Abstract
Striatin and S/G2 nuclear autoantigen (SG2NA) are related proteins that contain membrane binding domains and associate with protein phosphatase 2A (PP2A) and many additional proteins that may be PP2A regulatory targets. Here we identify a major member of these complexes as class II mMOB1, a mammalian homolog of the yeast protein MOB1, and show that its phosphorylation appears to be regulated by PP2A. Yeast MOB1 is critical for cytoskeletal reorganization during cytokinesis and exit from mitosis. We show that mMOB1 associated with PP2A is not detectably phosphorylated in asynchronous murine fibroblasts. However, treatment with the PP2A inhibitor okadaic acid induces phosphorylation of PP2A-associated mMOB1 on serine. Moreover, specific inhibition of PP2A also results in hyperphosphorylation of striatin, SG2NA, and three unidentified proteins, suggesting that these proteins may also be regulated by PP2A. Indirect immunofluorescence produced highly similar staining patterns for striatin, SG2NA, and mMOB1, with the highest concentrations for each protein adjacent to the nuclear membrane. We also present evidence that these complexes may interact with each other. These data are consistent with a model in which PP2A may regulate mMOB1, striatin, and SG2NA to modulate changes in the cytoskeleton or interactions between the cytoskeleton and membrane structures.
Protein phosphatase 2A (PP2A)1 is a heterotrimeric serine/threonine phosphatase that is critical to many cellular processes including development, neuronal signaling, cell cycle regulation, and viral transformation. PP2A also has been implicated in the development of some types of cancers, including human leukemias (1, 2), lung and colon cancers (3). The PP2A heterotrimer consists of a structural (A) subunit, a catalytic (C) subunit, and a regulatory (B-type) subunit. Recently, we have shown that S/G2 nuclear autoantigen (SG2NA) and striatin form stable complexes with the core A/C heterodimer of PP2A (4). SG2NA and striatin are highly related WD40 repeat proteins that bind to calmodulin in a calcium-dependent manner but bear little homology to known B-type subunits (4–6). Interestingly, SG2NA-PP2A and striatin-PP2A immune complexes contained calcium-independent, okadaic acid-sensitive phosphatase activity that was activated toward cdc2-phosphorylated histone H1 substrate (4). However, no known B-type subunits were detectable in immunoblots, silver stain, or Coomassie-stained gels of striatin and SG2NA immunoprecipitations, suggesting that SG2NA and striatin may represent a new family of PP2A regulatory subunits (4).
One of the characteristics of the striatin family, which includes striatin, SG2NA, and zinedin (7), is that each member contains multiple protein-protein interaction domains. These domains include a caveolin binding domain, a potential coiled-coil structure (7), a calmodulin binding domain, a membrane binding domain, and a WD repeat domain (6). Thus, these proteins may function as scaffolding proteins, assembling a large number of proteins into a complex with the PP2A A/C heterodimer.
SG2NA was originally isolated as an autoantigen in a human cancer patient (5). Immunofluorescence studies indicate that it is localized to the nucleus and that its expression peaks during the S and G2 phases of the cell cycle (5). However, more recent studies (7) indicate that SG2NA, like striatin (6, 7), is primarily localized to the cytosol and the membrane. Striatin has been detected by immunofluorescence throughout neuronal dendrites, especially in the post-synaptic densities of neuronal dendritic spines (6, 8). Moreover, striatin contains two polybasic domains that are absent in SG2NA and may facilitate association with the post-synaptic membrane (6). Down-regulation of striatin in vivo using antisense oligonucleotides results in decreased locomotor activity and reduced growth of dendrites in vitro (9). These data suggest that striatin targets PP2A to a cellular microenvironment in which it may play a role in the modulation of calcium-dependent neuronal signaling and possibly remodeling of the cellular cytoskeleton. Although striatin and SG2NA are most highly expressed in brain (6, 7), they have also been detected in many other tissues including liver (5), fibroblasts (4), and skeletal and cardiac muscle (4, 7).
Using affinity-purified antisera to SG2NA, we have immunopurified another member of the striatin-PP2A and SG2NA-PP2A complexes and identified it as the mammalian class II homolog of the yeast protein, MOB1. In Saccharomyces cerevisiae, MOB1 is an essential gene that is required for exit from mitosis, maintenance of cell ploidy, and possibly mitotic spindle pole body duplication (10). In Schizosaccharomyces pombe, MOB1 is required for cytokinesis and is localized to the spindle pole bodies throughout the cell cycle and to the medial ring during late mitosis (11, 12). Here we show that the mammalian class II homolog of MOB1 (mMOB1) is a member of striatin-PP2A and SG2NA-PP2A complexes and that striatin, SG2NA, and mMOB1 may be substrates of PP2A. Moreover, we also show by immunofluorescence microscopy that a subpopulation of striatin and SG2NA appear to colocalize with mMOB1 in the perinuclear region of murine fibroblasts.
EXPERIMENTAL PROCEDURES
Metabolic Labeling
NIH3T3 cells were metabolically labeled with [35S]methionine as previously described (4). Subconfluent 15-cm dishes of NIH3T3 cells were labeled for 4–6 h with 0.5 mCi/ml [32P]orthophosphate in phosphate-free Dulbecco's modified Eagle's medium supplemented with 0.5% dialyzed fetal calf serum. In the experiment shown in Fig. 3 using okadaic acid during labeling, cells were treated with 1 μm okadaic acid for 2 h before lysis. However, similar labeling was also observed using 500 nm okadaic acid in other experiments (not shown).
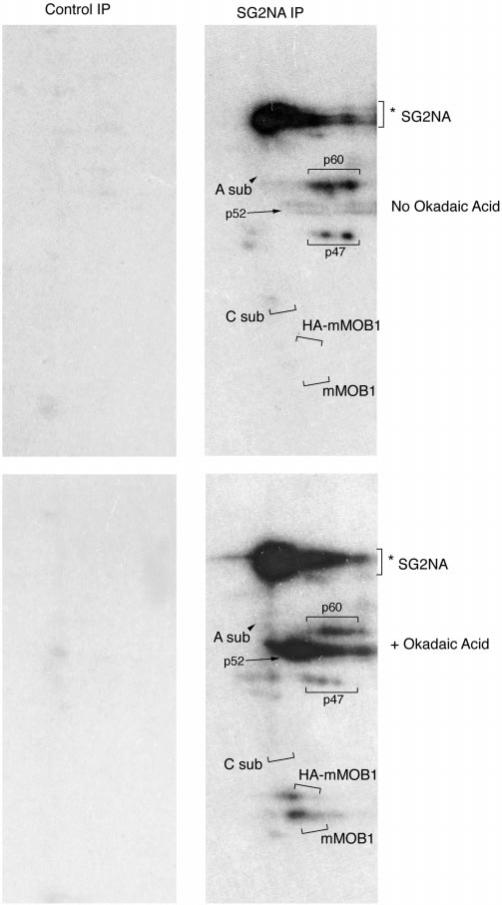
Fig. 3. PP2A inhibition in vivo by okadaic acid treatment results in phosphorylation of mMOB1 and HA-mMOB1 and hyperphosphorylation of SG2NA and three unidentified proteins of 47, 52, and 60 kDa.
NIH3T3 cells that stably express HA-mMOB1 were metabolically labeled with 32P and treated with 1 μm okadaic acid or left as untreated controls. Immunoprecipitations (IP) using preimmune antisera and anti-SG2NA antisera were prepared from radiolabeled cells and analyzed on SDS-PAGE, and phosphorylated proteins were detected by autoradiography. HA-mMOB1 and endogenous mMOB1 can be detected only in cells treated with okadaic acid. The migration positions of the various 35S-labeled proteins shown in Fig. 2 are indicated by brackets and arrows.
Immunoprecipitations
Cells were washed, whole cell lysates were prepared, and immunoprecipitations were performed as previously described (4).
Preparative Immunopurification
Using affinity-purified SG2NA antisera chemically cross-linked to protein A-Sepharose, samples were immunopurified from 28 15-cm dishes of NIH3T3 cells as described (13), except that immune complexes were prepared in a single batch immunoprecipitation instead of from multiple sequential immunoprecipitations. Protein complexes were eluted with 100 mm triethylamine (pH 11.4) and analyzed by two-dimensional gel electrophoresis as previously described (14). Preparative immunopurifications were visualized with Colloidal Blue stain (Novex, San Diego, CA). Control immunopurifications were performed with preimmune sera chemically cross-linked to protein A-Sepharose using two 15-cm dishes of cells analyzed by two-dimensional gel electrophoresis and visualized by silver stain.
Ion Trap Mass Spectrometry
Proteins isolated from two-dimensional gels were microsequenced by ion trap mass spectrometry as previously described (4).
Construction of a Stable Cell Line Expressing HA-tagged mMOB1
A full-length human cDNA clone of mMOB1 was obtained from the IMAGE consortium human germinal center B cell library of expressed sequence tags (ESTs) (GenBank™ accession number AA504251, IMAGE clone number 825396). The human and mouse class II homologs of MOB1 were 100% identical at the amino acid level. The mMOB1 cDNA was polymerase chain reaction-amplified from the EST clone using the 5′ primer (CGGCACGAGGCCATGGTC) and the 3′ primer (TCCCAGTCGACGTTGTAAAA). The 5′ primer created an NcoI site at the start ATG codon of the mMOB1 open reading frame, and the 3′ primer was complementary to the pT7T3D-Pac vector and created a SalI site at the 3′ end. The amplified mMOB1 cDNA was blunt-cloned into the PCRscriptSK+ vector (Stratagene) and DNA-sequenced. The mMOB1 cDNA was isolated from the PCRscriptSK+ vector using the introduced NcoI and SalI restriction sites. This fragment was then ligated into the BamHI and SalI sites of the pBABE_NEO mammalian expression vector together with a BamHI-NcoI double-stranded oligonucleotide linker encoding the hemagglutinin (HA) epitope tag followed by a thrombin protease cutting site (15). The resulting clone (HA-mMOB1) expressing a fusion protein with the HA tag at the amino terminus of mMOB1 was stably transfected into NIH3T3 cells and selected in Dulbecco's modified Eagle's medium, 10% calf serum supplemented with 400 μg/ml geneticin.
Antibodies
Lasergene DNASTAR Protean software was utilized to identify highly hydrophilic and antigenic sequences for selection of peptide antigens. Rabbit polyclonal mMOB1 antisera were generated using keyhole limpet hemocyanin (KLH)-conjugated peptide DP62 as an immunogen. Peptide DP62 (RNRPGTKAQDFYNWPDESFDEMDSTC) corresponds to residues 12–36 of class II mammalian MOB1 with an additional C-terminal cysteine for coupling to KLH. Peptides were conjugated to KLH using the Imject maleimide KLH conjugation kit (Pierce) according to the manufacturer's instructions. An anti-SG2NA mouse IgG1 monoclonal antibody (S-68) was generated commercially (Anogen, Toronto, Ontario, Canada) against a previously described KLH-linked SG2NA peptide antigen (4). Striatin and SG2NA antibodies are available from Upstate Biotechnology, Inc.
Immunofluorescence
Mouse fibroblasts were plated at 50% confluence and serum-starved (0.1% calf serum, Dulbecco's modified Eagle's medium) overnight to enhance the flattening and spreading of these cells. Coverslips were washed twice in phosphate-buffered saline (PBS) and fixed for 5 min in freshly prepared 1% paraformaldehyde, followed by 15 min in freshly prepared 2% paraformaldehyde.2 Coverslips were then washed 3 times in PBS and incubated in 50 mm NH4Cl in PBS for 15 min. The coverslips were then washed two times in PBS, and cells were permeabilized in 0.1% Triton X-100, PBS for 10 min. After washing three times in PBS, coverslips were blocked in PBS containing 10% horse serum, 3% BSA, and 3% nonfat dry milk. Primary antibodies were diluted into PBS containing 10% horse serum, 3% BSA. Affinity-purified anti-SG2NA and anti-striatin polyclonal antibodies were diluted 1:5000, and commercially obtained 16b12 (Berkeley Antibody Co.) specific for the HA-epitope tag was diluted 1:1000. Monoclonal SG2NA antibodies (S-68) were used at a 1:1000 dilution. For control experiments, affinity-purified polyclonal antibodies were preincubated with peptide antigens at 2 μg/ml, rocking at 4 °C in PBS, 10% horse serum, 3% BSA for 1 h. Coverslips were incubated in primary antibodies overnight at 4 °C. After primary antibody incubation, coverslips were washed three times in PBS, 3% BSA and then incubated with lissamine rhodamine-conjugated anti-rabbit secondary antibodies (1:2000) and fluorescein isothiocyanate-conjugated anti-mouse secondary antibodies (1:4000) (Jackson ImmunoResearch, West Grove, PA) in PBS, 3% BSA, 10% horse serum for 1 h at room temperature in the dark. Coverslips were washed twice with PBS, and DNA staining was performed with Hoescht dye (Molecular Probes, Eugene, OR). Immunofluorescence microscopy was performed on an Olympus BX-60 microscope with a Photometrix Quantix CCD camera. Confocal microscopy was performed on Zeiss Microsystems LSM510 microscope.
RESULTS
Identification of mMOB1 as a Member of the SG2NA-PP2A Complex
To investigate the composition of SG2NA-PP2A and striatin-PP2A complexes, NIH3T3 cells were metabolically labeled with [35S]methionine, and SG2NA complexes were immunoprecipitated from whole cell lysates. In addition to SG2NA, striatin, A subunit, and C subunit, several unidentified proteins were observed by two-dimensional gel analysis of 35S-labeled SG2NA and striatin immunoprecipitates (4). One component of these complexes that migrates as a doublet at ;24 kDa had previously been observed in PP2A (1d6) immunoprecipitations from metabolically labeled cells analyzed on two-dimensional gels.3 Preparative immunoprecipitation using affinity-purified SG2NA polyclonal antisera and two-dimensional gel electrophoresis followed by microsequencing of the 24-kDa protein by peptide ion trap mass spectrometry identified two peptides (IFSHAYFHHR and ILEPPEGQDEGVWK) that matched mammalian class II MOB1 (mMOB1) (10), a homolog of the yeast protein, MOB1 (Fig. 1A).
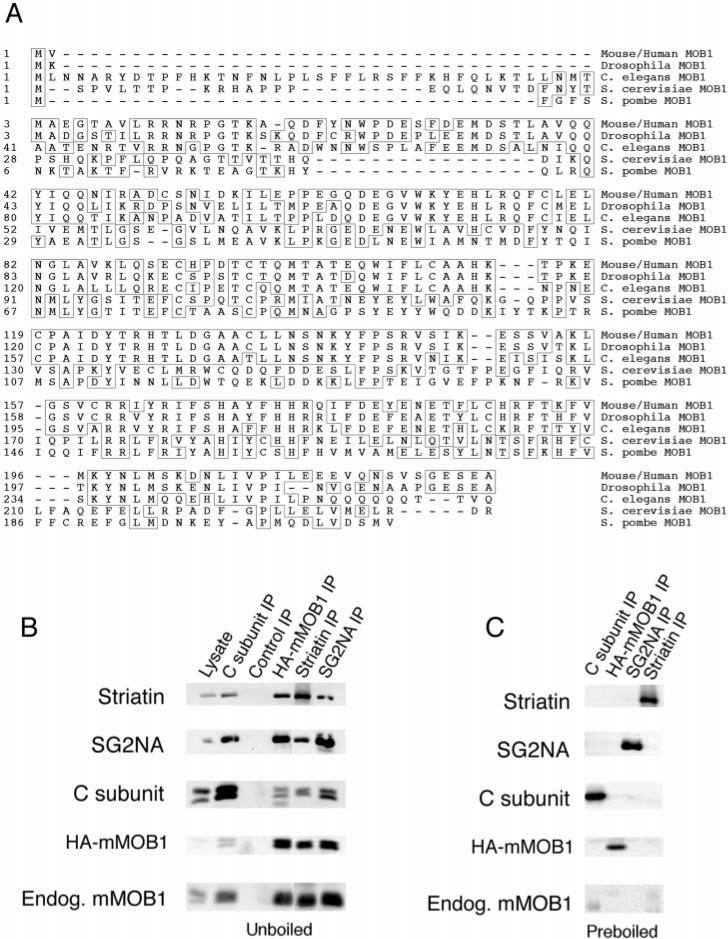
Fig. 1. SG2NA-PP2A and striatin-PP2A form stable complexes with mammalian class II MOB1.
A, amino acid sequence alignment of class II MOB1 from mouse/human, Drosophila, and C. elegans with S. cerevisiae and S. pombe MOB1. Human and mouse MOB1 are 100% identical at the amino acid level. Amino acids matching the consensus sequence are boxed. B, a whole cell lysate and the indicated immunoprecipitations (IP) were prepared from NIH3T3 cells that stably express HA-tagged mMOB1. Immune complexes were analyzed by SDS-PAGE, transferred to nitrocellulose, and sequentially probed by immunoblotting. Striatin, SG2NA, PP2A C subunit, HA-mMOB1, and endogenous (Endog.) mMOB1 were detected in 12CA5 immunoprecipitations of HA-mMOB1 but not in a control immunoprecipitation. Also, HA-mMOB1 and endogenous mMOB1 were detected in PP2A (1d6), striatin, and SG2NA immunoprecipitations but not in control immunoprecipitations. Control immunoprecipitations were performed using the 7-34-1 monoclonal antibody (American Type Culture Collection) directed against major histocompatibility complex class I swine leukocyte antigen. C, as an additional control, some lysates were preboiled in 0.5% SDS and 5 mm β-mercaptoethanol to disrupt complexes, and then parallel immunoprecipitations were prepared. The absence of co-immunoprecipitating proteins from preboiled lysates indicates that co-immunoprecipitations were not due to cross-reactive antibodies.
The S. cerevisiae protein, MOB1 (Mps One Binder), was first identified in a two-hybrid screen for substrates of the kinase MPS1 (10). The MPS1 kinase (16) carries out at least two functions during mitosis, regulation of spindle pole body duplication (17, 18) and activation of a mitotic checkpoint that monitors spindle integrity (19). S. cerevisiae MOB1 is an essential gene required for completion of mitosis and maintenance of cell ploidy (10). Although yeast MOB1 interacts physically with MPS1 and can be phosphorylated by MPS1 in vitro, MOB1 is not required for the mitotic checkpoint function of MPS1 (10). Although deletion of MOB1 in yeast is lethal, conditional mutants arrest in late mitosis as large budded cells with separated chromatin and long bipolar spindles. Also, some temperature-sensitive MOB1 mutants cause an increase in ploidy at permissive temperatures, suggesting that MOB1 may play a role in the spindle pole body duplication function of MPS1 (10). In both S. cerevisiae (20) and S. pombe (11, 12), MOB1 physically interacts with the DBF2/Sid2 kinase. Furthermore, MOB1 has been shown to be essential for cytokinesis in S. pombe. Also in fission yeast, MOB1 is localized to the spindle pole bodies throughout the cell cycle and to the medial ring during cytokinesis (11, 12).
Analysis of the EST data bases revealed that there are two divergent classes of homologs of yeast MOB1 in Caenorhabditis elegans, mouse, and humans (10), and the mammalian homolog identified here as a member of the PP2A complexes is the class II homolog of yeast MOB1. The class II homologs of yeast MOB1 in C. elegans, mouse, and humans are all more highly related to each other than they are to the class I homologs of MOB1 in each respective species. This is because different portions of the yeast MOB1 sequence are conserved in the two classes of MOB1 homologs (10).
Full-length cDNA clones of human and mouse mMOB1 were obtained from the IMAGE consortium collection of ESTs and completely sequenced. Other class II mMOB1 cDNA sequences have been deposited in GenBank™ under the various names of 2C4D (39), Prei3 (40, 41), Mob3 (accession number AB015441), and CGI-95 (42). The RIKEN definition for this gene product (41), Preimplantation protein 3 (Prei3), reflects the fact that mMOB1 was present in early mouse embryonic cDNA libraries. Sequence analysis showed that mMOB1 is 100% identical between human and mouse at the amino acid level and 95% identical at the DNA level. Computer analysis of the recently released Drosophila genomic sequence enabled identification of the MOB1 homolog in the fruit fly. BLAST comparison of the Drosophila and human MOB1 sequences showed that they are 80% identical and 87% similar at the amino acid level (Fig. 1A). This high level of conservation is consistent with a fundamentally important cellular role for MOB1.
To confirm the identification of mMOB1 as a member of striatin-PP2A and SG2NA-PP2A complexes, NIH3T3 cells were stably transfected with HA-tagged mammalian MOB1 (HA-mMOB1). Co-immunoprecipitation and immunoblotting (Fig. 1B) confirmed the HA-mMOB1/SG2NA-PP2A and HA-mMOB1/striatin-PP2A interactions. Immunoprecipitations with PP2A monoclonal antibody (1d6), striatin antisera, SG2NA antisera, and the 12CA5 anti-HA antibody all confirmed the HA-mMOB1-SG2NA-PP2A and HA-mMOB1-striatin-PP2A complexes. Interestingly, endogenous mMOB1 was detected in 12CA5 immunoprecipitations of HA-mMOB1, striatin was detected in SG2NA immunoprecipitations, and SG2NA was detected in striatin immunoprecipitations. A control immunoprecipitation did not precipitate striatin, SG2NA, PP2A C subunit, HA-mMOB1, or endogenous mMOB1, indicating that the observed immunoprecipitations are specific (Fig. 1B). Moreover, none of the members of these complexes were observed in 12CA5 immunoprecipitations from the parent NIH3T3 cell line that does not express HA-mMOB1 (not shown), indicating that the associations are specific. These data suggest that some component of these complexes could be a dimer or that more than one mMOB1 monomer could associate with striatin or SG2NA. Chemiluminescence quantitation of mMOB1, striatin, and SG2NA present in lysates and immunoprecipitates determined that approximately one-third of cellular mMOB1 is complexed with striatin and one-third of mMOB1 is complexed with SG2NA (data not shown). Whether the remaining third is free or complexed to other members of the striatin family could not be determined using the available reagents.
To ensure that the observed stable interactions between striatin and SG2NA could not be due to cross-reactive antibodies, lysates were preboiled in 0.5% SDS and 5 mm β-mercaptoethanol to disrupt complexes and denature proteins. The SDS was then diluted and absorbed into micelles with 1% Nonidet P-40, and then parallel immunoprecipitations were prepared (Fig. 1C). The only case in which there was occasional nonspecific sticking was a small amount of the endogenous mMOB1. However, the low levels detected in the preboiled immunoprecipitations cannot account for the much higher amounts of mMOB1 found in the immunoprecipitations from unboiled lysates. The absence of co-immunoprecipitating proteins from preboiled lysates indicates that each immunoprecipitating antibody is specific and that SG2NA-PP2A-mMOB1 and striatin-PP2A-mMOB1 complexes interact.
Post-translational Modification of mMOB1
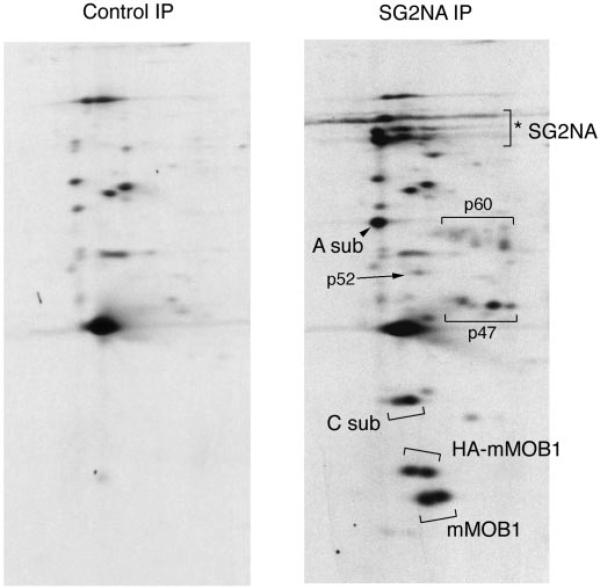
SG2NA immunoprecipitates were prepared from 35S-radiolabeled NIH3T3 cells stably expressing HA-mMOB1 (hereafter referred to as HA-mMOB1 cells) and analyzed on two-dimensional gels (Fig. 2). A second, higher molecular weight doublet produced by the epitope-tagged HA-mMOB1 was detected in addition to the 24-kDa doublet generated by the endogenous mMOB1. This observation demonstrates that the two spots detected at 24 kDa were different isoforms of mMOB1 and not produced by different, similarly migrating proteins. It also indicates that the doublet is not a product of alternative splicing, since both species can be generated from a cDNA, and instead must be due to a post-translational modification of mMOB1. This finding, together with earlier work demonstrating that yeast MOB1 can be phosphorylated by the MPS1 (10) and DBF2 (20) kinases raised the possibility that mMOB1 might be phosphorylated in vivo.
Fig. 2. Mammalian MOB1 is covalently modified in vivo.
NIH3T3 cells stably expressing HA-mMOB1 were metabolically labeled with [35S]methionine, and SG2NA and control immunoprecipitations (IP) were analyzed by two-dimensional gel electrophoresis. Isoelectric focusing was from right (basic end) to left (acidic end). Control immunoprecipitations were performed with preimmune sera from the same rabbit used to generate SG2NA antibodies. Both endogenous mMOB1 and HA-tagged mMOB1 and PP2A A subunit (A sub) and C subunit (C sub) are indicated. Also shown are the locations of unidentified members of the SG2NA-PP2A-MOB1 complexes that migrate at 47, 52, and 60 kDa, respectively. The multiple spots indicated by the SG2NA bracket (*) include SG2NA and striatin as well as possibly other striatin-SG2NA family members or alternatively spliced forms of SG2NA.
To begin the analysis of the role of phosphorylation in the regulation of SG2NA-PP2A complexes, HA-mMOB1 cells were metabolically labeled with either 35S or 32P, and SG2NA immunoprecipitates were prepared (Figs. 2 and 3). Both SG2NA (Fig. 3) and striatin (not shown) were phosphorylated in untreated cells and became hyperphosphorylated upon okadaic acid treatment. In addition to SG2NA, unknown 47, 52, and 60-kDa proteins present in SG2NA-PP2A-mMOB1 complexes were hyperphosphorylated upon okadaic acid treatment. Whereas SG2NA was easily detected in 32P-labeled cells, mMOB1 present in SG2NA-PP2A complexes was undetectable even upon long exposure and thus did not appear to be constitutively phosphorylated. This result indicates that the post-translational modification detected by 35S-labeling was not due to phosphorylation. However, when HA-mMOB1 cells were treated with okadaic acid at a concentration specific for PP2A inhibition (21, 22), both endogenous mMOB1 and HA-mMOB1 were detected by 32P-labeling (Fig. 3). The spots indicated as HA-mMOB1 and mMOB1 in Fig. 3 were confirmed by three criteria. First, the spot corresponding to HA-mMOB1 was not observed in the parent NIH3T3 cell line (not shown). Second, the migration positions on two-dimensional gels of HA-mMOB1 and mMOB1 from 32P-labeled cells corresponded to the expected position based on 35S-labeled immunoprecipitations. And third, the nitrocellulose membranes used to obtain the exposures shown in Fig. 3 were probed as immunoblots, and the mMOB1 and HA-mMOB1 spots were recognized with antibodies to mMOB1 (not shown). Thus, mMOB1 is both a member of the SG2NA-PP2A complex and may be an in vivo substrate of PP2A.
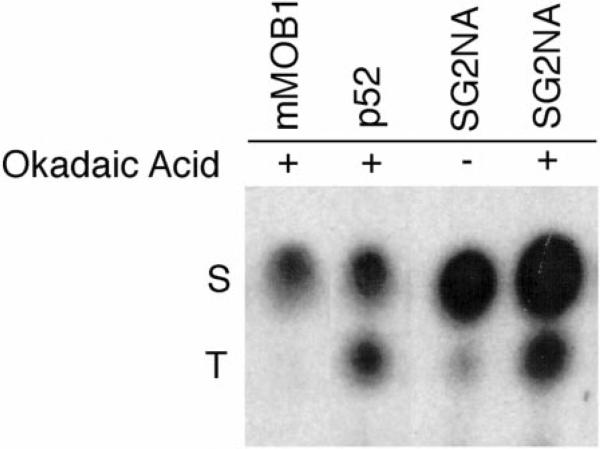
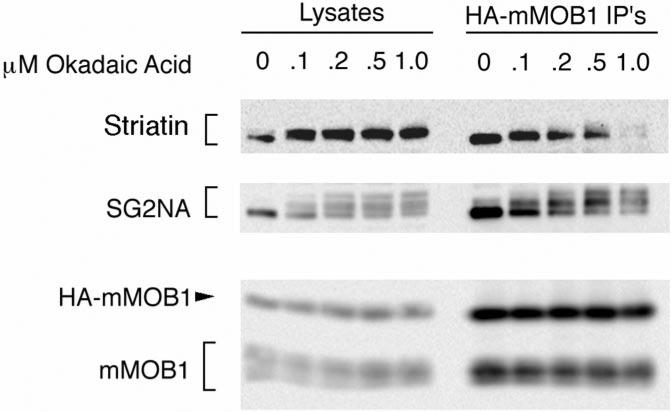
To examine the type of amino acid(s) in these proteins that is phosphorylated after okadaic acid treatment, phosphoamino acid analysis was performed on SG2NA complexes from 32P-labeled HA-mMOB1 cells (Fig. 4). Phosphoamino acid analysis demonstrated that although SG2NA and the unknown p52 protein are phosphorylated at both serine and threonine, mMOB1 is phosphorylated only on serine. Although SG2NA was phosphorylated primarily on serine in the absence of okadaic acid, it showed enhanced phosphorylation of both threonine and serine residues upon okadaic acid treatment. No tyrosine phosphorylation was observed for any of these proteins (data not shown). Because hyperphosphorylation of SG2NA results in a shift in its migration on SDS-PAGE, it was possible to monitor SG2NA phosphorylation in unlabeled NIH3T3 cells. Parallel dishes of NIH3T3 cells were treated with increasing okadaic acid concentrations, and HA-mMOB1 complexes were immunoprecipitated with the 12CA5 antibody and probed for the presence of SG2NA in immunoblots (Fig. 5). Partial phosphorylation of SG2NA was detected at an okadaic acid concentration of only 100 nm, and complete hyperphosphorylation was observed at 200 nm okadaic acid. Since this concentration of okadaic acid is specific for PP2A, SG2NA is likely a substrate of PP2A, not PP1. Because phosphorylation of mMOB1 does not result in a shift of its migration in SDS-PAGE gels, this assay could not determine what concentration of okadaic acid is minimally required to achieve phosphorylation of mMOB1.
Fig. 4. mMOB1 is phosphorylated only on serine residues, whereas SG2NA and p52 are phosphorylated on both serine and threonine.
NIH3T3 cells were labeled in vivo with [32P]orthophosphate and either treated (1) with 1 mm okadaic acid or left untreated (–). SG2NA complexes were immunoprecipitated and analyzed by SDS-PAGE, and phosphoamino acid analysis was performed as described (38). In the absence of okadaic acid, mMOB1 and p52 cannot be detected with 32P and, thus, are not shown. Phosphoserine residues (S) and phosphothreonine residues (T) are indicated on the left of the figure. No phosphotyrosine was detected in any of these proteins (not shown).
Fig. 5. SG2NA and striatin are substrates of PP2A.
NIH3T3 cells were treated with increasing concentrations of okadaic acid, and HA-mMOB1 complexes were immunoprecipitated (IP) and immunoblotted. Phosphorylation of SG2NA can be detected by a slower migration of SG2NA on SDS-PAGE. Partial phosphorylation of SG2NA can be detected with okadaic acid concentrations as low as 100 nm, and complete phosphorylation is observed at 200 nm, indicating that SG2NA may be a PP2A substrate. Striatin phosphorylation can also be detected at 100 nm okadaic acid. The migration of mMOB1 on SDS-PAGE is unaffected by phosphorylation.
Although striatin was also observed to supershift at 100 nm okadaic acid, the hyperphosphorylated form of striatin was not detected as well by Western blotting as it was by 32P-labeling. This could be due to the fact that the peptide antigen used to generate the striatin antisera is rich in serines. Thus, the striatin antisera may be specific for striatin that is not phosphorylated between Ser-373 and Ser-383.
Immunolocalization of Striatin, SG2NA, and HA-mMOB1
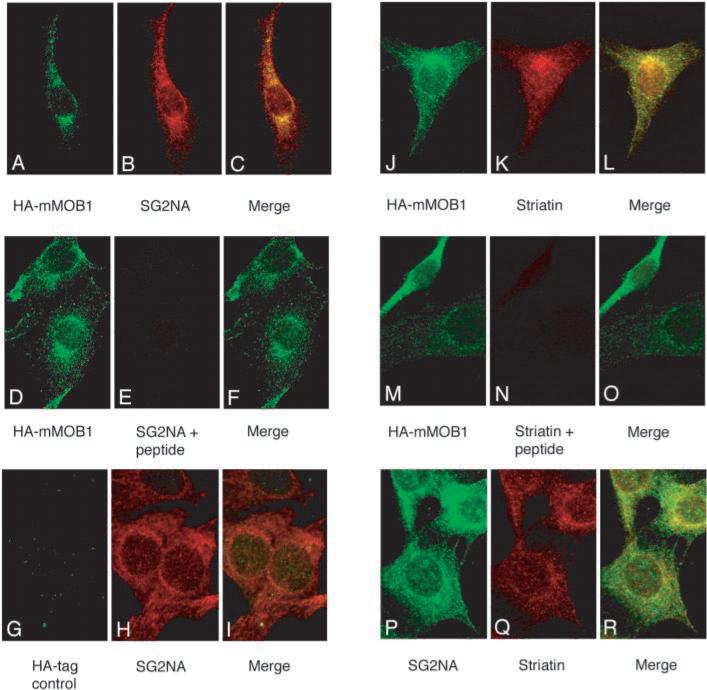
To investigate the subcellular localization of striatin, SG2NA, and HA-mMOB1, NIH3T3 and HA-mMOB1 cells were examined by indirect immunofluorescence (Fig. 6). Affinity-purified polyclonal rabbit antibodies against striatin, a monoclonal mouse antibody (16b12) that recognizes HA-tagged mMOB1, and both monoclonal and polyclonal antibodies to an identical peptide in SG2NA were used to study the subcellular distribution of striatin, SG2NA, and HA-mMOB1. Punctate staining was observed for all three proteins along the cellular membrane and throughout the cytoplasm, with high concentrations close to the nuclear membrane, possibly associated with the endoplasmic reticulum, golgi apparatus, or even the centrosome. Panels A, B, and C of Fig. 6 show the localization observed in HA-mMOB1 cells for HA-mMOB1 (green), SG2NA (red), and the merged images, respectively. The overall pattern of SG2NA and HA-mMOB1 fluorescence was highly similar for the two proteins; a subpopulation of SG2NA and HA-mMOB1 appear to colocalize, consistent with the observation that approximately one-third of cellular mMOB1 co-immunoprecipitates with SG2NA. The SG2NA staining pattern observed with both monoclonal (panel P) and polyclonal (panels B and H) antibodies to SG2NA showed an overall pattern very similar to that of HA-mMOB1 (panels A, D, J, and M) and showed the highest concentration of colocalization with HA-mMOB1 around the nuclear periphery (panel C). Panels D, E, and F show the fluorescence observed in HA-mMOB1 cells co-stained with the anti-HA monoclonal (16b12) antibody and affinity-purified SG2NA antibodies preincubated with SG2NA peptide. The lack of signal in panel E and the presence of an HA-mMOB1 signal in panel D indicate that the SG2NA signal is specific. Panels G, H, and I show the fluorescence observed in the parent NIH3T3 cell line that does not express HA-mMOB1 stained with 16b12 and SG2NA antibodies. The lack of signal in panel G demonstrates the specificity of the HA-mMOB1 signal in panels A, D, J, and M.
Fig. 6. A subpopulation of striatin, SG2NA, and HA-mMOB1 appear to colocalize around the nuclear periphery.
Murine fibroblasts were fixed in 2% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and stained for the presence of HA-mMOB1 (panels A, D, G, J, and M), SG2NA (panels B, E, H, and P), or striatin (panels K, N, and Q). Each set of three panels indicates the fluorescence observed using mouse monoclonal antibodies shown in green, rabbit polyclonal antibodies shown in red, and a third panel indicating the merge of the two signals. Panels A, B, and C are co-stained with 16b12 and affinity-purified SG2NA antibodies. Panels D, E, and F show the fluorescence observed in HA-mMOB1 cells co-stained with the 16b12 antibody and affinity-purified SG2NA antibodies preincubated with 2 μg/ml SG2NA peptide. The lack of signal in panel E and the presence of a HA-mMOB1 signal in panel D indicate that the SG2NA signal is specific. All panels used HA-mMOB1 cells except panels G, H, and I, which used the parental NIH3T3 cell line, demonstrating the specificity of the HA-mMOB1 signal. Panels J, K, and L are co-stained with 16b12 and affinity-purified striatin antibodies. In panels M, N, and O, the 16b12 and striatin antibodies were preincubated with 2 μg/ml striatin peptide. Panels P, Q, and R are co-stained with a monoclonal antibody to SG2NA (green) and affinity-purified striatin antibodies (red).
Co-staining of HA-mMOB1 with striatin is shown in panels J, K, and L. The striatin pattern indicated that striatin is present in the same regions of the cell as SG2NA and mMOB1 but was also present in much greater abundance in the nucleus. Confocal microscopy of single 0.4-μm thick sections through cell nuclei confirmed the presence of nuclear striatin (data not shown). The specificity of the striatin signal is demonstrated in panels M, N, and O, in which the 16b12 and striatin antibodies were preincubated with striatin peptide. Incubation of coverslips with secondary antibodies alone resulted in no observable signal (data not shown).
The observations that striatin could coimmunoprecipitate small amounts of SG2NA and vice versa (Fig. 1) as well as the similar staining pattern observed using polyclonal antibodies to striatin and SG2NA led us to investigate whether we could observe colocalization of these two related proteins. Using a monoclonal antibody to SG2NA (panel P) and affinity-purified polyclonal antibodies to striatin (panel Q), a subpopulation of striatin and SG2NA did appear to colocalize (panel R), particularly in the perinuclear region of these cells.
DISCUSSION
Here we have shown that mMOB1, the class II mammalian homolog of the yeast protein MOB1, physically associates with both striatin-PP2A and SG2NA-PP2A complexes and may be a PP2A substrate. Previous studies of MOB1 in both fission and budding yeast have demonstrated that MOB1 is a critical downstream target of the mitotic exit checkpoint (reviewed in Ref. 23). Although MOB1 has been shown to interact genetically with CDC5, CDC15, and LTE1 (10), it is not known if MOB1 interacts genetically with PP2A in yeast. Additional molecular evidence (12, 24) indicates that in yeast, MOB1 and the DBF2 kinase act downstream of a TEM1-LTE1-BUB2 pathway that controls cytokinesis and exit from mitosis (23, 25). When the spindle pole migrates into the daughter cell, the GTP exchange factor LTE1 (26) activates the GTPase TEM1 (23), which in turn activates the DBF2 kinase. Activated DBF2 binds to and phosphorylates MOB1 (20). Although MOB1 is probably not required for mitotic exit in S. pombe, MOB1 was shown to be essential for initiation of cytokinesis (11). MOB1 is localized to the spindle pole bodies in both budding and fission yeast during most of the cell cycle until cytokinesis, at which time both MOB1 and DBF2 relocalize to the bud neck in S. cerevisiae (24) and the medial ring in S. pombe (12). These data are suggestive of a model in which phosphorylated MOB1 facilitates cytoskeletal changes essential for initiation of cytokinesis and contraction of the medial ring.
When MOB1 was first cloned in S. cerevisiae (10), the authors noted that there were two classes of MOB1 homologs in higher eukaryotes. Thus, it is likely that the MOB1 gene was duplicated during evolution and that the two classes of MOB1 homologs diverged to carry out different function of yeast MOB1. Although we have identified the mammalian class II homolog of MOB1 as a member of striatin-PP2A and SG2NA-PP2A complexes, we do not yet have evidence what various functions the class I and class II homologs of MOB1 may perform in metazoans. It will be of keen interest to determine whether one or the other or both classes of MOB1 homologs are important for mitotic progression in mammals.
At the okadaic acid concentrations used to examine mMOB1 phosphorylation (0.5–1.0 μm), mammalian cells typically attempt to enter mitosis prematurely. Cells treated in this manner undergo morphologically normal chromosome condensation, nuclear lamina depolymerization, and centrosome separation in the absence of Cdc2 kinase activity (27). Additionally, cytoplasmic microtubules depolymerize, and the cells round up and become detached from the tissue culture plates. Okadaic acid can also arrest cells in mitosis by preventing the metaphase to anaphase transition (28). Thus, mMOB1 phosphorylation upon okadaic acid treatment could be a reflection of mitotic events rather than simply inhibition of PP2A.
During cytokinesis in mammalian cells, Rho-associated kinase phosphorylates intermediate filament proteins such as vimentin (29) and glial fibrillary acidic protein (GFAP) (30) at the medial ring. However, in preliminary immunofluorescence studies, the overall patterns of mMOB1, striatin, and SG2NA staining are not strikingly similar to those of microtubules, vimentin, or actin filaments.4 Although a subpopulation of SG2NA, striatin, and mMOB1 may be bound to filaments either along the cellular membrane or at the nuclear periphery, we were not able to clearly confirm or exclude this observation using the indirect immunofluorescence assay due to the high density of cytoskeletal fibers.
The hypothesis that striatin-PP2A-mMOB1 complexes function to regulate cytoskeletal changes is consistent with several lines of data taken from experiments in neuronal tissue. For example, striatin is highly abundant in the postsynaptic densities of neurons in rat brains (6, 8), and striatin antisense experiments in rat neuronal cell culture (9) resulted in a decrease in the number of observed neurites. Recently, it has been shown that Rho kinase phosphorylation of vimentin on Ser-71 and Ser-38 results in neurite retraction (31), whereas Rho kinase inhibitors produce irregular neurite outgrowth. Moreover, 20 nm okadaic acid treatment, which specifically inhibits PP2A, induces neurite retraction in neuroblastoma N2a cells (31). PP2A-specific concentrations of okadaic acid have also been shown to inhibit neurite outgrowth (32) and to induce axonal growth cone collapse (33) and axonal filopodial shortening (34). Finally, when a PP2A C subunit mutant (Y307E) that binds preferentially to striatin and SG2NA (35) is transfected into neuroblastoma cells, these cells form highly elongated, undifferentiated structures characteristic of hyperstabilized cytoskeletons.5 These data are consistent with a model in which striatin-PP2A-mMOB1 complexes function in neurons to maintain cytoskeletal structure and prevent neurite retraction. Although the B55 subunit of PP2A has been shown to target the A/C heterodimer toward vimentin (36), vimentin is phosphorylated at multiple sites (29), including cdc2 phosphorylation sites (37). We have shown previously that striatin and SG2NA, like Bα, can activate the A/C heterodimer toward cdc2-phosphorylated substrates (4). Thus, both the striatin family and the B55 family of PP2A subunits may play roles in regulation of vimentin phosphorylation.
The observation that the striatin-PP2A-mMOB1 and SG2NA-PP2A-mMOB1 complexes may interact with each other is intriguing. Because striatin and SG2NA have multiple protein-protein interaction motifs as well as a caveolin binding motif and membrane binding domains, one can envision a model in which these complexes could act as bridges to bring together cytoskeletal and membrane structures. PP2A might regulate such interactions or complex assembly by modulating the phosphorylation state of members of these complexes. Identification of additional members of these complexes and further analysis of the role of PP2A in them will likely provide new insights into the how these complexes may impact regulation of cell morphology and cell division.
Acknowledgments
We thank Danita Ashby, Marie Kozel, and Sameer Patel for technical assistance and Renee Robinson, Dan Kirby, and Kerry Pierce of the Harvard Microchemistry Facility for their expertise in high pressure liquid chromatography and mass spectrometry. We also thank Dr. Anita Corbett for critical reading of the manuscript. Finally, we thank Drs. Victoria Stevens, Egon Ogris, and Enrique Torres for technical advice. Under agreements between Upstate Biotechnology, Inc. and Emory University and Calbiochem and Emory University, David Pallas is entitled to a share of sales royalty received by the University from these companies. In addition, this same author serves as a consultant to Upstate Biotechnology, Inc. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict of interest policies.
Footnotes
This work was supported by National Institutes of Health Grant CA57327. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: PP2A, protein phosphatase 2A; SG2NA, S/G2 nuclear autoantigen, PBS, phosphate-buffered saline, BSA, bovine serum albumin; mMOB1, mammalian homolog MOB1; EST, expressed sequence tag; HA, hemagglutinin; KLH, keyhole limpet hemocyanin; PAGE, polyacrylamide gel electrophoresis.
T. Fellner and E. Ogris, unpublished information.
K. Conroy and D. C. Pallas, unpublished data.
Note Added in Proof—Concurrent with this study, Baillat et al. (43) identified the rat homolog of mMOB1, which they designated phocein. Using different approaches and reagents, they observed similar subcellular localization of mMOB1 and also showed that mMOB1 is a binding partner of striatin and SG2NA.
C. S. Moreno and D. C. Pallas, unpublished observations.
E. Sontag, personal communication.
REFERENCES
- 1.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Mol. Cell. Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li M, Makkinje A, Damuni Z. J. Biol. Chem. 1996;271:11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- 3.Wang SW, Esplin ED, Li JL, Huang L, Gazdar A, Minna J, Evans GA. Science. 1998;282:284–287. doi: 10.1126/science.282.5387.284. [DOI] [PubMed] [Google Scholar]
- 4.Moreno CS, Park S, Nelson K, Ashby DG, Hubalek F, Lane WS, Pallas DC. J. Biol. Chem. 2000;275:5257–5263. doi: 10.1074/jbc.275.8.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muro Y, Chan EK, Landberg G, Tan EM. Biochem. Biophys. Res. Commun. 1995;207:1029–1037. doi: 10.1006/bbrc.1995.1288. [DOI] [PubMed] [Google Scholar]
- 6.Castets F, Bartoli M, Barnier JV, Baillat G, Salin P, Moqrich A, Bourgeois JP, Denizot F, Rougon G, Calothy G, Monneron A. J. Cell Biol. 1996;134:1051–1062. doi: 10.1083/jcb.134.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castets F, Rakitina T, Gaillard S, Moqrich A, Mattei MG, Monneron A. J. Biol. Chem. 2000;275:19970–19977. doi: 10.1074/jbc.M909782199. [DOI] [PubMed] [Google Scholar]
- 8.Salin P, Kachidian P, Bartoli M, Castets F. J. Comp. Neurol. 1998;397:41–59. [PubMed] [Google Scholar]
- 9.Bartoli M, Ternaux JP, Forni C, Portalier P, Salin P, Amalric M, Monneron A. J. Neurobiol. 1999;40:234–243. [PubMed] [Google Scholar]
- 10.Luca FC, Winey M. Mol. Biol. Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou MC, Salek J, McCollum D. Curr. Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- 12.Salimova E, Sohrmann M, Fournier N, Simanis V. J. Cell Sci. 2000;113:1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- 13.Ogris E, Du X, Nelson KC, Mak EK, Yu XX, Lane WS, Pallas DC. J. Biol. Chem. 1999;274:14382–14391. doi: 10.1074/jbc.274.20.14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- 15.Ogris E, Gibson DM, Pallas DC. Oncogene. 1997;15:911–917. doi: 10.1038/sj.onc.1201259. [DOI] [PubMed] [Google Scholar]
- 16.Lauze E, Stoelcker B, Luca FC, Weiss E, Schutz AR, Winey M. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schutz AR, Winey M. Mol. Biol. Cell. 1998;9:759–774. doi: 10.1091/mbc.9.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schutz AR, Giddings TH, Jr., Steiner E, Winey M. J. Cell Biol. 1997;136:969–982. doi: 10.1083/jcb.136.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss E, Winey M. J. Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komarnitsky SI, Chiang YC, Luca FC, Chen J, Toyn JH, Winey M, Johnston LH, Denis CL. Mol. Cell. Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favre B, Turowski P, Hemmings BA. J. Biol. Chem. 1997;272:13856–13863. doi: 10.1074/jbc.272.21.13856. [DOI] [PubMed] [Google Scholar]
- 22.Yan Y, Mumby MC. J. Biol. Chem. 1999;274:31917–31924. doi: 10.1074/jbc.274.45.31917. [DOI] [PubMed] [Google Scholar]
- 23.Hoyt MA. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- 24.Frenz LM, Lee SE, Fesquet D, Johnston LH. J. Cell Sci. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- 25.Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. Mol. Biol. Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. Mol. Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- 27.Gowdy PM, Anderson HJ, Roberge M. J. Cell Sci. 1998;111:3401–3410. doi: 10.1242/jcs.111.22.3401. [DOI] [PubMed] [Google Scholar]
- 28.Vandre DD, Wills VL. J. Cell Sci. 1992;101:79–91. doi: 10.1242/jcs.101.1.79. [DOI] [PubMed] [Google Scholar]
- 29.Goto H, Kosako H, Tanabe K, Yanagida M, Sakurai M, Amano M, Kaibuchi K, Inagaki M. J. Biol. Chem. 1998;273:11728–11736. doi: 10.1074/jbc.273.19.11728. [DOI] [PubMed] [Google Scholar]
- 30.Yasui Y, Amano M, Nagata K, Inagaki N, Nakamura H, Saya H, Kaibuchi K, Inagaki M. J. Cell Biol. 1998;143:1249–1258. doi: 10.1083/jcb.143.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura Y, Hashimoto R, Amano M, Nagata KI, Matsumoto N, Goto H, Fukusho E, Mori H, Kashiwagi Y, Kudo T, Inagaki M, Takeda M. Genes Cells. 2000;5:823–837. doi: 10.1046/j.1365-2443.2000.00372.x. [DOI] [PubMed] [Google Scholar]
- 32.Giasson BI, Mushynski WE. J. Neurobiol. 1997;32:193–201. doi: 10.1002/(sici)1097-4695(199702)32:2<193::aid-neu4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama T, Goshima Y, Misu Y, Kato T. J. Neurobiol. 1999;41:326–339. [PubMed] [Google Scholar]
- 34.Cheng S, Mao J, Rehder V. Cell Motil. Cytoskeleton. 2000;47:337–350. doi: 10.1002/1097-0169(200012)47:4<337::AID-CM7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 35.Yu XX, Du X, Moreno CS, Green RE, Ogris E, Feng Q, Chou L, McQuoid MJ, Pallas DC. Mol. Biol. Cell. 2001;12:185–199. doi: 10.1091/mbc.12.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turowski P, Myles T, Hemmings BA, Fernandez A, Lamb NJ. Mol. Biol. Cell. 1999;10:1997–2015. doi: 10.1091/mbc.10.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ando S, Ikuhara T, Kamata T, Sasaki Y, Hisanaga S, Kishimoto T, Ito H, Inagaki M. J. Biochem. (Tokyo) 1997;122:409–414. doi: 10.1093/oxfordjournals.jbchem.a021768. [DOI] [PubMed] [Google Scholar]
- 38.Pallas D, Solomon F. Cell. 1982;30:407–414. doi: 10.1016/0092-8674(82)90238-0. [DOI] [PubMed] [Google Scholar]
- 39.Temeles GL, Ram PT, Rothstein JL, Schultz RM. Mol. Reprod. Dev. 1994;37:121–129. doi: 10.1002/mrd.1080370202. [DOI] [PubMed] [Google Scholar]
- 40.Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. Genome Res. 2000;10:1617–1630. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, et al. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 42.Lai CH, Chou CY, Chang LY, Liu CS, Lin W. Genome Res. 2000;10:703–713. doi: 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baillat G, Moqrich A, Castets F, Baude A, Bailly Y, Benmerah A, Monneron A. Mol. Biol. Cell. 2001;12:663–673. doi: 10.1091/mbc.12.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]