of the transgene was analyzed in the homozygotes by RNA blot analysis using CmKS cDNA as a probe (Fig. 1B). A 2.6-kb transcript was detected in RNA from the transgenic plants, whereas no bands were detectable in RNAs from wild-type and the ga2 mutant. These data confirmed that the CmKS gene was expressed in the transgenic plants.
Figure 1.
Expression of CmKS cDNA in the ga2 mutant. A, The ga2–1 mutant (ga2, an extreme dwarf), the ga2–1 mutant expressing CmKS cDNA (ga2/CmKS), and a wild-type plant (wt). The plants were 35 d old. B, Autoradiogram of an RNA blot showing expression of the CmKS cDNA. Poly(A+) RNAs (approximately 3 μg) isolated from wild type (wt), ga2, and the ga2–1 mutant transformed with pBI/KSB101 (lines 7–1-3, 17–1-4, and 18–1-6) were probed with radiolabeled CmKS cDNA and then reprobed with a cyclophilin cDNA (CyP).
Isolation of AtKS Genomic and cDNA Clones
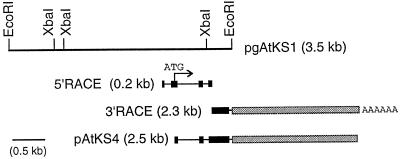
To determine whether the GA2 locus encodes AtKS and to study the regulation of AtKS expression in A. thaliana, we set out to isolate the AtKS gene. We first carried out DNA blot analysis of A. thaliana genomic DNA using the CmKS cDNA as a heterologous probe to determine the appropriate condition for cloning the AtKS gene (data not shown). Under the same low-stringency hybridization conditions, an A. thaliana genomic library was screened and two positive clones were isolated from 2 × 105 plaques. Restriction enzyme maps of these clones suggested that they were derived from a single locus. A 3.5-kb DNA fragment derived from one of the λ clones was subcloned into a plasmid vector (pgAtKS1; Fig. 2) and partially sequenced (data not shown). The predicted amino acid sequence showed significant sequence similarity to CmKS. Oligonucleotide primers based on the genomic DNA sequence were used to amplify 0.2- and 2.3-kb cDNA fragments by 5′ RACE and 3′ RACE, respectively (Fig. 2). Based on nucleotide sequence of the RACE products, a pair of primers was synthesized to amplify a 2.5-kb AtKS cDNA (Fig. 2; pAtKS4).
Figure 2.
Physical map of the AtKS gene. The top bar represents the genomic clone pgAtKS1 with the XbaI restriction map. The RACE products and pAtKS4 (containing the coding region) are cDNA clones. Horizontal thin lines in the cDNA clones show introns and black boxes show exons. Gray bars indicate the stretches of cDNA where a corresponding genomic sequence was not obtained in this study. The position of the first ATG codon and the direction of the open reading frame are indicated by an arrow.
The nucleotide sequence of the full-length AtKS cDNA was determined (accession no. AF034774). An open reading frame consisting of 2355 nucleotides (785 amino acids) was observed (Fig. 3). The predicted molecular mass of the translated product is 90 kD, and the deduced amino acid sequence shows 70% similarity (52% identity) to CmKS.
Figure 3.
Sequence alignment of plant terpene cyclases. Deduced amino acid sequences of CmKS (Yamaguchi et al., 1996), GA1 (A. thaliana CPS; Sun and Kamiya, 1994), and TEAS from Nicotiana tabacum (Facchini and Chappell, 1992) were compared with that of AtKS (GA2). Amino acids conserved between GA2 and at least one other cyclase are highlighted in reverse print. The amino acids that are proposed to be involved in the formation of the active site of TEAS (Starks et al., 1997) are indicated with asterisks below the sequence. The conserved DDXXD motif is indicated with a line with double arrowheads. The 32-amino acid stretch, which is highly conserved between GA2 and CmKS, is highlighted with a thick horizontal line. The position of the mutation in ga2–1 (Q to a stop codon) is shown with an arrow. Dots are gaps for optimization of alignments.
Functional Studies of the AtKS Protein
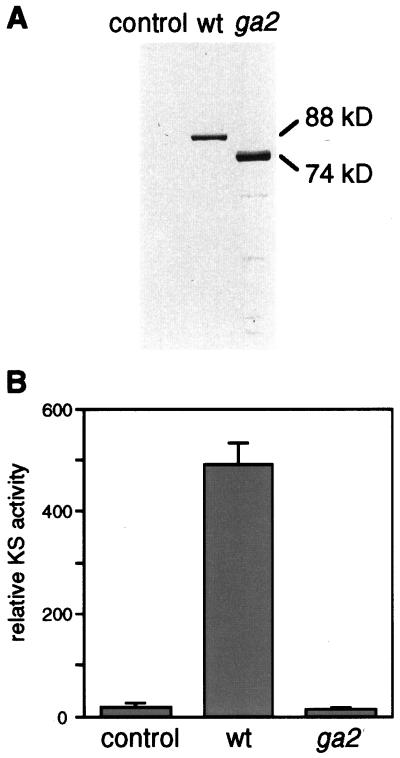
To verify that the isolated gene encodes KS, the full-length cDNA was overexpressed in E. coli and the enzymatic activity of the recombinant AtKS protein was tested in vitro. The 2.5-kb AtKS cDNA was subcloned into the expression vector pET28c (pET/AtKS), and a recombinant protein was produced as a fusion with His-T7-tag (His-T7-AtKS). Immunoblot analysis was performed to confirm the production of the fusion protein using a T7-tag antibody (Fig. 4A). In extracts of E. coli carrying the pET/AtKS, an 88-kD protein (His-T7-AtKS) was recognized by the antibody. No bands were detected in lysates of E. coli carrying the control plasmid pET28c.
Figure 4.
Functional studies of the AtKS gene from wild-type and the ga2–1 mutant. A, Immunoblot containing total cell lysates from E. coli carrying pET28c (control), pET/AtKS (wt), and pET/ga2 (ga2). The membrane was probed with an alkaline phosphate-conjugated T7-tag antibody. The observed molecular mass of each band is shown on the right. B, KS activities of soluble protein extracts (40 μg of protein each) prepared from E. coli indicated in A. Results are means ± se.
KS activity in extracts from E. coli was determined using [3H]CDP as the substrate (Yamaguchi et al., 1996). A significant amount of [3H]ent-kaurene (491 dpm) was detected when [3H]CDP was incubated with the E. coli extracts containing His-T7-AtKS. In contrast, [3H]ent-kaurene produced from the control extract of E. coli carrying pET28c was not above background level (23 dpm), indicating that the His-T7-AtKS possesses significant KS activity (Fig. 4B). The identity of the product was confirmed by GC-MS, using GGDP as a substrate in the presence of a recombinant CPS from pea (Ait-Ali et al., 1997; Kawaide et al., 1997; data not shown). ent-Kaurene was identified by full-scan GC-MS from an incubation mixture containing both CPS and His-T7-AtKS. ent-Kaurene was not detectable in the control reaction mixture containing CPS and extract of E. coli harboring pET28c. These data confirm that we had isolated a cDNA encoding KS from A. thaliana.
Identification of the ga2–1 Mutation
Previous biochemical studies (Zeevaart and Talon, 1992) and the result of the complementation test suggested that the GA2 locus could encode either KS or a regulator of KS gene expression. The possibility that the AtKS gene corresponds to the GA2 locus was examined. RNA-blot analysis indicated that the size and level of the AtKS transcript in the ga2–1 were comparable to those in wild-type plants (data not shown). From this observation, the GA2 gene is unlikely to encode a protein that regulates the transcription of the AtKS gene.
The AtKS cDNA isolated from the ga2–1 mutant was inserted into the E. coli expression vector pET28 (pET/ga2), and the recombinant fusion protein was produced. By immunoblot analysis, a 74-kD protein was detected in lysates of E. coli carrying pET/ga2, whereas the observed mass of His-T7-AtKS was 88 kD (Fig. 4A). KS activity in E. coli extracts containing the truncated recombinant protein from the ga2–1 was as low as the control E. coli extract (Fig. 4B). DNA sequence analysis further revealed that a base substitution of C-2099 to T is present in the AtKS cDNA from the ga2–1 mutant. This mutation changes a Gln-678 codon (CAA) to a stop (TAA, Fig. 3) codon. The calculated mass of the truncated protein from the ga2–1 was 77 kD. This was consistent with the result of the immunoblot analysis (Fig. 4B), and we concluded that the AtKS gene corresponds to the GA2 locus. Therefore, the AtKS gene will be referred to as the GA2 gene.
A database search revealed that a BAC end sequence contains a sequence identical to GA2. Because the GA2 gene appeared to be a single-copy gene in the haploid genome (see below), the BAC is most likely to contain the GA2 locus. According to hybridization data (http://cbil.humgen.upenn.edu/~atgc/physical-mapping/physmaps.html), the BAC clone is located at the bottom of chromosome I. These data coincide with the genetic mapping data of the GA2 locus to the same region of chromosome I (Koornneef and van der Veen, 1980), supporting the conclusion that we have isolated the GA2 locus.
DNA and RNA Blot Analysis
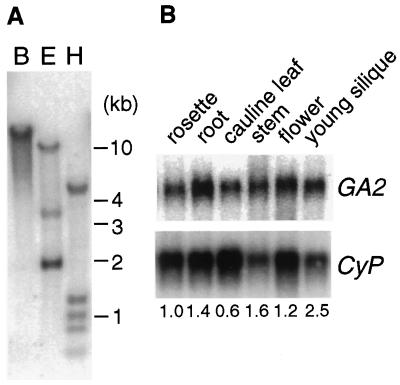
To examine the presence of GA2 homologs in A. thaliana, genomic DNA blot analyses were carried out using the GA2 cDNA as a probe under low- and high-stringency conditions. The result of the experiment under low-stringency conditions is shown in Figure 5A, and an identical banding pattern was observed under high-stringency conditions (data not shown). These results suggest that no closely related sequences exist in A. thaliana.
Figure 5.
Genomic DNA and RNA blot analyses of the GA2 gene. A, Autoradiogram of a DNA blot containing A. thaliana genomic DNA digested with BamHI (B), EcoRI (E), or HindIII (H). Hybridization was carried out under low-stringency conditions using radiolabeled GA2 cDNA as a probe. B, Autoradiogram of an RNA blot containing poly(A+) RNAs purified from 300 μg of total RNAs isolated from different tissues. Aerial tissues (rosette) and roots were harvested from 14-d-old plants grown on agar medium. Cauline leaves, main stems (stem), flower clusters, and young siliques (which contain globular or heart-shaped embryos) were harvested from 35-d-old plants on soil. The membrane was probed with radiolabeled GA2 cDNA and then stripped and reprobed with radiolabeled cyclophilin cDNA (CyP). Numbers below the blot indicate the relative amount of GA2 mRNA standardized by the relative levels of cyclophilin mRNA (“rosette” was arbitrarily set as 1.0).
Tissue distribution of the GA2 transcripts was studied by RNA blot analysis (Fig. 5B). The GA2 mRNA was detected in all tissues that we examined. A cDNA for cytosolic cyclophilin (Lippuner et al., 1994) was used as a probe to standardize RNA loadings. The mRNA level of GA2 was approximately 4 times higher in young siliques than in cauline leaves in 35-d-old plants.
Primary Structure of GA2
The predicted amino acid sequence of GA2 showed significant homology to other terpene cyclases with enzymatic activities distinct from KS. These cyclases include GA1 (A. thaliana CPS; 55% similarity, 31% identity; Sun and Kamiya, 1994), abietadiene synthase from Abies grandis (55% similarity, 34% identity; Vogel et al., 1996), taxadiene synthase from Taxus brevifolia (54% similarity, 32% identity; Wildung and Croteau, 1996), and TEAS ( 50% similarity, 25% identity; Facchini and Chappell, 1992). The sequence alignment of GA2, CmKS, GA1, and TEAS is shown in Figure 3.
A putative transit peptide sequence is present in the N terminus of GA2, as was also observed in the CmKS (Yamaguchi et al., 1996). The first 44 amino acids are rich in hydroxylated amino acids (25% are Ser or Thr), and the predicted pI of this stretch is 10.3, which agrees with the common characteristics of transit peptides that target proteins into plastids (Keegstra et al., 1989).
DISCUSSION
Our results show that expression of the CmKS cDNA was able to complement the A. thaliana ga2 mutant. We isolated and characterized an A. thaliana cDNA encoding KS and demonstrated that the KS cDNA from the ga2–1 mutant contains a nonsense mutation. These data allow us to conclude that the GA2 gene encodes KS and confirm that a mutation in KS causes the GA-deficient phenotype of the ga2 mutant.
To complement the ga2 mutant, the CmKS cDNA was expressed under the regulation of the cauliflower mosaic virus 35S promoter with dual enhancers and tobacco etch virus-nontranslated region, which often results in overexpression (Carrington and Freed, 1990; Sun and Kamiya, 1994). Overproduction of the KS protein could increase GA biosynthesis and, consequently, result in a phenotype that is similar to wild-type plants treated with an excess amount of GAs. However, phenotypes of the four transgenic lines were almost indistinguishable from the wild-type plants. Immunoblot analysis using a specific antibody to the CmKS suggested that the levels of CmKS in the transgenic plants were much lower than in the C. maxima endosperm (data not shown), which contains an exceptionally high level of GA biosynthetic activity (Graebe, 1987). We have not determined the level of heterologous CmKS in the transgenic ga2 mutants in comparison with the endogenous AtKS in wild-type plants. However, the CmKS may not have been overproduced to such an extent that it increases the active GA concentration above wild-type level. Alternatively, a higher level of KS protein may not significantly increase the amount of active GAs if other enzymes in the GA biosynthetic pathway are limited, or genes for those enzymes are regulated to control the level of active GAs. In fact, several genes encoding enzymes catalyzing later steps of GA biosynthesis are negatively regulated by GA activity (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995).
Comparison of the two KS proteins from C. maxima and A. thaliana indicates highly conserved amino acid stretches. One stretch of 32 amino acids (amino acid 599–630 in Fig. 3) contains the DDXXD motif, a putative binding site for the metal ion-diphosphate complex (Chappell, 1995; McGarvey and Croteau, 1995). Recently, the x-ray crystal structure of TEAS was reported (Starks et al., 1997). TEAS catalyzes the conversion of prenyl diphosphate to a cyclic hydrocarbon and contains the DDXXD motif. The x-ray structures with substrate analogs indicated that, in fact, the D residues of the DDXXD motif bind directly to the Mg2+-diphosphate complex. It is likely that the DDXXD motif in KS is also involved in binding to the diphosphate group of the substrate.
The sequence data showed that the ga2–1 allele has a single-base substitution that introduces a premature stop codon at position 756 (Fig. 3). Thus, the GA2 protein from the ga2–1 mutant is missing 108 amino acids at the C terminus. The x-ray crystal structure of TEAS indicated that, in addition to the DDXXD motif, several other amino acid residues at the C terminus are also involved in binding to the substrate, including aromatic amino acid residues at positions 832 and 839 and E-761, D-837, and T-840 near the C terminus (Fig. 3; Starks et al., 1997). These amino acid residues are conserved in both KS proteins but are missing in the truncated protein in the ga2–1 mutant. The absence of these amino acid residues and a possible conformational change resulting from the truncation of 108 amino acids could be the cause of the loss of enzymatic activity.
The ga2–1 mutant still contains low levels of GAs (Zeevaart and Talon, 1992), although the recombinant AtKS protein from the ga2–1 mutant was not functional in vitro and, presumably, not in vivo. The presence of GAs in the ga2–1 mutant implies the existence of another enzyme that can catalyze the same reaction. However, we were unable to detect any GA2 homologs by DNA blot analysis under low-stringency conditions using the GA2 cDNA as a probe. Under the same hybridization conditions, the CmKS cDNA, which has 61% nucleotide sequence identity to the GA2 cDNA, was able to detect the GA2 DNA. Therefore, the cyclase that exhibits KS activity in the ga2–1 mutant would have lower sequence similarity than CmKS.
The ga2–1 mutant is a severe GA-deficient mutant with a phenotype that is a nongerminating, male-sterile, extreme dwarf (Koornneef and van der Veen, 1980). The phenotype suggests that the GA2 gene may be responsible for the formation of ent-kaurene throughout the life cycle of the plant. The GA2 transcripts were detected in every tissue examined in this study (Fig. 5B). From biochemical studies of CPS and KS activities in Marah macrocarpus, it was proposed that CPS and KS might interact to efficiently catalyze the two-step cyclization reaction (Duncan and West, 1981). Therefore, expression of genes encoding CPS and KS may be similarly regulated during plant development. Expression of the GA1 gene is limited to specific cell types, even though the GA1 transcripts are present in all organs (Silverstone et al., 1997), in which the GA2 mRNAs were detected in this study. In addition, the GA1 transcript levels were high in young siliques and were very low in cauline leaves (Silverstone et al., 1997). This tendency was also observed in the relative levels of the GA2 mRNA (Fig. 5B). However, the GA2 mRNA levels did not change as much as the GA1 mRNA levels (Fig. 5B; Silverstone et al., 1997). In chloroplast lysates from apical shoots of pea, significant KS activity was detected, but CPS activity was absent in the same lysate (Railton et al., 1984). These results imply that CPS and KS may not necessarily be present at the same level during plant development. Further expression studies of the GA2 gene using reporter genes or in situ RNA hybridization will reveal whether expression of the GA2 gene is limited to the same cells as it is in the GA1 gene.
ACKNOWLEDGMENTS
We thank Dr. Aron Silverstone (Duke University, Durham, NC) for fruitful discussions and critical reading of the manuscript. We also thank Drs. Tahar Ait-Ali, Gerard Bishop, Mariken Rebers, Maria Smith (RIKEN, Saitama, Japan), Dr. Tomonobu Toyomasu (Yamagata University, Tsuruoka, Japan), and Dr. Andy Phillips (IACR-Long Ashton Research Station, Bristol, UK) for helpful comments concerning the manuscript. The authors are grateful to Ms. Yukiji Tachiyama (RIKEN) for technical assistance with DNA sequencing and Ms. Elzbieta Krol (Duke University) for help with plant transformation. We also thank Prof. C.S. Gasser (University of California, Davis) for the cyclophilin cDNA.
Abbreviations:
- BAC
bacterial artificial chromosome
- CDP
copalyl diphosphate
- CPS
copalyl diphosphate synthase
- GGDP
geranylgeranyl diphosphate
- KS
ent-kaurene synthase
- RACE
rapid amplification of cDNA ends
- TEAS
tobacco 5-epi-aristolochene synthase
- YAC
yeast artificial chromosome
LITERATURE CITED
- Aach H, Böse G, Graebe JE. ent-Kaurene biosynthesis in a cell-free system from wheat (Triticum aestivum L.) seedlings and the localisation of ent-kaurene synthetase in plastids of three species. Planta. 1995;197:333–342. [Google Scholar]
- Ait-Ali T, Swain SM, Reid JB, Sun T-p, Kamiya Y. The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J. 1997;11:443–454. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers E, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- Bensen RJ, Zeevaart JAD. Comparison of ent-kaurene synthetase A and B activities in cell-free extracts from young tomato fruits of wild-type and gib-1, gib-2 and gib-3 tomato plants. J Plant Growth Regul. 1990;9:237–242. [Google Scholar]
- Carrington JC, Freed DD. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Bowman JL, DeJohn AW, Lander ES, Meyerowitz EM. Restriction fragment length polymorphism linkage map for Arabidopsis thaliana. Proc Natl Acad Sci USA. 1988;85:6856–6860. doi: 10.1073/pnas.85.18.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J. The biochemistry and molecular biology of isoprenoid metabolism. Plant Physiol. 1995;107:1–6. doi: 10.1104/pp.107.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H-H, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CH, Coolbaugh RC. ent-Kaurene biosynthesis in cell-free extracts of excised parts of tall and dwarf pea seedlings. Plant Physiol. 1986;80:544–548. doi: 10.1104/pp.80.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot F, Fouilloux E, Dron M, Lafleuriel J, Picard G, Billault A, Paslier DL, Cohen D, Chabouté M-E, Durr A and others. The CIC library: a large insert YAC library for genome mapping in Arabidopsis thaliana. Plant J. 1995;8:763–770. doi: 10.1046/j.1365-313x.1995.08050763.x. [DOI] [PubMed] [Google Scholar]
- Duncan JD, West CA. Properties of kaurene synthetase from Marah macrocarpus endosperm. Evidence for the participation of separate but interacting enzymes. Plant Physiol. 1981;68:1128–1134. doi: 10.1104/pp.68.5.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini PJ, Chappell J. Gene family for an elicitor-induced sesquiterpene cyclase in tobacco. Proc Natl Acad Sci USA. 1992;89:11088–11092. doi: 10.1073/pnas.89.22.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graebe JE. Gibberellin biosynthesis and control. Annu Rev Plant Physiol. 1987;38:419–465. [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hedden P, Phinney BO. Comparison of ent-kaurene and ent-isokaurene synthesis in cell-free systems from etiolated shoots of normal and dwarf-5 maize seedlings. Phytochemistry. 1979;18:1475–1479. [Google Scholar]
- Kawaide H, Imai R, Sassa T, Kamiya Y. ent-Kaurene synthase from the fungus Phaeosphaeria sp. L487. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Olsen LJ, Theg SM. Chloroplastic precursors and their transport across the envelope membranes. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:471–501. [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Lippuner V, Chou IT, Scott SV, Ettinger WF, Theg SM, Gasser CS. Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J Biol Chem. 1994;269:7863–7868. [PubMed] [Google Scholar]
- McGarvey DJ, Croteau R. Terpenoid metabolism. Plant Cell. 1995;7:1015–1026. doi: 10.1105/tpc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TC, Moore JA. Induction of ent-kaurene biosynthesis by low temperature in dwarf peas. J Plant Growth Regul. 1991;10:91–95. [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Railton ID, Fellows B, West CA. ent-Kaurene synthesis in chloroplasts from higher plants. Phytochemistry. 1984;23:1261–1267. [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC. Nuclear transport of plant potyviral proteins. Plant Cell. 1990;2:987–998. doi: 10.1105/tpc.2.10.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, Murfet IC, Reid JB. Gibberellin mutants. Physiol Plant. 1997;100:550–560. [Google Scholar]
- Saito T, Abe H, Yamane H, Sakurai A, Murofushi N, Takio K, Takahashi N, Kamiya Y. Purification and properties of ent-kaurene synthase B from immature seeds of pumpkin. Plant Physiol. 1995;109:1239–1245. doi: 10.1104/pp.109.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Silverstone AL, Chang C-W, Krol E, Sun T-p. Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 1997;12:9–19. doi: 10.1046/j.1365-313x.1997.12010009.x. [DOI] [PubMed] [Google Scholar]
- Sponsel VM, Schmidt FW, Porter SG, Nakayama M, Kohlstruk S, Estelle M. Characterization of new gibberellin-responsive semidwarf mutants of Arabidopsis. Plant Physiol. 1997;115:1009–1020. doi: 10.1104/pp.115.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starks CM, Back K, Chappell J, Noel JP. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- Sun T-p, Goodman HM, Ausubel FM. Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell. 1992;4:119–128. doi: 10.1105/tpc.4.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel BS, Wildung MR, Vogel G, Croteau R. Abietadiene synthase from grand fir (Abies grandis). cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase involved in resin acid biosynthesis. J Biol Chem. 1996;271:23262–23268. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- Wildung MR, Croteau R. A cDNA clone for taxadiene synthase, the diterpene cyclase that catalyzes the committed step of taxol biosynthesis. J Biol Chem. 1996;271:9201–9204. doi: 10.1074/jbc.271.16.9201. [DOI] [PubMed] [Google Scholar]
- Xu Y-L, Li L, Wu K, Peeters AJM, Gage DA, Zeevaart JAD. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Saito T, Abe H, Yamane H, Murofushi N, Kamiya Y. Molecular cloning and characterization of a cDNA encoding the gibberellin biosynthetic enzyme ent-kaurene synthase B from pumpkin (Cucurbita maxima L.) Plant J. 1996;10:203–213. doi: 10.1046/j.1365-313x.1996.10020203.x. [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD, Gage DA. ent-Kaurene biosynthesis is enhanced by long photoperiods in the long-day plants Spinacia oleracea L. and Agrostemma githago L. Plant Physiol. 1993;101:25–29. doi: 10.1104/pp.101.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Talon M (1992) Gibberellin mutants in Arabidopsis thaliana. In CM Karssen, LC van Loon, D Vreugdenhil, eds, Progress in Plant Growth Regulation. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 34–42