Abstract
Background:
Diabetes mellitus has become a global epidemic illness and poses a threat for development of resistant bacterial infections.
Aim:
This study was aimed to know the bacteriological and resistance profile of isolates obtained from diabetic patients.
Materials and Methods:
The bacterial isolates obtained from various samples of diabetic patients admitted in medicine department in 6-month period were identified and tested for antibiotic susceptibility. The extended spectrum beta-lactamases (ESβL), AmpC, and metallo-beta-lactamases (MβL) enzymes were detected in gram-negative bacilli. Methicillin, macrolide-lincosamide-streptogramin (MLS), and linezolid resistance in Staphylococcus spp. were detected. High-level aminoglycoside resistance (HLAR) in Enterococcus spp. was also tested.
Results:
In all, 38 of 125 diabetic patients (30.4%) had bacterial infection, 18 patients had wound infections, 18 had urinary tract infections (UTIs), and 2 had respiratory tract infections. Escherichia coli among gram-negative bacteria and Staphylococcus aureus among gram-positive bacteria were the predominant pathogens. 32.5% gram-negative bacilli were AmpC producers, 37.5% were MβL producers, and 40% were ESβL producers. Methicillin and MLS resistance was found in 50% and 33.3% isolates of Staphylococcus spp., respectively. HLAR resistance was alarming in Enterococcus spp. Polymyxin among gram-negative bacteria and vancomycin for gram-positive bacteria were the last resort with highest susceptibility rates to treat infections among diabetic patients.
Conclusion:
Resistant bacterial infections in diabetic patients are common. The presence of various resistance mechanisms in isolates of our study shows that therapeutic failure can occur if empirical prescription is unsubstantiated.
Keywords: Antibiotic resistance, Diabetes mellitus, Therapeutic failure
Introduction
Diabetes mellitus has become a global epidemic illness[1] and poses a threat for development of resistant bacterial infections. Diabetic patients are more prone to life-threatening infections than nondiabetic patients;[2–4] therefore, they have more exposure to antibiotics. Diabetic patients have greater problems with healing of infections because of reduced blood supply, which affects the body's ability to fight infection.[5] When a diabetic patient contracts pneumonia, wound infections, and urinary tract infections, the illness is often more frequent than in nondiabetic patients.[2,5] The studies discussing the profile and resistance mechanisms in isolates obtained from diabetic patients are regularly needed to decide empirical therapies in such high-risk patients.
Therefore, this study was attempted to know the bacteriological profile and associated resistance in isolates from diabetic patients.
Materials and Methods
This prospective study was conducted on pus, urine, and sputum/blood samples obtained from consecutive diabetic patients (n=125) admitted to Medicine Department of Govt. Medical College and Sushila Tiwari Hospital, Haldwani, Uttarakhand, from January to June 2009. The mean age of diabetic patients was 42.3 years, and sex ratio male:female was 1.4:1. The patients included in the study were those of type 2 diabetes mellitus who had poor control of glycemia (assessed on basis of repeated fasting plasma glucose levels), history of infection, hospitalization, and antibiotics exposure (either complete or incomplete course) in last 3 months putting them at increased risk of wound infection, urinary tract infection (UTI), and pneumonia.
Thirty-five pus (25 pedal wounds and 10 nonpedal wounds), 65 urine, and 25 sputum/blood samples were collected and transported within 30 minutes to microbiology laboratory. Pus samples were cultured on blood agar and MacConkey agar plates, blood/sputum on blood and chocolate agar, and urine was cultured on Cysteine lactose electrolyte deficient (CLED) medium as per standard protocol.[6] The bacterial isolates obtained from various specimens were identified[6] and tested for antimicrobial susceptibility testing as per our institutional antibiotic policy for both first line and second line of antibiotics (Hi Media Laboratories, India) by Kirby Bauer disc diffusion method.[7] Multidrug resistance was defined as resistance to three or more groups of drugs.
Wound infection was defined as evidence of microorganism(s) from pus culture.[8] A diagnosis of pneumonia was made by sputum culture and by isolation of a compatible organism from blood.[6] Presence of organism(s) of at least 105 cfu/mL urine specimens with clinical symptoms indicated UTI.[6]
Detection of resistance mechanisms by phenotypic methods
ES0βL, AmpC, and MβL detection
The gram-negative bacilli leading to resistance for third-generation cephalosporins, cephamycins, and carbapenems on routine screening were also phenotypically tested for detection of extended spectrum beta- lactamase (ESβL), AmpC, and metallo-beta-lactamase (MβL) enzymes, respectively. Briefly, Mueller-Hinton agar plates were prepared and inoculated with standardized inoculums to form a lawn culture. The combined disc methods were used to confirm above resistance mechanisms as described elsewhere.[9–11] Discs of ceftazidime (30 μg) and ceftazidime clavulanate (30/10 μg) for ESβL detection, cefoxitin, and cefoxitin with cloxacillin (30 + 500 μg, prepared in house) for AmpC and imipenem (30 μg) and imipenem with Ethylene diaminetetra acetic acid (EDTA) (30 μg+5 μg, prepared in house) for MβL detection were used. The increase in inhibition zone of ceftazidime clavulanate disc ≥5 mm than the ceftazidime disc alone was considered ESβL positive.[9] If the increase in inhibition zone with cefoxitin and cloxacillin disc was ≥5 mm than the cefoxitin disc alone, it was considered AmpC positive (slight modification of Ruppé et al.).[10] The increase in inhibition zone with imipenem and EDTA disc ≥7 mm than the imipenem disc alone was considered MβL positive.[11]
Methicillin resistance
The methicillin resistance in Staphylococcus spp. was tested by cefoxitin disc (30 μg) as documented in Clinical and Laboratory Standard Institute (CLSI).[12] Cefoxitin disc diffusion of ≤21 mm for S. aureus and ≤24 mm for coagulase negative Staphylococcus (CNS) was reported as methicillin resistant.
Linezolid resistance
Agar dilution method was done to confirm linezolid resistance (MIC ≥8 μg/ml) in Staphylococcus spp.[13]
High-level aminoglycoside resistance
High content gentamicin disc (120 μg) was used to detect high-level aminoglycoside resistance (HLAR) in Enterococcus spp. The whole zone diameter of ≤6 mm was considered resistant.[14]
Macrolide–lincosamide–streptogramin resistance
Staphylococcus spp. with erythromycin resistance (ER-R) were tested for inducible clindamycin resistance using D test keeping 15-mm inter-disc distance as per CLSI recommendation. Briefly, erythromycin (15 μg) disc was placed at distance of 15 mm (edge to edge) from clindamycin dics (2 μg) on Mueller-Hinton agar plate previously inoculated with 0.5 McFarland bacterial suspensions. Following overnight incubation at 37°C, flattening of zone around clindamycin in area between the two discs indicated inducible clindamycin resistance[15]. In our study, two different phenotypes were appreciated after testing as follows:
MLSB inducible phenotype (MLSBi): Isolates resistant to erythromycin while being sensitive to clindamycin and giving D-shaped zone of inhibition around clindamycin with flattening toward erythromycin disc.
MLSB constitutive phenotype (MLSBc): Isolates resistant to both clindamycin and erythromycin.
Results
Of 125 diabetic patients, 38 (30.4%) were infected. The male:female ratio of infections was 25:13 (1.92:1). Of 35 patients, 18 (51.4%) had wound infections, 18 of 65 (27.7%) had UTI, and 2 of 25 (8%) had pneumonia. No patient had concomitant infection.
Sixty-three bacterial species were isolated from 38 samples. Of 63 bacterial isolates, 40 (63.5%) were gram-negative and 23 (36.5%) were found to be gram-positive. Two bacterial species were isolated from sputum samples (n=2; monomicrobial infection), 26 bacterial species from urine samples (n=18; 10 were monomicrobial infection while 8 were bimicrobial), while 35 bacterial species were isolated from pus sample (n=18; 3 were monomicrobial infections, 13 were bimicrobial, and 2 were polymicrobial with 3 organisms in each sample). Table 1 shows the bacteriological profile of isolates.
Table 1.
Profile and frequency distribution of gram-positive and gram-negative bacterial isolates obtained from various sites
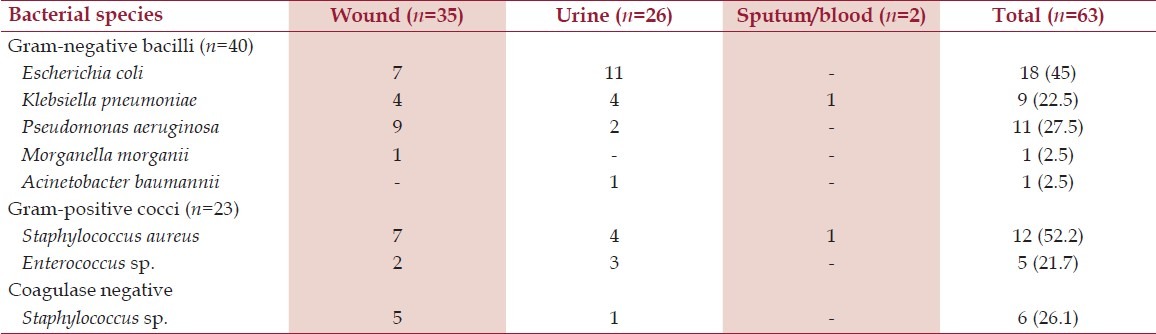
The resistance rates of gram-negative bacilli and gram-positive cocci to various antibiotics are shown in Table 2. The various drug-resistance mechanisms investigated among gram-negative and gram-positive bacteria are shown in Table 3.
Table 2.
Antibiotic-resistance rates of various bacterial isolates obtained from diabetic patients (n=63)
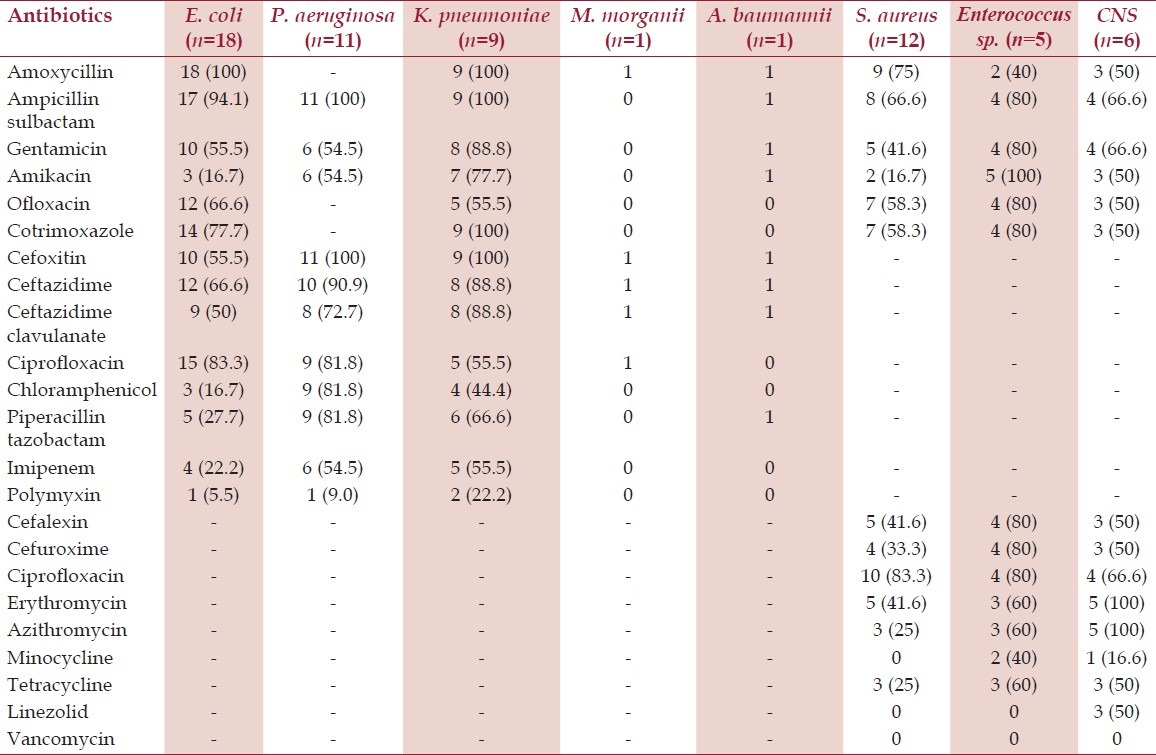
Table 3.
Detection of various drug-resistance mechanisms in bacteria isolated from diabetic patients

Discussion
In all, the prevalence of infections in diabetic patients was 30.4% with an average of 1.65 organisms per case as compared with nondiabetic population having infection rates of 21.2% with an average 1.2 organisms per case (Singhai, unpublished data). To have further insight, the infection rates and average number of organisms in diabetic and nondiabetic population in various infections have been compared in Table 4. A study on diabetic wound infections has reported 1.52 organism per case,[16] while another reported 2.3 organism per case;[17] however, in our study, 1.9 organism per case was found from wound infections. This reinforces the concept of polymicrobial infection in diabetic patients and was seen in our study also.
Table 4.
Infection rates and average number of organisms in diabetic and nondiabetic patients with various infection

The number of gram-negative as compared with gram-positive bacterial isolates was high in our study. E. coli among gram-negative and S. aureus among gram-positive cocci were the most common pathogens. Other studies from India have also reported gram-negative bacilli as predominant pathogen in diabetic infections.[4,16,17] The anaerobic bacteria also form major pathogens of such infections but could not be assessed because of lack of facilities. Beta-hemolytic Streptococcus spp., an important pathogen of wound infections ecology in diabetic patients was not found in our study, which may be attributed to antibiotic usage for infections in past.
The overall susceptibility rates to various antibiotics especially amoxicillin, third-generation cephalosporins, fluoroquinolones tested among gram-negative bacilli and macrolides, amoxycillin, first-generation cephalosporins, fluoroquinolones among gram-positive cocci were low. The other studies on infections among diabetic patients have also reported high resistance rates to various antibiotics among gram-negative and gram-positive bacteria.[18–20] Multidrug resistance was found in 82.5% (33/40) among gram-negative and 60.8% (14/23) in gram-positive bacteria.
Another interesting finding of this study was low resistance level to piperacillin/tazobactam and the high resistance level to ceftazidime/clavulanate, which may be attributed either to hyperproduction of AmpC or presence of inhibitor-resistant beta-lactamases (especially in E. coli) masking the detection of ESβL by clavulanate but remaining susceptible to inhibition by tazobactam which is in accordance with Bradford et al.[21] When we probed in-depth beta-lactams resistance, 32.5% were AmpC producers, 37.5% were MβL producers, and 40% were ESβL producers among gram-negative bacilli. In our hospital, we have found presence of AmpC, MβL, and ESβL enzymes (28%, 33.5%, and 35%) in gram-negative bacterial strains obtained from clinical specimens of nondiabetic patients slightly low as compared with diabetic patients (Rawat, unpublished data). The focus on detection of the forementioned resistance mechanisms despite of increasing reports on the cephalosporin, cephamycin, and carbapenem-resistance genes from various parts of our country was due to presence of ESβL, AmpC, and MβL in our area as reported by Shahid et al.;[22] however, we could not confirm the molecular epidemiology because of lack of molecular setup. The concurrent mechanisms of resistance occurred only in four isolates. Each of the two Klebsiella pneumoniae were ESβL and AmpC coproducers, while one E. coli and one Pseudomonas aeruginosa, each produced AmpC and MβL enzymes simultaneously [Figure 1]. AmpC, ESβL, and MβL producing strains were high in our study population and were in accordance to studies on gram-negative bacilli isolated from diabetic patients.[23] The major concern is emergence of resistance to third- and fourth-generation of cephalosporins and even more alarming is carbapenem-resistance surfacing nowadays.[24,25] The carbapenems are often considered a “drug of choice” and are increasingly used in empirical therapy.[25] However, recent emergence of resistance to this group of antibiotic leaves little options for treating such life-threatening infections as seen in our study group.
Figure 1.
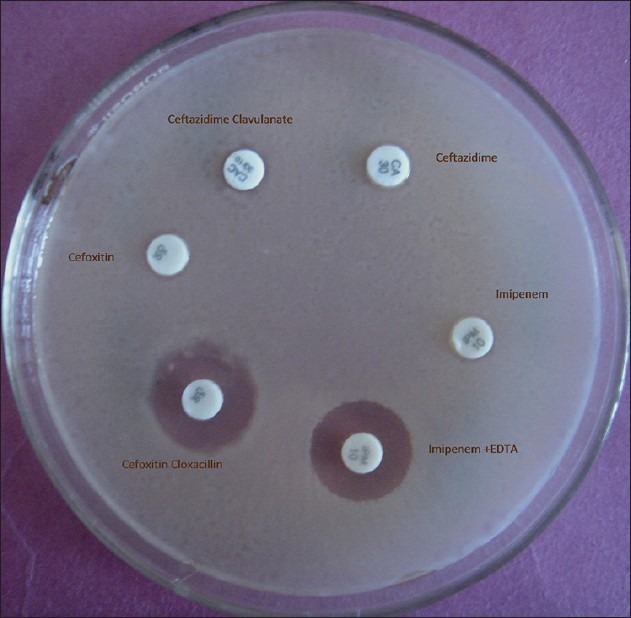
Detection of AmpC and MBL production in E. coli
The excellent coverage and good oral bioavailability of clindamycin makes it effective therapy against infections in diabetic patients by Staphylococcus spp.[26] However, important issue with clindamycin use is a risk of clinical failure during therapy due to inducible clindamycin resistance.[26] MLS resistance was found in 33.3% of S. aureus and CNS. In our previous study, 38.4% MLS resistance was found in gram-positive cocci.[15] Failure to identify MLSBi may lead to clinical failure of clindamycin therapy, and therefore, its detection is advisable. The true incidence of clindamycin resistance depends on the patient population studied and needs to be assessed to guide the clinicians.[26] Among Staphylococcus spp., methicillin resistance was found in 50% S. aureus and 50% CNS, while 50% CNS were linezolid resistant. Methicillin-resistant Staphylococcus spp. (MRS) strains with intrinsic resistance to methicillin and all beta-lactam antibiotics are emerging pathogens in hospitalized diabetic patients.[27] Linezolid, a novel synthetic antibiotic, is considered one of the few effective ways to treat severe methicillin-resistant infections in critically ill patients. Recently, linezolid resistance is reported in our country which may be a consequence of prolonged and injudicious use of this drug[27,28] and is an imperative threat to lose an effective and safe drug for treating MRS. HLAR in Enterococcus spp. was also alarming (60%) in our study.
Polymyxin among gram-negative bacilli and vancomycin among gram-positive cocci were the last resorts found to treat multidrug-resistant infections. Our study warrants an urgent need of screening of antibiotic resistance in high-risk population and queries unsubstantiated empirical prescription of antibiotics, which is a common practice in our country.
The detection of various resistance mechanisms by phenotypic methods are easy to interpret, reproducible, and inexpensive and can be included as routine testing protocol. The routine reporting of resistant phenotypes in a target population would allow the clinician to re-valuate their empirical therapy polices.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
References
- 1.Devraj S, Goyal R, Jialal I. London: Touch Briefings; 2008. Inflammation, oxidative stress and metabolic syndrome, US endocrinology; pp. 32–7. [Google Scholar]
- 2.Muller LM, Gorter KJ, Hak E, Goudzwaard WL, Schellevis FG, Hoepelman AI, et al. Increased risk of common infections in patients with type 1 and type 2 diabetes mellitus. Clin Infect Dis. 2005;41:281–8. doi: 10.1086/431587. [DOI] [PubMed] [Google Scholar]
- 3.Marble A, White HJ, Fernald AT. The nature of the lowered resistance to infection in diabetes mellitus. J Clin Invest. 1938;17:423–30. doi: 10.1172/JCI100969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zubair M, Abida M, Jamal A. Clinicobacteriology and risk factors for the diabetic foot infection with multidrug resistant microorganisms in North India. Biol Med. 2010;2:22–34. [Google Scholar]
- 5.Luo G, Spellberg B, Gebremariam T, Bolaris M, Lee H, Fu Y, et al. Diabetic murine models for Acinetobacter baumannii infection. J Antimicrob Chemother. 2012;67:1439–45. doi: 10.1093/jac/dks050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colle JG, Miles RS, Watt B. Tests for the identification of bacteria. In: Collee JG, Marmion BP, Fraser AG, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology. 14th ed. New Delhi: Churchill Livingstone; 2006. pp. 131–45. [Google Scholar]
- 7.Performance standards for antimicrobial susceptibility test. no. 1. Vol. 1. Pennsylvania, USA: Clinical and Laboratory Standard Institute; 2007. Clinical and Laboratory Standard Institute. M2 A9. [Google Scholar]
- 8.Apelqvist J, Bakker K, van Houtum WH, Schaper NC. The development of global consensus guidelines on the management of the diabetic foot.Prepared by the International Working Group on the Diabetic Foot. Diabetes Metab Res Rev. 2008;2(Suppl 1):S116–93. doi: 10.1002/dmrr.832. [DOI] [PubMed] [Google Scholar]
- 9.Screening and confirmatory test for ESBLs. no. 1. Vol. 27. Pennsylvania, USA: Clinical and laboratory Standard Institute; 2007. Clinical and Laboratory Standard Institute. M2 A9. [Google Scholar]
- 10.Ruppé E, Bidet P, Verdet C, Arlet G, Bingen E. First detection of the Ambler class C 1 AmpC beta-lactamase in Citrobacter freundii by a new, simple double-disk synergy test. J Clin Microbiol. 2006;44:4204–7. doi: 10.1128/JCM.00918-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawat V. Study of mettalo-β-talactamses production in noscomial nil fermenter from gram-negative bacilli in a tertiary care centre. Int J App Basic Med Res. 2011;1:139–40. doi: 10.4103/2229-516X.91166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898. no. 1. Vol. 27. USA: Clinical and laboratory Standard Institute; 2007. Clinical and Laboratory Standard Institute. Disc diffusion test for prediction of mec A mediated resistance in Staphylococci; sixteenth informational supplement. M2 A9. [Google Scholar]
- 13.940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898. no. 1. Vol. 27. USA: Clinical and laboratory Standard Institute; 2007. Clinical and Laboratory Standard Institute. MIC interpretive standards (μg/ml) for Staphylococcal spp. M2 A9. [Google Scholar]
- 14.940 West Valley Road, Suite 1400, Wayne, Pennysylvania 19087-1898. no. 1. Vol. 27. USA: Clinical and Laboratory Standard Institute; 2007. Clinical and Laboratory Standard Institute Screening test for high level aminoglycoside resistance. M2 A9. [Google Scholar]
- 15.Rawat V. Inducible clindamycin resistance among gram-positive cocci in a tertiary care centre. J Pure Appl Microbiol. 2012;6:455–8. [Google Scholar]
- 16.Bansal E, Garg A, Bhatia S, Attri AK, Chander J. Spectrum of microbial flora in diabetic foot ulcers. Indian J Pathol Microbiol. 2008;51:204–8. doi: 10.4103/0377-4929.41685. [DOI] [PubMed] [Google Scholar]
- 17.Gadepalli R, Dhawan B, Sreenivas V, Kapil A, Ammini AC, Chaudhry R. A clinico-microbiological study of diabetic foot ulcers in an Indian tertiary care hospital. Diabetes Care. 2006;29:1727–32. doi: 10.2337/dc06-0116. [DOI] [PubMed] [Google Scholar]
- 18.Motta RN, Oliveira MM, Magalhães PS, Dias AM, Aragão LP, Forti AC, et al. Plasmid-mediated extended-spectrum beta-lactamase-producing strains of Enterobacteriaceae isolated from diabetes foot infections in a Brazilian diabetic center. Braz J Infect Dis. 2003;7:129–34. doi: 10.1590/s1413-86702003000200006. [DOI] [PubMed] [Google Scholar]
- 19.Varaiya A, Kulkarni M, Bhalekar P, Dogra J. Incidence of metallo-beta-lactamase-producing Pseudomonas aeruginosa in diabetes and cancer patients. Indian J Pathol Microbiol. 2008;51:200–3. doi: 10.4103/0377-4929.41683. [DOI] [PubMed] [Google Scholar]
- 20.Sotto A, Lina G, Richard JL, Combescure C, Bourg G, Vidal L, et al. Virulence potential of Staphylococcus aureus strains isolated from diabetic foot ulcers: A new paradigm. Diabetes Care. 2008;31:2318–24. doi: 10.2337/dc08-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford PA. Extended-spectrum beta-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–51. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahid M, Umesh, Sobia F, Singh A, Khan HM, Malik A, et al. Molecular epidemiology of carbapenem resistant Enterobacteriaceae from a North Indian Tertiary Hospital. NZ J Med Lab Sci. 2012;66:5–7. [Google Scholar]
- 23.Umadevi S, Kumar S, Joseph NM, Easow JM, Kandhakumari G, Srirangaraj S, et al. Microbiological study of diabetic foot infections. Indian J Med Spec. 2011;2:12–7. [Google Scholar]
- 24.Lee JH, Bae IK, Lee SH. New definitions of extended-spectrum β-lactamase conferring worldwide emerging antibiotic resistance. Med Res Rev. 2012;32:216–32. doi: 10.1002/med.20210. [DOI] [PubMed] [Google Scholar]
- 25.Shahid M, Singhai M, Malik A, Shukla I, Khan HM, Shujatullah F, et al. In vitro efficacy of ceftriaxone/sulbactam against Escherichia coli isolates producing CTX-M-15 extended-spectrum beta-lactamase. J Antimicrob Chemother. 2007;60:187–8. doi: 10.1093/jac/dkm131. [DOI] [PubMed] [Google Scholar]
- 26.Gadepalli R, Dhawan B, Mohanty S, Kapil A, Das BK, Chaudhry R. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus. Indian J Med Res. 2006;123:571–3. [PubMed] [Google Scholar]
- 27.Peer MA, Nasir RA, Kakru DK, Fomda BA, Bashir G, Sheikh IA. Sepsis due to linezolid resistant Staphylococcus cohnii and Staphylococcus kloosii: First reports of linezolid resistance in coagulase negative staphylococci from India. Indian J Med Microbiol. 2011;29:60–2. doi: 10.4103/0255-0857.76527. [DOI] [PubMed] [Google Scholar]
- 28.Kalawat U, Sharma KK, Reddy S. Linezolid-resistant Staphylococcus spp.at a tertiary care hospital of Andhra Pradesh. Indian J Med Microbiol. 2011;29:314–5. doi: 10.4103/0255-0857.83923. [DOI] [PubMed] [Google Scholar]


