Abstract
Third generation aromatase inhibitors (AI) have shown good clinical efficacy in comparison to the anti-estrogen tamoxifen. The steroidal AI, exemestane (EXE) has previously been shown to act as an androgen, but this report demonstrates the estrogen-like activity of EXE. Based on genome-wide microarray analysis, high correlation was seen between EXE-Only (EXE O, hormone-free) and hormone-containing AI-resistant lines. In addition, the top regulated genes in the EXE O lines were mostly estrogen-responsive genes. This estrogen-like activity of EXE was further validated using estrogen receptor (ER) activity assays, where in comparison to 17β-estradiol (E2), EXE was able to induce ER activity, though at a higher concentration. Also, this EXE-mediated ER activity was blocked by the ER antagonist ICI as well as the ERα-specific antagonist methyl-piperidino-pyrazole (MPP). Similarly, EXE was able to induce proliferation of breast cancer cell lines, MCF-7 and MCF-7aro, as well as activate transcription of known estrogen-responsive genes, i.e., PGR, pS2 and AREG. These results suggest that EXE does have weak estrogen-like activity.
Keywords: Aromatase inhibitors, Exemestane, Estrogen receptor
Introduction
Hormone-responsive breast cancers represent approximately 60% of pre-menopausal and 75% of post-menopausal patients, and depend on the steroid hormone estrogen for cancer cell survival and proliferation. It has been shown that estrogen is produced locally within the tumor by aromatase [1–3] and that aromatase expression and activity are markedly elevated in breast carcinomas [4–8]. Treatment of hormone-dependent breast cancers has relied on the use of anti-estrogens, such as tamoxifen, which antagonize estrogen receptor (ER) function and subsequently minimize hormone-dependent cell growth. In addition to this treatment strategy, the use of aromatase inhibitors (AIs) have shown excellent clinical efficacy in post-menopausal breast cancer patients by suppressing whole-body estrogen synthesis. Significant increases in disease-free survival (DFS), lengthened time to disease recurrence, and a decrease in the incidence of contralateral breast cancers have been shown in multiple clinical trials demonstrating the benefit of AIs versus tamoxifen [9–11]. The FDA-approved third generation AIs are structurally different and include the steroidal inhibitor, exemestane (EXE), as well as non-steroidal inhibitors, letrozole (LET) and anastrozole (ANA).
Several unique properties have been reported regarding EXE that differentiate this AI from its non-steroidal counterparts. Largely due to its steroidal structure that resembles the androgen substrate of aromatase, EXE acts as a mechanism-based inhibitor that binds to the active site of aromatase, resulting in inactivation of the enzyme over a long period of time [12]. Furthermore, Wang and Chen [13] have described that EXE acts as an aromatase destabilizer that degrades the enzyme in a proteosome-dependent manner. In addition to its action as an aromatase inhibitor/destabilizer, EXE has been shown to have androgen-like properties and can bind to androgen receptor (AR), though at differing reported affinities [14, 15]. Results from clinical trials have shown that EXE can be used as second-line therapy after acquired resistance develops to non-steroidal AIs in the metastatic setting [16], suggesting that a lack of cross-resistance exists between steroidal and non-steroidal AIs. Also, EXE may have unique effects that differ from non-steroidal AIs, with reports from clinical trials suggesting differences in terms of bone formation marker profiles [17].
To further investigate the unique properties of EXE, we report genome-wide microarray analysis performed with EXE-resistant breast cancer cell lines in comparison to parental MCF-7aro (aromatase overexpressed). A positive correlation was seen between hormone-containing resistant lines and hormone-free EXE O cells, suggesting a hormone-like property of this AI. Top regulated genes from the EXE O lines were primarily estrogen-responsive genes. Additional functional analysis was performed to assess ER activity, which was enhanced using EXE and blocked by the ER antagonist ICI as well as the ERα-selective antagonist MPP. Lastly, EXE was able to drive breast cancer cell proliferation and induce transcription of known estrogen-responsive genes, at micromolar concentrations. Overall, this data suggests which EXE has weak estrogen-like properties that may explain key differences in the effects of this compound on breast cancer treatment.
Materials and methods
Cell culture and compounds
MCF7 cells were cultured in MEM medium and T47D cells were cultured in RPMI1640 medium and supplemented with 10% fetal bovine serum (FBS), 2 mM L-Glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin–streptomycin and 1% non-essential amino acids (NEAA). MCF-7 cells (ER+) that stably overexpress the aromatase gene (MCF-7aro) were previously generated in this lab and used for endocrine resistant cell line production [18]. MCF-7aro cells were routinely cultured in MEM containing 10% FBS, 2 mM L-Glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin–streptomycin and 100 μg/ml G418. Resistant cell lines were maintained in phenol red-free MEM containing 10% charcoal/dextran-treated FBS with identical supplements as parental MCF-7aro cells. Resistant lines were continuously cultured in the presence of the appropriate inhibitor (1 μM EXE and 1 μM ANA), with or without the addition of 1 nM testosterone (T). In addition, MCF-7aro/ERE cells which stably express aromatase as well as an estrogen response element (ERE)-containing luciferase reporter, were generated in this laboratory and described previously (Lui et al. in press). Medium was replenished twice weekly and cells were grown at 37°C with 5% CO2. EXE was a gift from Pharmacia Italia S.P.A. (Nerviano, Italy) and ANA was provided by AstraZeneca Pharmaceuticals (Macclesfield, UK). Testosterone (T) and 17β-estradiol (E2) were purchased from Sigma (Sigma chemical, St. Louis, MO) while ICI (ICI 182780 or fulvestrant), MPP (methyl-piperidino-pyrazole) and PHTPP (4-[2-Phenyl-5,7-bis(trifluoromethyl)pyrazolo[1,5-a]pyrimidin-3-yl]phenol) were purchased from Tocris (Ellisville, MO).
Microarray analysis
The Affymetrix GeneChip Human Genome U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA) were used to define gene expression profiles in each cell line. One-cycle target labeling and hybridization were performed following manufacturer's recommendation (Affymetrix, Santa Clara, CA) at the University of California, Irvine microarray core facility. To ensure the high quality of the whole microarray process, a set of quality assessment steps were applied to the data using an R/Bioconductor package developed internally. All subsequent analysis was performed using Partek Genomics Suite, version 6.3 beta (Partek Inc,. St. Louis MO). Raw Affymetrix intensity measurements of all probe sets were background-corrected, normalized, and summarized into gene expression level measurements using Robust Multichip Average (RMA). One-way ANOVA was performed to select 5007 target genes that are differentially expressed between resistant cell lines and MCF-7aro cell lines, based on a fold-change criteria of ±1.2-fold and a P-value < 0.05. Similarity matrix, based on correlation coefficient calculations, was done using Partek Genomics Suite. Correlation coefficients range from 0.5 to 0.98. In addition, estrogen-inducibility was determined by two different estrogen-responsive gene databases, KBERG1 and ERTargetDB2.
ER activity assays
T47D and MCF-7 cells were seeded in 12 well plates at 8 × 104 cells/well in phenol red-free RPMI1640 and MEM medium respectively, supplemented with 10% charcoal/dextran-treated FBS, 24 h prior to transfection. Cells were transfected with 1.5 μl of lipofectamine 2000 (Invitrogen) and 0.2 μg of pGL3(ERE)3 luciferase reporter plasmid in 0.5 ml of Opti-MEM medium. After a 4 h incubation, 1 ml of MEM or RPMI1640 (10% charcoal/dextran-treated FBS) medium containing DMSO (vehicle control), 0.1 nM E2, or 10 nM, 100 nM, 1 μM EXE were added to each well. Luciferase activity was assayed 24 h after transfection. For MCF-7aro/ERE, approximately 1 × 104 cells were seeded in 96-well plates containing 200 μl of phenol red-free MEM, supplemented with 10% charcoal/dextran-treated FBS. On the following day, cells were washed once with PBS and supplemented with T, E2 or EXE at indicated concentrations and cells were further incubated for 24 h. For all ER activity assays, cells were lysed with 1× reporter lysis buffer (Promega, Madison, WI), protein concentration was determined using the Bradford method [19] and used to normalize luciferase response as determined by the Luciferase Assay System (Promega, Madison, WI). For the MCF-7aro/ERE screening system cell lines, luciferase units were read using a robotic injector and the Victor2 V plate reader (Perkin Elmer, Waltham, MA), while all other luciferase assays were read on a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). All experimental conditions were performed in triplicate.
Cell proliferation and quantitative real-time PCR analysis
For cell proliferation assays, MCF-7 or MCF-7aro cells were cultured in phenol red-free MEM supplemented with 10% charcoal/dextran-treated FBS for 2 days before being plated into 96-well plates (5000 cells/well). Cells were then treated in phenol red-free medium with various compounds for 4 days. The CellTiter 96 Aqueous One Proliferation Assay kit (Promega, Madison, WI) was used according to the manufacturer's protocol, to measure cell proliferation. For real-time PCR of estrogen-responsive genes, total RNA was extracted from MCF-7aro cells using TRIzol reagent (Invitrogen, Carlsbad, CA). About 5 μg of total RNA was used for reverse transcription, using 100 ng random primer (Invitrogen, Carlsbad, CA), 0.5 mM dNTP mix, 40 U AMV-RT and supplied reaction buffer (Life Sciences Inc, St. Petersburg, FL) and RNase Inhibitor (Promega, Madison, WI) for 45 min at 42°C. Subsequently, 1 μl cDNA was used for real-time PCR using iQ5 SYBR Green Supermix (Bio-Rad, Hercules, CA) on an iCycler IQ5 PCR machine (Bio-Rad, Hercules, CA). All real-time PCR reactions were done in triplicate and the housekeeping gene β-actin was used as a reference. Gene-specific primer sequences used were: PGR, 5′-AGGTCTACCCGCCCTATCTC-3′ (forward) and 5′-CAG CTCCCACAGGTAAGGAC-3′ (reverse); pS2, 5′-TCCCCTGGTGCTTCTATCC-3′ (forward) and 5′-AGCCGAGCTCTGGGACTAA-3′ (reverse).
Results
Microarray analysis of exemestane and anastrozole-resistant cell lines
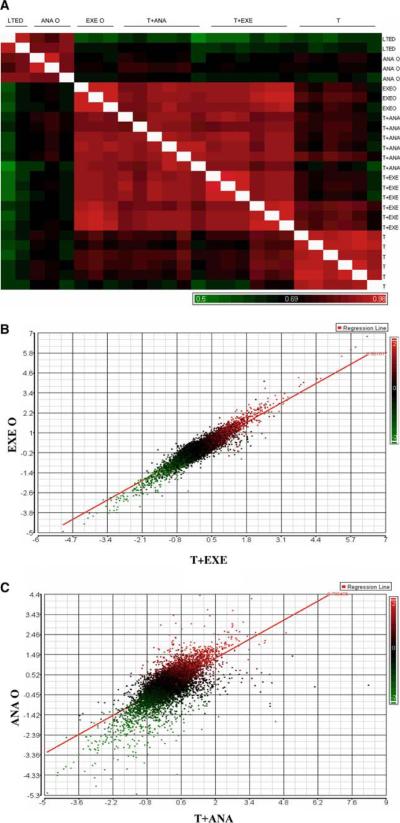
MCF-7aro cells that are resistant to EXE and ANA were generated in our laboratory and described previously [20, 21]. The EXE-resistant lines were cultured continuously in the presence of T (T+EXE R) or absence of T (EXE O). Similar conditions were used for the ANA-resistant lines, T+ANA R and ANA O, with and without T, respectively. Microarray analysis was performed using the MCF-7aro parental cells as a control for the resistant lines, in addition to the testosterone-only (T-only) lines cultured continuously in the presence of hormone. The long-term estrogen deprived (LTEDaro) cells generated in our laboratory were also used for comparison to the resistant lines, as a hormone-free model system. Results from microarray analysis are displayed as a similarity matrix, which demonstrates unbiased correlation coefficients for all cell lines (Fig. 1a). More correlated lines of ~0.7 or higher are indicated in red, with less correlation shown in black and green. The T+EXE R and T+ANA R lines were highly correlated, but unexpectedly the EXE O lines demonstrated high correlation with the T-containing resistant lines. In contrast, the ANA O lines were only highly correlated with themselves as well as the LTEDaro system. Furthermore, the T-only lines demonstrated correlation with T+EXE R, T+ANA R as well as EXE O, though at a decreased level than cell lines that have acquired resistance. Similarly, correlation coefficients displayed graphically with a regression line, for T+EXE R versus EXE O (Fig. 1b) and T+ANA R versus ANA O (Fig. 1c), demonstrated high correlation between EXE O and T-containing resistant lines.
Fig. 1.
Similarity matrix of resistant cell lines. (a) EXE and ANA-resistant lines, as well as T-Only (T) and LTEDaro controls, were normalized with parental MCF-7aro and the correlation coefficients of 5007 significant genes were displayed as a similarity matrix, using Pearson's correlation. Significant genes were selected based on a fold-change criteria of ±1.2-fold and a P-value < 0.05. Correlation coefficients ranged between 0.5 to 0.98, with red indicating good correlation and green representing less correlated lines. Additionally, correlation coefficients were displayed graphically using Partek Genomics Suite for T + EXE R versus EXE O (b) and T + ANA R versus ANA O (c). Correlation between T + EXE R and EXE O is 0.901017 and correlation between T+ANA R and ANA O lines is 0.703406
Further analysis of microarray data demonstrated a large proportion of highly statistically significant estrogen-responsive genes playing a role in the T-only as well as T+EXE R and T+ANA R lines [22]. Tables 1 and 2 list top 20 up and down-regulated genes from hormone-free EXE O and ANA O lines, respectively. Of these top 20 gene lists, estrogen-responsive genes are highly represented in the EXE O lines much more than the ANA O lines, and these estrogen-responsive genes in EXE O and ANA O lines do not greatly overlap. A select group of estrogen-responsive genes were found in both EXE O and ANA O lists, including TFF1, PBK, MYBL1 and TOP2A, though PBK and TOP2A did not make the top 20 EXE O list. Also, fold-changes of these genes, especially in the up-regulated groups, are much greater in the EXE O lines as compared to ANA O. As an example, MYBL1 is up-regulated 41.1-fold in EXE O lines and only 7.2-fold in ANA O lines. Interestingly, known estrogen-responsive genes found in the EXE O lines (GJA1, PDZK1 and MGP) were not significantly up-regulated or even down-regulated (MGP, −2.1) in the ANA O lines. Additionally, significantly regulated genes from EXE O lines were not known androgen-regulated genes, with the exception of KLK11. Estrogen-responsive genes were observed in ANA O lines, though these cell lines do not correlate with other hormone-containing lines in the similarity matrix (Fig. 1). The ANA O lines do contain a constitutively active ERα (data not shown), similar to the LTEDaro lines [22] which would explain this result. Based on genome-wide unbiased analysis using microarray, the correlation of hormone-free EXE O lines with hormone-containing cell lines, as well as the high proportion of known estrogen-responsive genes in the EXE O lines suggests a hormone-like property of EXE.
Table 1.
EXE O top 20 up- and down-regulated genes
| Up-regulated |
Down-regulated |
||||
|---|---|---|---|---|---|
| Gene symbol | Fold-change | Estrogen inducible | Gene symbol | Fold-change | Estrogen inducible |
| GJA1 | 66.9 | ✔ | SPANXA1 | −25.9 | |
| PDZK1 | 66.7 | ✔ | PCSK2 | −23.2 | |
| MGP | 65.1 | ✔ | PMP22 | −18.2 | ✔ |
| MYBL1 | 41.1 | ✔ | ITGB6 | −16.9 | |
| EGR3 | 24.7 | ✔ | DKK1 | −16.0 | ✔ |
| RHOBTB3 | 17.6 | ✔ | SOX11 | −15.7 | |
| KLK11 | 17.2 | EPAS1 | −14.1 | ||
| SLC1A1 | 17.1 | ✔ | TAGLN | −13.5 | ✔ |
| SGK3 | 15.9 | PSD3 | −12.7 | ||
| AREG | 14.3 | ✔ | SGCG | −12.4 | |
| CA2 | 13.4 | ✔ | SEPP1 | −11.9 | ✔ |
| AMPD1 | 13.3 | APOD | −11.0 | ✔ | |
| TMEM164 | 10.1 | L1CAM | −10.9 | ✔ | |
| PTGES | 9.8 | ✔ | NT5E | −10.5 | |
| FHL1 | 9.4 | ✔ | TFPI | −10.0 | ✔ |
| NPY1R | 9.2 | ✔ | AQP3 | −10.0 | ✔ |
| TFF1 | 8.6 | ✔ | FBN2 | −9.6 | |
| GREB1 | 8.0 | ✔ | TIMP3 | −9.5 | ✔ |
| GFRA1 | 7.8 | ✔ | PLS3 | −9.3 | |
| PLOD2 | 7.7 | ✔ | TNS3 | −8.9 | |
Table 2.
ANA O top 20 up- and down-regulated genes
| Up-regulated |
Down-regulated |
||||
|---|---|---|---|---|---|
| Gene symbol | Fold-change | Estrogen inducible | Gene symbol | Fold-change | Estrogen inducible |
| CYP26B1 | 19.6 | SPANXA1 | −38.0 | ||
| RLN2 | 11.7 | PCSK2 | −26.8 | ||
| GALNT12 | 9.8 | APOD | −19.4 | ✔ | |
| PKIA | 8.5 | ✔ | PLS3 | −19.4 | |
| TFF1 | 8.2 | ✔ | SOX11 | −19.3 | |
| DLG7 | 8.1 | ITGB6 | −18.9 | ||
| MYBL1 | 7.2 | ✔ | WISP2 | −18.6 | ✔ |
| DBF4 | 7.1 | TAGLN | −15.6 | ✔ | |
| ECT2 | 6.4 | SEPP1 | −14.7 | ✔ | |
| PBK | 6.1 | ✔ | MAOB | −13.9 | |
| CDKN2C | 5.9 | ✔ | NT5E | −12.1 | |
| B3GALNT1 | 5.8 | ATF3 | −11.8 | ✔ | |
| SLC9A6 | 5.8 | EPAS1 | −11.8 | ||
| CDKN3 | 5.8 | FBN2 | −11.6 | ||
| BUB1 | 5.8 | ✔ | FHL2 | −11.0 | ✔ |
| TOP2A | 5.7 | ✔ | EFEMP1 | −10.2 | ✔ |
| RABEP1 | 5.7 | L1CAM | −9.5 | ✔ | |
| AGR2 | 5.6 | ✔ | SNAI2 | −8.9 | ✔ |
| RET | 5.6 | ✔ | NUAK1 | −8.8 | |
| KCNJ8 | 5.6 | ✔ | COL5A1 | −8.4 | ✔ |
Functional analysis of EXE as a potential ERα ligand
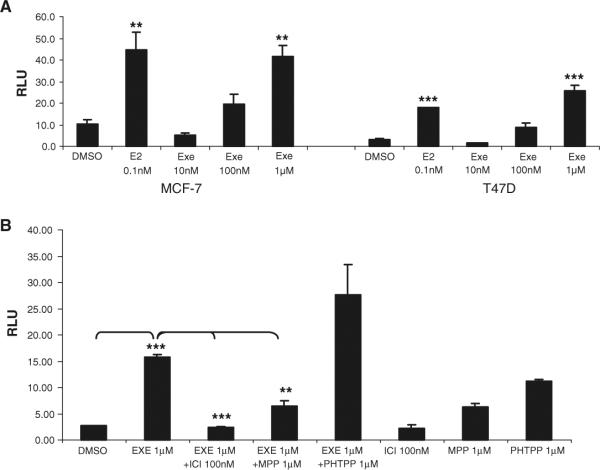
To functionally validate the estrogen-like effect of EXE, ER activity was evaluated using an estrogen response element (ERE) luciferase reporter that was transfected into MCF-7 and T47D breast cancer cell lines. These cell lines contain very low levels of aromatase activity, but endogenously express ERα, while ERβ is expressed moderately in T47D with low expression levels in MCF-7. In comparison to the ER ligand E2, EXE was shown to behave like an estrogen in both types of cells (Fig. 2a), especially at the 1μM concentration typically used in cell culture [23, 24]. In addition, ER activity was inhibited using the pure ER antagonist ICI (Fig. 2b). Similarly, EXE-mediated ER activity was inhibited using the ERα-specific ligand MPP, but not with the ERβ-selective antagonist, PHTPP (Fig. 2b). Interestingly, ER activity was increased in the presence of PHTPP, which could be a compound specific effect, especially because MCF-7 express low levels of ERβ. Our results suggest that under our assay conditions, PHTPP has agonistic activity on ER.
Fig. 2.
EXE-mediated ER activity in MCF-7 and MCF-7aro lines. (a) ER activity, measured by endogenous ER binding to a pGL3-ERE luciferase reporter, was performed in the MCF-7 and T47D cell lines, using DMSO as a vehicle control for E2 and EXE treatment. Luciferase activity was normalized with total protein levels and indicated as relative luciferase units (RLU). All samples were performed in triplicate with standard deviation shown. For statistical analysis, samples were compared to DMSO treated using Student's T-test, ** indicates P-value ≤ 0.01 and *** indicates statistical significance at a P-value ≤ 0.001. (b) EXE-mediated ER activity in the presence of the ER antagonist ICI, as well as the ERα-specific antagonist (MPP) and ERβ-selective antagonist (PHTPP). For statistical analysis, samples were compared to 1 μM EXE or DMSO control as shown using Student's T-test, ** indicates P-value ≤ 0.01 and *** indicates statistical significance at a P-value ≤ 0.001
Dose–response evaluation of EXE, in comparison to T and E2, was performed in the MCF-7aro/ERE stable cell line that is a dual reporter system of both aromatase activity as well as ERα functionality (Lui et al. in press). In this system, T is converted to E2 by aromatase, which then activates luciferase activity mediated by the stably expressed ERE, making this screening system very sensitive to T and E2 treatment. Dose–response of both T and E2 were evaluated in this system, ranging from 0.1 fM to 1 nM concentrations (Fig. 3). Both T and E2 displayed typical sigmoidal dose–response curves, with an increase in activity beginning at 1 pM and a plateau in activity seen at 0.1 nM. Since this is not a ligand binding assay, Ki or Kd values of the ligands cannot be determined. However, the MCF-7aro/ERE screen system is very sensitive, as indicated by highly effective response at low concentrations of hormone (Fig. 3). The dose-–response of EXE was similarly evaluated in this MCF-7aro/ERE system, ranging from concentrations of 0.1 nM and plateauing at 0.1 μM (Fig. 3). Based on these dose–response curves, the effective agonist concentration of EXE is 1000 times less potent than both T (once converted to E2) and E2 supplemented directly into the system. These experiments further suggest that EXE does act as a weak agonist of ERα.
Fig. 3.
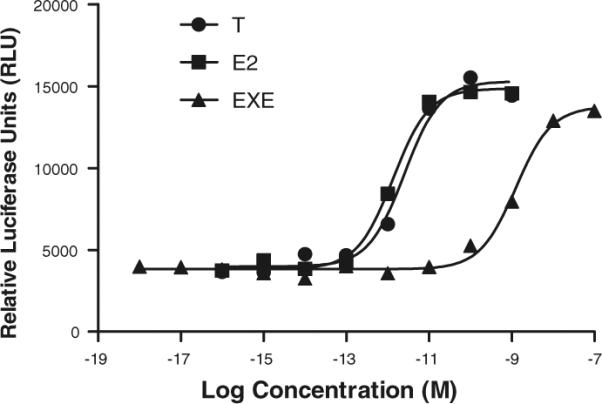
Dose-response curves of T, E2 and EXE in MCF-7aro/ERE lines. Dose–response was determined for T, E2 and EXE in the MCF-7 aromatase and ERE stable luciferase reporter cell line, MCF-7aro/ERE. Cells were treated with T, E2 and EXE at indicated concentrations for 24 h. Luciferase substrate (Promega, Madison, WI) was mixed with cell lysate using a robotic injector and luciferase units were determined using the Victor2 V plate reader (Perkin Elmer, Waltham, MA). Luciferase activity was standardized based on the protein concentration and shown as relative luciferase units (RLU)
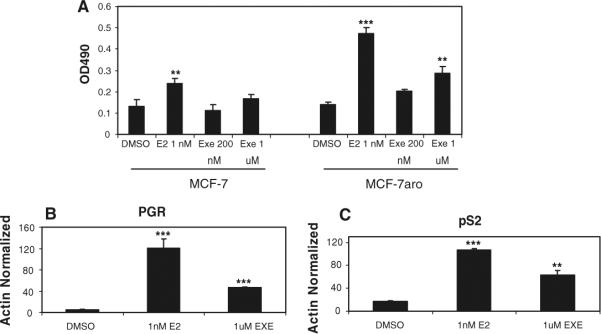
The agonistic properties of EXE were further evaluated by cell proliferation assays as well as the transactivation of ERα transcriptional targets, in comparison to E2. Figure 4a showed the E2-dependent growth of MCF-7 and MCF-7aro cells at a 1 nM concentration, in comparison to EXE-dependent cell growth at 1 μM. Additionally, MCF-7aro cells were treated with either DMSO, 1 nM E2, or 1 μM EXE for 48 h, with subsequent quantitative real-time PCR performed to assess mRNA levels of known estrogen-responsive genes. The E2-mediated induction of PGR (Fig. 4b) and pS2 (TFF1, Fig. 4c) transcript levels was observed, with a weaker yet clear estrogen-like effect of EXE on these same genes. Similarly, Wang et al. [25] have previously reported that EXE can also induce transcript levels of amphiregulin (AREG), another known estrogen-responsive gene. The effects of EXE as a weak estrogenic compound, in addition to its known ability to act as a steroidal aromatase inhibitor and aromatase destabilizer, establish a unique property of this compound to target both aromatase as well as ERα.
Fig. 4.
EXE-dependent functional analysis in MCF-7 and MCF-7aro lines. (a) Cell proliferation assays were performed in the MCF-7 and MCF-7aro cell lines using the CellTiter 96 aqueous one proliferation assay kit in 96-well plate format. After being cultured in hormone-deprived media for 2 days, 5000 cells were cultured for 4 days in the presence of DMSO vehicle control, E2 or EXE at indicated concentrations. All samples shown were performed in triplicate, with standard deviation shown. For statistical analysis, samples were compared to DMSO treated using Student's T-test, ** indicates P-value ≤ 0.01 and *** indicates statistical significance at a P-value ≤ 0.001. Quantitative real-time PCR was performed with E2 and EXE treated MCF-7aro cells. Cells were hormone depleted and treated with DMSO, E2 or EXE for 2 days prior to harvesting total RNA. Expression of estrogen-responsive genes, PGR (b) and pS2 (c), was normalized with the beta-actin housekeeping gene. All samples were performed in triplicate with standard deviation shown. For statistical analysis, samples were compared to DMSO treated using Student's T-test, ** indicates P-value ≤ 0.01 and *** indicates statistical significance at a P-value ≤ 0.001
Discussion
AIs have shown good clinical efficacy versus the anti-estrogen tamoxifen, in the treatment of post-menopausal and hormone-dependent breast cancers. Being a steroidal AI that is distinctive from the non-steroidal inhibitors, we further investigated the unique properties of EXE on T47D, MCF-7 and derivative cell lines. Based on microarray analysis, a high correlation was observed between EXE O and hormone-containing resistant lines, T + EXE R and T + ANA R, in addition to moderate correlation with T-only lines. Top regulated genes from EXE O lines were mainly estrogen-responsive, with only one known androgen-regulated gene observed (KLK11). Further functional analysis demonstrated that EXE can activate ER activity, though at a lower potency than E2, and this activity can be blocked by ER antagonists. Furthermore, EXE was able to induce breast cancer cell proliferation as well as transcription of known estrogen-responsive genes by quantitative real-time PCR. This data suggests that the steroidal AI EXE does have weak estrogen-like properties.
Previous reports with EXE have suggested an androgenic effect of this inhibitor, though a discrepancy in binding affinity of EXE to AR were shown [14, 15]. Of interest, Ariazi et al. [14] looked at the active metabolite of EXE, 17-hydroexemestane, and its binding affinity to AR, which may account for the affinity differences reported. Based on our microarray data, androgen-regulated genes did not play a major role in EXE O lines, as estrogen-responsive genes were significantly differentially regulated. Furthermore, EXE-mediated induction of ER activity was inhibited by an ERα-specific antagonist, MPP [26], and not by an ERβ-selective antagonist, PHTPP [27]. Also, EXE-mediated ER activity was not blocked when the nonsteroidal AIs LET and ANA were used, confirming that aromatase is not involved (data not shown). It is also important to note that Ariazi et al. [14] did observe very weak binding of 17-hydroexemestane to ERα, with an IC50 of 2.12 × 10−5 mol/l, compared with an IC50 of 1.33 × 10−9 mol/l for E2. Though the competitive binding assay used in the Ariazi et al. study differs from our approach and does not allow for direct comparison of results, similar findings were obtained from our luciferase-based MCF-7aro/ERE screening system. We observed a plateau in luciferase-driven activity that is 1000 times more potent for E2 than for EXE. Based on our current approach using a cell culture model system, it is difficult to differentiate the action of EXE versus its metabolite 17-hydroexemestane. Yet, gene expression profiles of resistant cell lines as well as EXE-dependent ER activation in parental MCF-7 and T47D responsive cell lines suggests that the estrogen-like effect of EXE is similar in both cell types.
The weak estrogenic properties of EXE may have critical clinical implications. One of the major side-effects associated with AI treatment is loss in bone mineral density (BMD) and increases in bone fracture rates [28–30]. These effects have been attributed to whole-body suppression of estrogen synthesis by AIs, as estrogens have a protective effect in bone. Interestingly, recent reports reveal that the effects of EXE treatment on bone are less severe than nonsteroidal AI therapy, and that rates of bone fractures were lower in EXE-treated patients [9, 31]. Goss et al. [17] compared short-term EXE versus letrozole and anastrozole treatment on bone turnover markers, and observed high levels of procollagen type I N-terminal propeptide (PINP), a serum marker of bone formation, in EXE-treated patients after 24 weeks, but not in non-steroidal AI treated patients. Goss et al. [17] also comment that previous clinical trials that assess loss in BMD and bone fracture rate after AI-treatment are compared with tamoxifen-treated patients. The concerns of Goss et al. are that tamoxifen is an agonist in bone and therefore has protective properties, suggesting that the effects of tamoxifen and AIs are not comparable. To support this clinical data, animal studies showed that ovariectomized rats treated with exemestane and 17-hydroexemestane, had significantly increased BMD, increased strength of cortical and trabecular bone, and decreased markers of bone resorption [32]. In contrast, ovariectomized rats receiving letrozole or no AI had significant loss of BMD, accompanied by decreasing bone strength and increasing bone resorption markers [33].
The protective effects of estrogen on bone have been well characterized. Loss of estrogen stimulates both osteoblast as well as osteoclast action, resulting in bone remodeling at a rate where bone resorption is faster than bone formation [34–36]. In addition, estrogen as well as ERα, have been shown to play critical roles in osteoblast progenitor cell self-renewal [37, 38]. An important study looked at the effect of EXE directly on osteoblast and osteoblast-like cell lines, and determined that EXE mediated cell proliferation effects similar to that of an androgen [39]. Interestingly, these effects were suggested to be AR-dependent as well as AR-independent, because an AR-specific antagonist was not able to inhibit high dose EXE-mediated cell growth. Microarray analysis of E2, EXE and DHT treated hFOB osteoblast lines was also performed in this study, with EXE-treated cells clustering more closely to DHT-treated cells [39]. It is important to note in these studies that the hFOB cell line is AR and ERβ positive but ERα negative. Therefore, it is difficult to compare these results with our own, in order to determine if EXE has any weak estrogenic properties in osteoblast cell lines.
Based on microarray studies and estrogen-responsive gene profiles from EXE O lines, we have observed an estrogen-like property of the steroidal AI, EXE. Functional analysis confirmed that an induction of ER activity was EXE-mediated and could be blocked by an ER antagonist ICI as well as an ERα-specific antagonist MPP. In addition, EXE was able to induce breast cancer cell proliferation as well as transactivation of mRNA levels of estrogen-responsive genes. This data suggests that EXE does have weak estrogen-like properties, which can potentially explain certain key differences observed clinically between steroidal and non-steroidal AIs. In addition to previous reports on the unique properties of EXE, these findings provide further information that differentiates EXE from other non-steroidal AIs currently used in the clinic.
Acknowledgement
S. Masri was supported by NIH pre-doctoral training fellowship CA123691 and S. Chen by NIH grants CA044735 and ES08258.
Footnotes
References
- 1.Santner SJ, Chen S, Zhou D, Korsunsky Z, Martel J, Santen RJ. Effect of androstenedione on growth of untransfected and aromatase-transfected MCF-7 cells in culture. J Steroid Biochem Mol Biol. 1993;44(4–6):611–616. doi: 10.1016/0960-0760(93)90267-z. doi:10.1016/0960-0760(93)90267-Z. [DOI] [PubMed] [Google Scholar]
- 2.Yue W, Wang JP, Hamilton CJ, Demers LM, Santen RJ. In situ aromatization enhances breast tumor estradiol levels and cellular proliferation. Cancer Res. 1998;58(5):927–932. [PubMed] [Google Scholar]
- 3.Miller WR, O'Neill J. The importance of local synthesis of estrogen within the breast. Steroids. 1987;50(4–6):537–548. doi: 10.1016/0039-128x(87)90037-7. doi: 10.1016/0039-128X(87)90037-7. [DOI] [PubMed] [Google Scholar]
- 4.Bulun SE, Price TM, Aitken J, Mahendroo MS, Simpson ER. A link between breast cancer and local estrogen biosyn-thesis suggested by quantification of breast adipose tissue aromatase cytochrome P450 transcripts using competitive polymerase chain reaction after reverse transcription. J Clin Endocrinol Metab. 1993;77(6):1622–1628. doi: 10.1210/jcem.77.6.8117355. doi:10.1210/jc.77.6.1622. [DOI] [PubMed] [Google Scholar]
- 5.Esteban JM, Warsi Z, Haniu M, Hall P, Shively JE, Chen S. Detection of intratumoral aromatase in breast carcinomas. An immunohistochemical study with clinicopathologic correlation. Am J Pathol. 1992;140(2):337–343. [PMC free article] [PubMed] [Google Scholar]
- 6.James VH, McNeill JM, Lai LC, Newton CJ, Ghilchik MW, Reed MJ. Aromatase activity in normal breast and breast tumor tissues: in vivo and in vitro studies. Steroids. 1987;50(1–3):269–279. doi: 10.1016/0039-128x(83)90077-6. doi:10.1016/0039-128X(83)90077-6. [DOI] [PubMed] [Google Scholar]
- 7.Lu Q, Nakmura J, Savinov A, Yue W, Weisz J, Dabbs DJ, et al. Expression of aromatase protein and messenger ribonucleic acid in tumor epithelial cells and evidence of functional significance of locally produced estrogen in human breast cancers. Endocrinology. 1996;137(7):3061–3068. doi: 10.1210/endo.137.7.8770932. doi:10.1210/en.137.7.3061. [DOI] [PubMed] [Google Scholar]
- 8.Santen RJ, Martel J, Hoagland M, Naftolin F, Roa L, Harada N, et al. Stromal spindle cells contain aromatase in human breast tumors. J Clin Endocrinol Metab. 1994;79(2):627–632. doi: 10.1210/jcem.79.2.8045987. doi: 10.1210/jc.79.2.627. [DOI] [PubMed] [Google Scholar]
- 9.Coombes RC, Hall E, Gibson LJ, Paridaens R, Jassem J, Delozier T, et al. A randomized trial of exemestane after two to three years of tamoxifen therapy in postmenopausal women with primary breast cancer. N Engl J Med. 2004;350(11):1081–1092. doi: 10.1056/NEJMoa040331. doi: 10.1056/NEJMoa040331. [DOI] [PubMed] [Google Scholar]
- 10.Goss PE, Ingle JN, Martino S, Robert NJ, Muss HB, Piccart MJ, et al. Randomized trial of letrozole following tamoxifen as extended adjuvant therapy in receptor-positive breast cancer: updated findings from NCIC CTG MA.17. J Natl Cancer Inst. 2005;97(17):1262–1271. doi: 10.1093/jnci/dji250. [DOI] [PubMed] [Google Scholar]
- 11.Howell A, Cuzick J, Baum M, Buzdar A, Dowsett M, Forbes JF, et al. Results of the ATAC (Arimidex, Tamoxifen, alone or in combination) trial after completion of 5 years' adjuvant treatment for breast cancer. Lancet. 2005;365(9453):60–62. doi: 10.1016/S0140-6736(04)17666-6. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 12.Brueggemeier RW. Aromatase, aromatase inhibitors, and breast cancer. Am J Ther. 2001;8(5):333–344. doi: 10.1097/00045391-200109000-00007. doi:10.1097/00045391-200109000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Chen S. Aromatase destabilizer: novel action of exemestane, a food and drug administration-approved aromatase inhibitor. Cancer Res. 2006;66(21):10281–10286. doi: 10.1158/0008-5472.CAN-06-2134. doi:10.1158/0008-5472.CAN-06-2134. [DOI] [PubMed] [Google Scholar]
- 14.Ariazi EA, Leitao A, Oprea TI, Chen B, Louis T, Bertucci AM, et al. Exemestane's 17-hydroxylated metabolite exerts biological effects as an androgen. Mol Cancer Ther. 2007;6(11):2817–2827. doi: 10.1158/1535-7163.MCT-07-0312. doi:10.1158/1535-7163.MCT-07-0312. [DOI] [PubMed] [Google Scholar]
- 15.di Salle E, Ornati G, Giudici D, Lassus M, Evans TR, Coombes RC. Exemestane (FCE 24304), a new steroidal aromatase inhibitor. J Steroid Biochem Mol Biol. 1992;43(1–3):137–143. doi: 10.1016/0960-0760(92)90198-r. doi: 10.1016/0960-0760(92)90198-R. [DOI] [PubMed] [Google Scholar]
- 16.Lonning PE, Bajetta E, Murray R, Tubiana-Hulin M, Eisenberg PD, Mickiewicz E, et al. Activity of exemestane in metastatic breast cancer after failure of nonsteroidal aromatase inhibitors: a phase II trial. J Clin Oncol. 2000;18(11):2234–2244. doi: 10.1200/JCO.2000.18.11.2234. [DOI] [PubMed] [Google Scholar]
- 17.Goss PE, Hadji P, Subar M, Abreu P, Thomsen T, Banke-Bochita J. Effects of steroidal and nonsteroidal aromatase inhibitors on markers of bone turnover in healthy postmenopausal women. Breast Cancer Res. 2007;9(4):R52. doi: 10.1186/bcr1757. doi:10.1186/bcr1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun XZ, Zhou D, Chen S. Autocrine and paracrine actions of breast tumor aromatase. A three-dimensional cell culture study involving aromatase transfected MCF-7 and T-47D cells. J Steroid Biochem Mol Biol. 1997;63(1–3):29–36. doi: 10.1016/s0960-0760(97)00068-x. doi:10.1016/S0960-0760(97)00068-X. [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Masri S, Hong Y, Wang X, Phung S, Yuan YC, Wu X. New experimental models for aromatase inhibitor resistance. J Steroid Biochem Mol Biol. 2007;106(1–5):8–15. doi: 10.1016/j.jsbmb.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, Masri S, Wang X, Phung S, Yuan YC, Wu X. What do we know about the mechanisms of aromatase inhibitor resistance? J Steroid Biochem Mol Biol. 2006;102(1–5):232–240. doi: 10.1016/j.jsbmb.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masri S, Phung S, Wang X, Wu X, Yuan YC, Wagman L, et al. Genome-wide analysis of aromatase inhibitor-resistant, tamoxifen-resistant, and long-term estrogen-deprived cells reveals a role for estrogen receptor. Cancer Res. 2008;68(12):4910–4918. doi: 10.1158/0008-5472.CAN-08-0303. doi:10.1158/0008-5472.CAN-08-0303. [DOI] [PubMed] [Google Scholar]
- 23.Itoh T, Karlsberg K, Kijima I, Yuan YC, Smith D, Ye J, et al. Letrozole-, anastrozole-, and tamoxifen-responsive genes in MCF-7aro cells: a microarray approach. Mol Cancer Res. 2005;3(4):203–218. doi: 10.1158/1541-7786.MCR-04-0122. [DOI] [PubMed] [Google Scholar]
- 24.Kijima I, Itoh T, Chen S. Growth inhibition of estrogen receptor-positive and aromatase-positive human breast cancer cells in monolayer and spheroid cultures by letrozole, anastrozole, and tamoxifen. J Steroid Biochem Mol Biol. 2005;97(4):360–368. doi: 10.1016/j.jsbmb.2005.09.003. doi:10.1016/j.jsbmb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Masri S, Phung S, Chen S. The role of amphiregulin in exemestane-resistant breast cancer cells: evidence of an autocrine loop. Cancer Res. 2008;68(7):2259–2265. doi: 10.1158/0008-5472.CAN-07-5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington WR, Sheng S, Barnett DH, Petz LN, Katzenellenbogen JA, Katzenellenbogen BS. Activities of estrogen receptor alpha- and beta-selective ligands at diverse estrogen responsive gene sites mediating transactivation or transrepression. Mol Cell Endocrinol. 2003;206(1–2):13–22. doi: 10.1016/s0303-7207(03)00255-7. doi:10.1016/S0303-7207(03)00255-7. [DOI] [PubMed] [Google Scholar]
- 27.Compton DR, Sheng S, Carlson KE, Rebacz NA, Lee IY, Katzenellenbogen BS, et al. Pyrazolo[1, 5-a]pyrimidines: estrogen receptor ligands possessing estrogen receptor beta antagonist activity. J Med Chem. 2004;47(24):5872–5893. doi: 10.1021/jm049631k. doi:10.1021/ jm049631k. [DOI] [PubMed] [Google Scholar]
- 28.Chien AJ, Goss PE. Aromatase inhibitors and bone health in women with breast cancer. J Clin Oncol. 2006;24(33):5305–5312. doi: 10.1200/JCO.2006.07.5382. doi:10.1200/JCO.2006.07.5382. [DOI] [PubMed] [Google Scholar]
- 29.Lonning PE. Bone safety of aromatase inhibitors versus tamoxifen. Int J Gynecol Cancer. 2006;16(Suppl 2):518–520. doi: 10.1111/j.1525-1438.2006.00685.x. doi: 10.1111/j.1525-1438.2006.00685.x. [DOI] [PubMed] [Google Scholar]
- 30.Perez EA. Safety profiles of tamoxifen and the aromatase inhibitors in adjuvant therapy of hormone-responsive early breast cancer. Ann Oncol. 2007;18(Suppl 8):viii26–viii35. doi: 10.1093/annonc/mdm263. [DOI] [PubMed] [Google Scholar]
- 31.Coleman RE, Banks LM, Girgis SI, Kilburn LS, Vrdoljak E, Fox J, et al. Skeletal effects of exemestane on bone-mineral density, bone biomarkers, and fracture incidence in postmenopausal women with early breast cancer participating in the Intergroup Exemestane Study (IES): a randomised controlled study. Lancet Oncol. 2007;8(2):119–127. doi: 10.1016/S1470-2045(07)70003-7. doi:10.1016/S1470-2045(07)70003-7. [DOI] [PubMed] [Google Scholar]
- 32.Goss PE, Qi S, Josse RG, Pritzker KP, Mendes M, Hu H, et al. The steroidal aromatase inhibitor exemestane prevents bone loss in ovariectomized rats. Bone. 2004;34(3):384–392. doi: 10.1016/j.bone.2003.11.006. doi: 10.1016/j.bone.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Goss PE, Qi S, Cheung AM, Hu H, Mendes M, Pritzker KP. Effects of the steroidal aromatase inhibitor exemestane and the nonsteroidal aromatase inhibitor letrozole on bone and lipid metabolism in ovariectomized rats. Clin Cancer Res. 2004;10(17):5717–5723. doi: 10.1158/1078-0432.CCR-04-0438. doi:10.1158/1078-0432.CCR-04-0438. [DOI] [PubMed] [Google Scholar]
- 34.Eriksen EF, Hodgson SF, Eastell R, Cedel SL, O'Fallon WM, Riggs BL. Cancellous bone remodeling in type I (post-menopausal) osteoporosis: quantitative assessment of rates of formation, resorption, and bone loss at tissue and cellular levels. J Bone Miner Res. 1990;5(4):311–319. doi: 10.1002/jbmr.5650050402. [DOI] [PubMed] [Google Scholar]
- 35.Jilka RL, Takahashi K, Munshi M, Williams DC, Roberson PK, Manolagas SC. Loss of estrogen upregulates osteoblasto-genesis in the murine bone marrow. Evidence for autonomy from factors released during bone resorption. J Clin Invest. 1998;101(9):1942–1950. doi: 10.1172/JCI1039. doi:10.1172/JCI1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parfitt AM, Mathews CH, Villanueva AR, Kleerekoper M, Frame B, Rao DS. Relationships between surface, volume, and thickness of iliac trabecular bone in aging and in osteoporosis. Implications for the microanatomic and cellular mechanisms of bone loss. J Clin Invest. 1983;72(4):1396–1409. doi: 10.1172/JCI111096. doi:10.1172/JCI111096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Gregorio GB, Yamamoto M, Ali AA, Abe E, Roberson P, Manolagas SC, et al. Attenuation of the self-renewal of transit-amplifying osteoblast progenitors in the murine bone marrow by 17 beta-estradiol. J Clin Invest. 2001;107(7):803–812. doi: 10.1172/JCI11653. doi: 10.1172/JCI11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dang ZC, van Bezooijen RL, Karperien M, Papapoulos SE, Lowik CW. Exposure of KS483 cells to estrogen enhances osteogenesis and inhibits adipogenesis. J Bone Miner Res. 2002;17(3):394–405. doi: 10.1359/jbmr.2002.17.3.394. doi:10.1359/jbmr.2002.17.3.394. [DOI] [PubMed] [Google Scholar]
- 39.Miki Y, Suzuki T, Hatori M, Igarashi K, Aisaki KI, Kanno J, et al. Effects of aromatase inhibitors on human osteoblast and osteoblast-like cells: a possible androgenic bone protective effects induced by exemestane. Bone. 2007;40(4):876–887. doi: 10.1016/j.bone.2006.11.029. doi:10.1016/ j.bone.2006.11.029. [DOI] [PubMed] [Google Scholar]