Abstract
Background and Aims
Legumes overcome nitrogen limitations by entering into a mutualistic symbiosis with N2-fixing bacteria (rhizobia). Fully compatible associations (effective) between Trifolium spp. and Rhizobium leguminosarum bv. trifolii result from successful recognition of symbiotic partners in the rhizosphere, root hair infection and the formation of nodules where N2-fixing bacteroids reside. Poorly compatible associations can result in root nodule formation with minimal (sub-optimal) or no (ineffective) N2-fixation. Despite the abundance and persistence of strains in agricultural soils which are poorly compatible with the commercially grown clover species, little is known of how and why they fail symbiotically. The aims of this research were to determine the morphological aberrations occurring in sub-optimal and ineffective clover nodules and to determine whether reduced bacteroid numbers or reduced N2-fixing activity is the main cause for the Sub-optimal phenotype.
Methods
Symbiotic effectiveness of four Trifolium hosts with each of four R. leguminosarum bv. trifolii strains was assessed by analysis of plant yields and nitrogen content; nodule yields, abundance, morphology and internal structure; and bacteroid cytology, quantity and activity.
Key Results
Effective nodules (Nodule Function 83–100 %) contained four developmental zones and N2-fixing bacteroids. In contrast, Sub-optimal nodules of the same age (Nodule Function 24–57 %) carried prematurely senescing bacteroids and a small bacteroid pool resulting in reduced shoot N. Ineffective-differentiated nodules carried bacteroids aborted at stage 2 or 3 in differentiation. In contrast, bacteroids were not observed in Ineffective-vegetative nodules despite the presence of bacteria within infection threads.
Conclusions
Three major responses to N2-fixation incompatibility between Trifolium spp. and R. l. trifolii strains were found: failed bacterial endocytosis from infection threads into plant cortical cells, bacteroid differentiation aborted prematurely, and a reduced pool of functional bacteroids which underwent premature senescence. We discuss possible underlying genetic causes of these developmental abnormalities and consider impacts on N2-fixation of clovers.
Keywords: Trifolium subterraneum, T. purpureum, T. polymorphum, Rhizobium, symbiosis, bacteroid, clover, compatibility, effective, nitrogen fixation, nodule morphology
INTRODUCTION
Symbiotic di-nitrogen (N2) fixation by crop and forage legumes via their association with root nodule bacteria (collectively called rhizobia) plays a critical role in agricultural systems throughout the world (Herridge et al., 2008; Unkovich et al., 2009). The Trifolium genus (clovers) is one of the largest in the Fabaceae family and is grown widely in improved pasture systems throughout cool temperate regions. In many cases, soils with a history of hosting Trifolium spp. have large and symbiotically diverse populations of Rhizobium leguminosarum bv. trifolii (R. l. trifolii) strains that can infect and nodulate clover. The efficiency of symbiosis established by different combinations of clover hosts (Trifolium spp.) and strains of R. l. trifolii can vary widely from 10 % to 130 % when compared with an effective host–strain combination (Rys and Bonish, 1981; Slattery and Coventry, 1995; Denton et al., 2000; Ballard et al., 2002; Drew et al., 2011). Whilst effective inoculant strains of R. l. trifolii have been identified for commercially grown species of clover, the abundant and competitive rhizobia that are naturalized in many soils can reduce the impact of an inoculant strain by occupying a significant portion of the nodules (Yates et al., 2011).
Clovers inoculated with compatible and effective strains of rhizobia form indeterminate nodules which have been well characterized and described in the literature. Effective nodules contain different morphological regions with the meristem at the distal part of the nodule and the senescent zone at the proximal end adjacent to the root (Vasse et al., 1990; Timmers et al., 2000). Bacteroids at different stages of development occupy the different nodule zones (Vasse et al., 1990) (see schematic representation in Fig. 1). By comparison, morphological examination of sub-optimal nodules and their comparison to effective nodules is underrepresented in the literature, despite their frequency in agricultural systems. The abundance of naturally occurring sub-optimal interactions is even overlooked by researchers who use model organisms which are poorly matched for N2-fixation as the benchmark for symbiotic success (Terpolilli et al., 2008). Sub-optimal symbiotic interactions often develop seemingly inconsistent phenotypes, which, in part, may be due to the pleomorphic nature of plants within the ILRC taxonomic clade (Sprent, 2007). Morris and Djordjevic (2006) demonstrated this well when they spot-inoculated a sub-optimal R. l. trifolii strain on clover and identified four unique developmental responses in nodule and lateral root primordia including: effective nodules (5 %), aberrant nodules (25 %), lateral roots (50 %) and hybrid structures devoid of bacterial colonization (20 %).
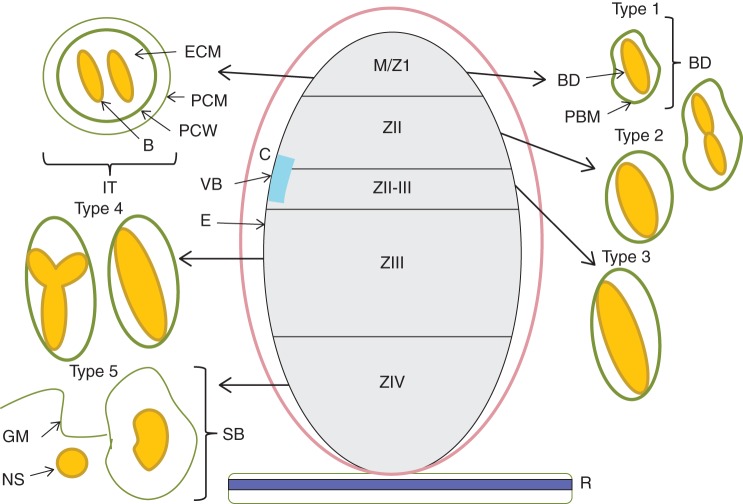
Fig. 1.
Internal nodule structure and stages of bacteroid differentiation. Indeterminate nodules arise from the root (R) cortex via a persistent meristem. The morphological regions include zone I where infection threads (IT) and meristematic cells (M) are found, the pre-fixation (ZII), inter-fixation (ZII–III), nitrogen-fixation (ZIII) and senescent (ZIV) zones. Type 1 bacteroids (BD) are newly released from the infection thread surrounded by a symbiosome (peribacteroid) membrane (PBM) and include those undergoing division, type 2 bacteroids occupying zone II elongate without dividing, type 3 bacteroids in the interzone II–III cease elongation, type 4 N2-fixing bacteroids occupy distal zone III (the common Y and maximally elongated rod shapes are shown); type 5 bacteroids of proximal zone III undergo senescence and ghost membranes (GM) of plant and bacterial origin are deposited within the plant cells. Transverse section of an infection thread shows the bacterial cells (B) within the infection thread lumen [the host cells' extracellular matrix (ECM)] enveloped by both a membrane derived from the plant cell wall (PCW) and the plant cell membrane (PCM). The proximity of cortical cells (C) to vascular bundles (VB) to epidermal cells (E) is shown.
Significant research efforts have focussed on identifying genetic control for specificity of nodulation (Hirsch, 1992; Perret et al., 2000), early stages of infection thread development and occupation (Oldroyd and Downie, 2008), and development or maintenance of the nitrogen-fixing form of rhizobia (Vasse et al., 1990; Prell et al., 2010). However, genetic regulation of compatibility for N2-fixation is not yet clear, nor do we understand why incompatible R. l. trifolii strains commonly infect Trifolium spp. Miller et al. (2007) started to address these questions using R. l. trifolii strains which nodulated both Caucasian clover (T. ambiguum) and white clover (T. repens) but were only effective with one of these hosts. They identified an intergenic region (111 bp) of nifH-fixA which determines the Fix+ phenotype on Caucasian clover, possibly by interaction at this site with a host-specific signal. However, we could not identify this same intergenic sequence when aligning the nifH-fixA intergenic sequences of R. l. trifolii WSM1325 and R. l. trifolii WSM2304 (data not presented) suggesting that it is specific to Caucasian clover nodulating strains. Furthermore, genetic regulation of the sub-optimal phenotypes where bacteroids can fix nitrogen is likely to be under a more complicated genetic regulation.
The genome sequences of R. l. trifolii strains will assist with these investigations. Four R. l. trifolii strains were selected for use in this study, and of these the complete genome sequences of WSM1325 and WSM2304 are publicly available (Reeve et al., 2010a, b), whilst the genomes of R. l. trifolii strains SRDI565 and SRDI943 are in the post-draft sequencing stage (W. G. Reeve, pers. comm., May 2012). These strains in combination with four clover genotypes have been used in this present study to produce the full range of symbiotic outcomes and nodule phenotypes, i.e. ineffective through to effective symbioses that are common place in the field (Howieson et al., 2005; Yates et al., 2008; Drew and Ballard, 2010).
Here we present the first detailed histological assessment of sub-optimal clover nodules and their bacteroids. We aimed to determine whether reduced N2-fixation in sub-optimal nodules was due to a smaller pool of terminally differentiated bacteroids. Additionally, we aimed to investigate whether one or more developmental abnormalities led to the development of ineffective clover nodules and whether these are conserved across Trifolium species.
MATERIALS AND METHODS
Rhizobium and plant species
Four Rhizobium leguminosarum bv. trifolii (R. l. trifolii) strains [WSM1325, WSM2304, SRDI943 (formerly V2-2) and SRDI565 (formerly N8-J) (Drew and Ballard, 2010)] were selected for this study based on their compatibility with different Trifolium genotypes. The four plant hosts used were the annual Trifolium subterraneum ‘Campeda’, Trifolium subterraneum ‘Clare’ and Trifolium purpureum ‘Paratta’, and the perennial Trifolium polymorphum. Information on the genotypes and origins of the clover hosts used in these experiments are summarized in Table 1.
Table 1.
Genotypes and origins of clover species used in these experiments
| Species | Subspecies | Subgenus* | Chromosome number* | Ploidy | Origin | Growth habit | Reference |
|---|---|---|---|---|---|---|---|
| T. subterraneum ‘Campeda’ | subterraneum | Trichocephalum | 2n = 16 | Diploid | Sardinia | Annual | Italy ISpICF, 2000 |
| T. subterraneum ‘Clare’ | brachycalycinum | Trichocephalum | 2n = 16 | Diploid | Mediterranean | Annual | Gladstones and Collins, 1983 |
| T. purpureum ‘Paratta’ | Trichocephalum | 2n = 14 | Aneuploid | Greece | Annual | Anon, 1972 | |
| T. polymorphum (acc. # 087102) | Involucrarium | 2n = 16,32 | Polyploid | Uruguay | Perennial | Labandera and Vincent, 1975 |
Seed preparation
Seeds were surface-sterilized in 80 % (v/v) ethanol for 15 s, followed by 3 % (w/v) sodium hypochlorite solution for 3 min and then thoroughly rinsed with sterile deionized water. Surface-sterilized seeds were stored at 4 °C on 1·5 % (w/v) agar plates for 2 d and then placed in an incubator at 25 °C for 16 h for the T. subterraneum cultivars to germinate and for 2 d for T. purpureum and T. polymorphum to germinate.
Bioassay and growth conditions
Germinated seeds were planted into sterilized (121 °C, 15 min) 60-cm3 plastic screw-cap vials filled with 30 g of vermiculite moistened with 31 mL of nitrogen-free nutrient solution (McKnight, 1949) and 4 mL of starter N solution (70 mg N L−1 as NH4NO3). Vial lids contained two small holes, one for the plant and the other for addition of the inoculant and water. Roots were contained within vials, while shoots were exposed to the atmosphere. Vials were placed in plastic holders which restricted light to the root zone. Plants were grown in a greenhouse (12 h day/night cycle, 25 °C day, 15 °C night) for 32 d and watered aseptically as required with sterile deionized water. See ‘Experimental design’ for details of sample size.
Inoculum preparation and inoculation
Pure cultures of R. l. trifolii were grown on Yeast Mannitol Agar (Vincent, 1970) at 28 °C for 3 d and suspended in sterile 0·152 m NaCl solution to achieve a concentration of approx. 109 cells/mL (OD600nm 1·0) prior to 100-fold dilution in sterile saline to approx. 107 cells/mL as confirmed by viable cell counts. Each plant was inoculated with 1 mL of the diluted cells of the appropriate culture 3 d after planting. Uninoculated controls (–N and +N) received 1 mL of 0·152 m NaCl at the time of inoculation of the treatments.
Experimental design
Data from three experiments are presented. The plants in each experiment were arranged in a completely randomized block design. The first experiment consisted of four clover genotypes with each of six treatments including four Rhizobium strains, an uninoculated control (uninoculated) and uninoculated nitrogen-fed control (+N), and five replicates of each of these treatments. The +N control received 4 mL of 0·112 m NH4NO3 weekly from 1 week after inoculation. The plants from this experiment were used for analysis of all plant measurements, nodule function and nodule microscopy. The second experiment consisted of two clover genotypes with the greatest variability in nodule effectiveness (T. subterraneum ‘Campeda’ and T. purpureum) with each of the four Rhizobium strains. The same +N and uninoculated controls as described above were also applied here and there were five replicates of all treatments. Bacteroid and acetylene reduction data are presented from this experiment. The third experiment consisted of two genotypes (T. subteranneum ‘Campeda’ and ‘Clare’) with R. l trifolii strains WSM1325 and SRDI565, with four replicates of each for confocal microscopy. Trifolium subterraneum ‘Clare’–SRDI565 was selected due to its ineffective-vegetative phenotype. Representative treatment images for T. subterraneum ‘Clare’ are presented since the replicates within a treatment behaved similarly.
Plant measurements
Five weeks after inoculation, shoots were removed from each plant (n = 5), dried at 60 °C for 48 h and weighed. After removal of nodules, roots were dried as above and weighed.
Leaf chlorophyll readings were measured using a SPAD 502 chlorophyll meter and used to calculate shoot N (Wood et al., 1993; Bullock and Anderson, 1998). Four readings were taken on the two oldest leaves of each plant and the mean value recorded. Shoots from plants in replicate 2 were selected to provide a range of chlorophyll values for each clover genotype and were analysed for nitrogen by the combustion technique using an Elementar Instrument. A standard curve was generated for leaf chlorophyll versus shoot [N] for each plant genotype (R2 = 0·87–0·96), and later the [N] for the remaining plant shoots (replicates 1, 3, 4 and 5) was estimated by regression. Dry matter and [N] for shoots were used as an index of nitrogen fixation, hereinafter referred to as fixed Nshoot:
Fixed Nshoot = ([N]shoot × SDW)treatment – ([N]shoot × SDW)uninoculated (1)
where uninoculated is the mean of uninoculated treatments for each plant genotype.
Nodule number, size and function
Representative plants were chosen from each plant–Rhizobium combination and photographed. The following was conducted for all treatments: nodules were removed, counted and dried (by desiccation at 20 °C within microfuge tubes which were filled with silica desiccating beads) prior to weighing. Mean weight per nodule was calculated by dividing total nodule weight by total nodule number for each plant. Nodule Function was calculated by dividing the Fixed Nshoot derived from eqn (1) by the total nodule mass per plant in milligrams (data in Table 2).
Table 2.
Parameters analysed to compare relative effectiveness of R. leguminosarum bv. trifolii strains on each clover genotype as listed
| Plant | Strain | Shoot d. wt (mg per plant) | Root d. wt (mg per plant) | Shoot [N] (mg g−1) | Nodule no. per plant | Total nodule mass (mg per plant) | Nodule size (mg per nodule) |
|---|---|---|---|---|---|---|---|
| T. subterraneum | +N | 217d | 135e | 44·0gh | – | ||
| ‘Campeda’ | WSM1325 | 178c | 79c | 43·8gh | 71de | 8·0f | 121bc |
| SRDI565 | 132b | 67bc | 40·0fg | 79e | 9·3g | 118bc | |
| SRDI943 | 50a | 30a | 21·8c | 166g | 3·4d | 20a | |
| WSM2304 | 27a | 16a | 12·0a | 135f | 2·6cd | 19a | |
| Uninoculated | 27a | 31a | 11·9a | – | |||
| T. subterraneum | +N | 246e | 110d | 39·5f | – | ||
| ‘Clare’ | WSM1325 | 253e | 105d | 42·8fgh | 72de | 6·8e | 96b |
| SRDI565 | 33a | 22a | 13·0a | 0 (pseudo)a | 0 (pseudo)a | 0 (pseudo)a | |
| SRDI943 | 149b | 62b | 39·1ef | 75·8e | 6·4e | 85b | |
| WSM2304 | 32a | 18a | 12·0a | 77·2e | 1·0b | 13a | |
| Uninoculated | 35a | 29a | 11·8a | – | |||
| T. purpureum | +N | 76b | 34c | 44·5h | – | ||
| WSM1325 | 60b | 27bc | 35·6e | 21b | 2·0c | 98b | |
| SRDI565 | 27a | 16ab | 31·1d | 55cd | 1·9c | 38a | |
| SRDI943 | 62b | 27bc | 38·9ef | 14ab | 2·2c | 165d | |
| WSM2304 | 10a | 7a | 18·0bc | 50c | 0·4ab | 8a | |
| Uninoculated | 11a | 11a | 17·5b | – | |||
| T. polymorphum | +N | 35c | 21d | 56·0j | – | ||
| WSM1325 | 5a | 4ab | 19·2bc | 22b | 0·4ab | 20a | |
| SRDI565 | 5a | 2a | 19·2bc | 27b | 0·4ab | 15a | |
| SRDI943 | 5a | 4ab | 19·2bc | 26b | 0·4ab | 15a | |
| WSM2304 | 18b | 13c | 49·5i | 14ab | 2·0c | 143cd | |
| Uninoculated | 5a | 8b | 18·7bc | – | |||
| P-value (l.s.d.) | |||||||
| Plant | <0·001 (10·8)* | n.s. | <0·001 (1·4) | <0·001 (9) | <0·001 (0·4) | 0·001 (17) | |
| Strain | <0·001 (18·64)* | <0·001 (9·9)* | <0·001 (1·7) | <0·001 (9) | <0·001 (0·4) | <0·001 (17) | |
| Plant × strain | <0·001 (23·4)* | <0·001 (14·0)* | <0·001 (3·5) | <0·001 (18) | <0·001 (0·8) | <0·001 (34) | |
Abbreviations: Shoot [N], nitrogen concentration in the shoots; +N, nitrogen-fed control; pseudo, pseudonodule, a type of nodule stalled during organogenesis.
* For shoot and root dry weights plant species were analysed separately due to large differences in species size. Different letters indicate significant differences between strains within a plant species. Significance values for T. subterraneum (‘Campeda’ and ‘Clare’) are in the table. Trifolium purpureum; Strain, P < 0·001, l.s.d. = 22·8; T. polymorphum; Strain, P < 0·001, l.s.d = 5·4. For all remaining parameters different letters indicate significant differences between Plant*Strain combinations.
Microscopy
Five representative, fresh nodules from each Rhizobium–clover combination in replicate 4 were selected from across the root system and put aside for microscopy. Observations of nodule morphology and bacteroid ultrastructure were made on a minimum of three nodules of each Rhizobium–clover combination; images that best represent each strain on T. subterraneum ‘Campeda’ are presented here. The nodules were immediately fixed in 3 % (v/v) gluteraldehyde in 0·025 m phosphate buffer upon harvesting, followed by secondary fixation in 2 % (w/v) osmium tetroxide in 0·025 m phosphate buffer. The tissues were then dehydrated in an ascending series of acetone, infiltrated in increasing concentrations of Spurr's resin in acetone, then embedded in fresh Spurr's epoxy resin (Spurr, 1969). For light microscopy, 1-μm-thick sections were cut and stained with a mixture of 1 % (w/v) methylene blue and 1 % (w/v) azur II (Richardson et al., 1960), then examined and photographed on an Olympus BX 51 microscope fitted with an Olympus DP-70 camera. For electron microscopy, 80- to 90-nm-thick sections were cut and stained with saturated aqueous uranyl acetate and lead citrate (Venable and Coggeshall, 1965) and examined and photographed on a Philips CM100 bio twin transmission electron microscope. Length and width of symbiosomes (differentiated bacteria enclosed in intracellular compartments) were determined from images at a low magnification (×2000–4000).
For confocal microscopy, longitudinal sections of nodules (attached to a root) were cut by hand using single-edge razor blades, and stained with 5 µm SYTO13 (Haynes et al., 2004) in DMSO, followed by 0·05 % (w/v) Calcofluor White in distilled water (fluorescent stain for cellulose), each for 5 min. Samples were rinsed in deionized water, transferred to slides, and a coverslip placed on top prior to immediate examination using a Leica TCS SP2 confocal laser scanning microscope (CLSM) in both fluorescence and transmitted light modes. Fluorescence was collected between 420 and 470 nm (blue) with 405 nm excitation for Calcofluor White, and between 500 and 550 nm (green) with 488-nm excitation for SYTO13. Extended focal series were compiled using Leica LCS-Lite software. The confocal images presented here are representative of replicate samples (eight nodules from four plants).
Bacteroid abundance
Nodules from the second experiment of T. subterraneum ‘Campeda’ and T. purpureum plants (nplant = 5) inoculated with the same R. l. trifolii strains were collected intact and stored in a cold microfuge tube at 4 °C for 24 h prior to bacteroid isolation. To avoid damage to small nodules, approx. 0·2 cm of root tissue either side of the nodule was left attached. A crude bacteroid extract was prepared according to Riley and Dilworth (1985) with slight adaptations. In brief, nodules were macerated in 1 mL of cold 0·1 m sodium phosphate buffer (pH 6·8) and the macerate was centrifuged at 100 g for 15 min. The supernatant was collected and centrifuged at 4300 g for 15 min; the resulting pellet was resuspended in 100 µL of cold 0·1 m sodium phosphate buffer (pH 6·8) and frozen at –20 °C. Samples were thawed at 4 °C and the total number of bacteroids in the 100-μL sample was counted in a Hawksley–Thoma cell counting chamber using a phase-contrast filter on an Olympus BX 51 microscope at ×400 magnification. Bacteroids were differentiated from vegetative rhizobia by comparing them with the reference images which were of free-living (culture-grown) R. l. trifolii strains at the same magnification. Vegetative-state bacteria were distinguished from bacteroids by crude estimation of the width of the cell; here we used the observation that the width of a vegetative-state bacterium is approx. one-third of the width of a differentiated bacteroid. It is yet to be shown if R. leguminosarum bv. trifolii bacteroids are predominately viable like B. japonicum or mostly terminally differentiated as is the case with R. leguminosarum bv. viciae strain TOM bacteroids (van den Bos and Broughton, 1981), a visual method for distinguishing vegetative bacteria from bacteroids was used in this study.
Acetylene reduction
Acetylene reduction was used to detect nitrogenase activity in nodules, specifically to determine whether ineffective nodules were capable of N2-fixation. The reduction of acetylene to ethylene was measured by gas chromatography (Shimadzu GC-8A, Kyoto, Japan) equipped with a 0·3-cm OD stainless steel column packed with 80- to 100-mesh activated molecular sieve; the column was maintained at 135 °C and the flame-ionization detector and injection port at 150 °C. Dry H2 was supplied at 70 mL min−1, air (medical grade) at 500 mL min−1 and carrier gas (N2) at 170 mL min−1. The output was analysed with a Shimadzu Integrator (Shimadzu model C-RIA Chromatopac) and peak area calibrated against an ethylene in air standard at 33 nmol mL−1. Plants of each Rhizobium treatment (n = 5) were removed from their growth vial and the vermiculite was gently loosened from the roots. The intact plants were immediately placed in 50-mL glass vials and closed with re-sealable rubber caps. Acetylene (5 mL) was injected into the vial and gas samples (1 mL) were taken after both 10 (T10) and 20 (T20) min. All nodules were collected and weighed as previously described.
Statistical analyses
Comparisons between treatments were made using ANOVA (GenStat 11, VSN International Ltd). All data were assessed for homoscedascity, non-correlations of means and variances and normality of distribution. Any deviations were corrected by logarithmic transformations prior to ANOVA where necessary. Duncan's pair-wise comparison (P < 0·05) test was used to establish which treatment means were significantly different. Where appropriate simple linear regression analysis was used to assess the relationship between variables. All references to significance in the text imply statistical significance at P < 0·05.
Amplification and phylogenetic comparison of 16S rRNA
An internal region of 1150 bp of the 16S ribosomal RNA (rRNA) sequence was amplified by PCR using conditions as described by Laguerre et al. (1996). Two sets of primers were used in separate PCR reactions to obtain sequences from both strands: 16S rRNA universal forward AGAGTTTGATCCTGGCTCAG with 16S rRNA universal reverse ACGGATACCTTGTTACGACTT (Weisburg et al., 1991) and FGPS1490 TGCGGCTGGATCACCTCCTT (Navarro et al., 1992) with FGPS6 GGAGAGTTAGATCTTGGCTCAG (Laguerre et al., 1996). 16S rRNA gene fragments from each strain were aligned by ClustaW. Kimura two-parameter distances were derived from the aligned sequences. A bootstrap analysis was performed with 1000 replicates in order to construct a consensus tree using the neighbor-joining method for each gene alignment separately. All alignments and phylogenetic analyses were performed using MEGA, version 5·0. The internal 16S rRNA sequences for both Rhizobium leguminosarum bv. trifolii strains SRDI565 and SRDI943 were submitted to GenBank (NCBI) and given accession numbers JN585113 and JN585114, respectively.
RESULTS
Phylogenetic comparison of rhizobial strains used in this experiment
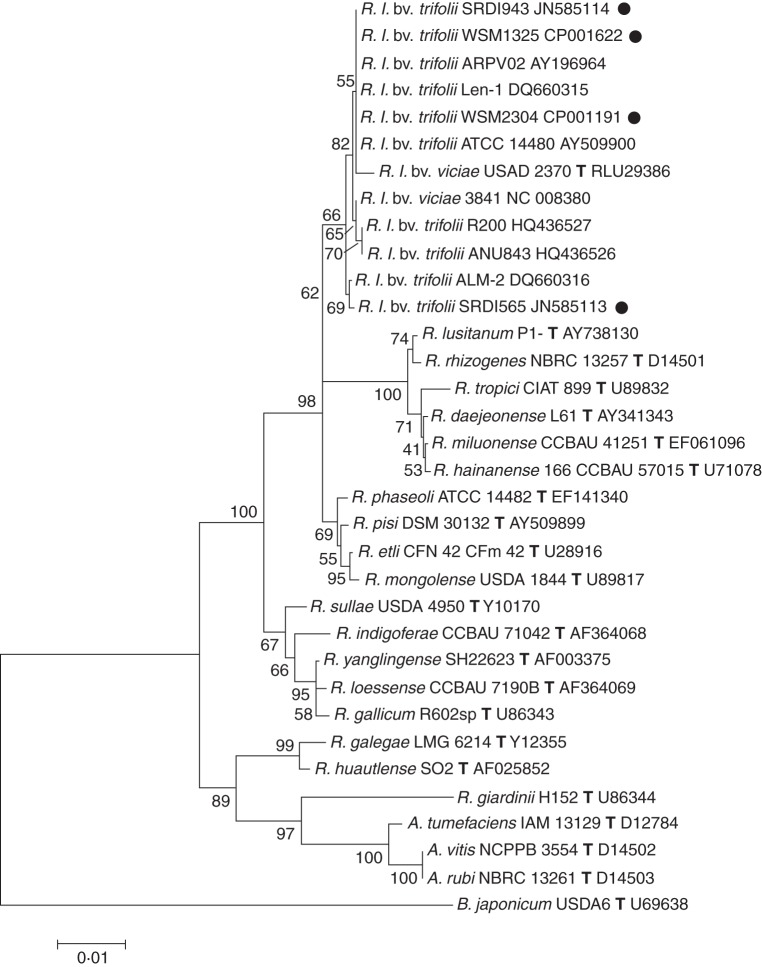
A partial 16S rRNA phylogenetic comparison of each of the four rhizobial strain used in this study was conducted to determine their phylogenetic similarity and to confirm that strains SRDI565 and SRDI943 group with other R. leguminosarum strains. The results demonstrated that they do in fact group within the same clade, which includes other identified Rhizobium leguminosarum strains and the type strain R. leguminosarum bv. viciae USDA 2370. Strains WSM1325, WSM2304 and SRDI943 share 100 % similarity within the partial rRNA gene region (1150 bp) examined yet they differ by 3 bp from SRDI565, 5 bp from R. leguminosarum bv. viciae USDA 2370 and 1 bp from the common laboratory strain R. leguminosarum bv. viciae 3841 (Fig. 2).
Fig. 2.
Phylogenetic tree showing the relationship of R. leguminosarum bv. trifolii strains WSM1325, WSM2304, SRDI943 and SRDI565 used here (marked with black circles) with a selection of type strains of Rhizobiaceae and additional Rhizobium strains. An internal region of 1150 bp of each 16S rRNA gene was aligned by ClustaW. All nucleotide sites were informative and there were no gap-containing sites. Kimura two-parameter distances were derived from the aligned sequences and a bootstrap analysis was performed with 1000 replicates in order to construct a consensus tree rooted to B. japonicum USDA6 using the neighbor-joining method for each gene alignment separately. Phylogenetic analysis was performed using MEGA, version 5·0. R. l. = R. leguminosarum; bv. = biovar; T = type strain.
Nitrogen fixation responses
The nitrogen-fed treatment (+N) produced the greatest shoot and root biomass, whilst the uninoculated treatment produced the lowest biomass for all clover genotypes. Trifolium subterraneum ‘Campeda’ inoculated with either WSM1325 or SRDI565 produced significantly more biomass than the uninoculated treatment (Table 2). Trifolium subterraneum ‘Clare’ produced most growth when inoculated with WSM1325 or SRDI943 (Table 2). Similarly, T. purpureum produced most growth with WSM1325 or with SRDI943 yet no growth response was evident with SRDI565 above that of the uninoculated treatment (Table 2). Trifolim polymorphum was the only genotype to show a growth response with strain WSM2304 (Table 2).
The nitrogen concentration [N] in the shoot followed similar trends to the biomass data (Table 2), as did the Fixed Nshoot (see eqn 1), reflecting the N-limited media in which the plants were grown. Although T. polymorphum produced less biomass due to its small seed size, [N] in the shoots of T. polymorphum inoculated with WSM2304 was consistent with the values for other plant genotypes tested.
Nodule number, size and function
Plants that were not inoculated with rhizobia (+N and uninoculated controls) did not produce nodules, indicating that the growth system remained free of contamination. All plants inoculated with R. l. trifolii formed nodules to varying degrees (Table 2). The combination of T. subterraneum ‘Clare’ and SRDI565 resulted in the production of a large number of small nodules that could not be counted accurately. In all combinations total nodule mass per plant was positively correlated with plant biomass (R2 = 0·71) and Fixed Nshoot (R2 = 0·69); however, only a weak correlation was observed between nodule number and plant biomass (R2 = 0·05), and there was no significant correlation between nodule number and Fixed Nshoot. Notably the two inoculation treatments that produced low biomass of T. subterraneum ‘Campeda’ (SRDI943 and WSM2304) and T. purpureum (SRDI565 and WSM2304) had at least twice the number of nodules compared with that of the WSM1325 inoculation treatment, which produced the highest biomass.
Nodule function (Nshoot mg nodule dry weight−1) was also calculated for each treatment (Fig. 3) and then converted to a percentage (NF %) of the best performing strain within a host genotype (Table 3); this parameter enabled a comparison of symbiotic efficiency between the inoculation treatments. Trifolium subterraneum ‘Clare’ inoculated with WSM1325 had the greatest nodule function of all the plant–Rhizobium combinations tested (Fig. 3); however, the NF of T. subterraneum ‘Clare’–SRDI943 as a percentage of that with WSM1325 was only 54 % (Table 3). WSM2304 was the only strain that produced N2-fixing nodules on T. polymorphum but the level of nodule function was only comparable to the Sub-optimal combinations of SRDI565 with T. subterraneum ‘Campeda’ and with T. purpureum (Fig. 3). The range of symbiotic effectiveness was most apparent when intra-host comparisons were made within T. subterraneum ‘Campeda’ and T. purpureum. NF % of strains on T. subterraneum ‘Campeda’ ranged from 100 % (WSM1325) to 57 % (SRDI565), 24 % (SRDI943) and ineffective (WSM2304). In T. purpureum nodule function ranked the strains differently: 100 % (SRDI943), 83 % (WSM1325), 30 % (SRDI565) and ineffective (WSM2304). NF (Fig. 3) indicates that WSM1325 was equally effective on these two hosts whilst strain SRDI943 showed the greatest variability across the four hosts.
Fig. 3.
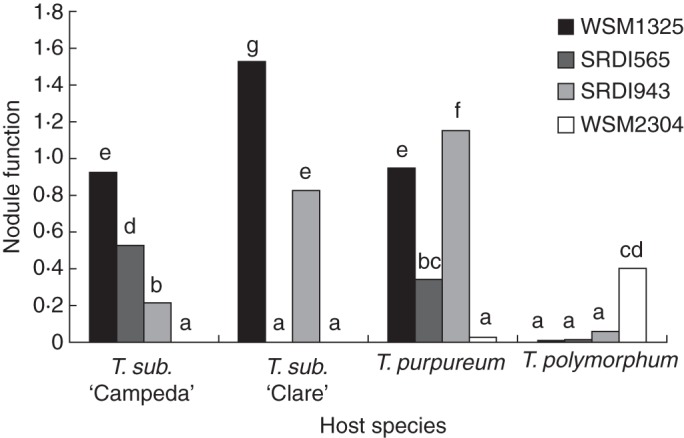
Nodule function expressed as Fixed Nshoot mg nodule dry weight−1 for four clover genotypes each inoculated with one of four R. leguminosarum bv. trifolii strains (WSM1325, SRDI565, SRDI943, WSM2304). Columns with different letters are significantly different. T. sub. = T. subterraneum.
Table 3.
Summarized morphological characteristics observed in nodules of Trifolium spp. 32 d after inoculation with a strain of R. leguminosarum bv. trifolii
| Plant | Strain | NF (% of reference strain)* | Morphological characteristics |
||
|---|---|---|---|---|---|
| Zones of differentiation (I–IV)† | Observed characteristics‡ | Effectiveness classification§ | |||
| T. subterraneum ‘Campeda’ | WSM1325 | 100e | I, II, II–III, III | IT, b, BD, NS | E |
| SRDI565 | 57d | I, II, II–III, III, IV | IT, b, BD, NS | S | |
| SRDI943 | 24bc | No | IT, b, BD#, NV | S | |
| WSM2304 | –1a | No | IT, b, BD#, NS | ID | |
| T. subterraneum ‘Clare’ | WSM1325 | 100e | I, II, II–III, III | IT, b, BD, NS | E |
| SRDI565 | 0a | No | IT, b, P | IV | |
| SRDI943 | 54d | I, II, II-III, III | IT, b, BD, NS | S | |
| WSM2304 | –2a | No | IT, b, BD#, NV | ID | |
| T. purpureum | WSM1325 | 83e | I, II, II–III, III | IT, b, BD, NS | E |
| SRDI565 | 30c | I, II, II–III, III, IV | IT, b, BD, NS | S | |
| SRDI943 | 100e | I, II, II–III, III | IT, b, BD, NS | E | |
| WSM2304 | 2a | I, II¶, IV | IT, b, BD#, NV | ID | |
| T. polymorphum | WSM1325 | 3a | No | IT, b, NS | IV |
| SRDI565 | 5ab | No | IT, b, BD#, NS | ID | |
| SRDI943 | 15abc | No | IT, b, NS | IV | |
| WSM2304 | 100e | I, II, II–III, III | IT, b, BD, NS | E | |
* NF (Nodule function), Fixed Nshoot mg−1 nodule d. wt expressed as percentage of most effective strain on that host. Different letters indicate treatments are significantly different across both plant species (P < 0·05, Duncan's multiple range test).
† Nodule zones: I, Meristem; II, pre-fixation; II–III, inter-fixation; III, fixation; IV, senescent.
‡ IT, Infection threads; b, bacteria; BD, bacteroids; NS, nodule shape stable; NV, nodule shape variable; P, pseudonodule.
§ E, Effective; S, sub-optimal; ID, ineffective-differentiated; IV, ineffective-vegetative.
¶ Presence of zone varies with sample.
# Bacteroids only in some plant cells, intracellular density variable.
Nodule morphology and bacteroid ultrastructure
Nodules were classified according to the consistency of nodule shape on the root system as well as the presence of the following: nodule zones (1–IV) (Hirsch, 1992), infection threads (IT), vegetative-state bacteria (b) and bacteroids (BD) (Table 3). This allowed nodules from each host–Rhizobium combination to be assigned to one of four groups, namely: Effective, Sub-optimal, Ineffective-differentiated and Ineffective-vegetative (Table 3). Hereinafter the phenotype group name is capitalized to distinguish it from general descriptors.
Effective nodules (E)
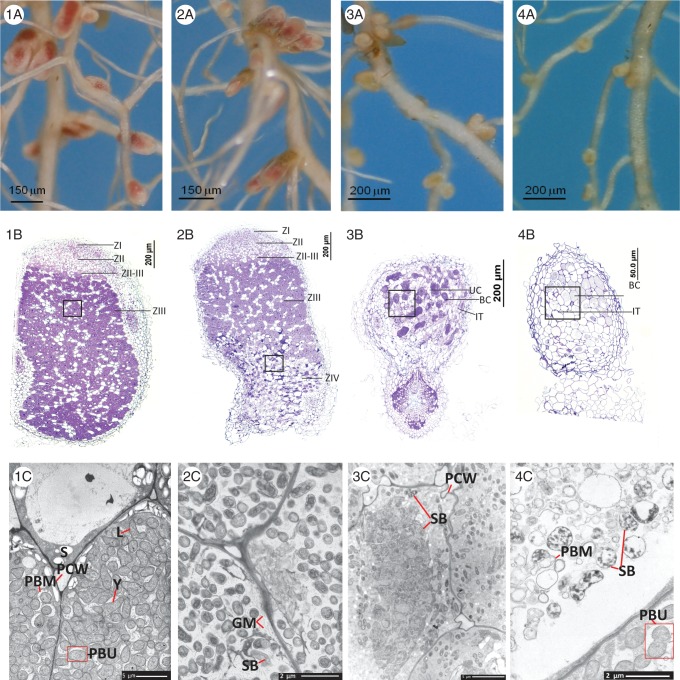
Nodules from the five Effective combinations of T. subterraneum ‘Campeda’–WSM1325, T. subterraneum ‘Clare’–WSM1325, T. purpureum–SRDI943, T. purpureum–WSM1325 and T. polymorphum–WSM2304 (Table 3) were consistent in both shape and size across the root system; the common appearance of these nodules was that they were elongated, increasing in width at the end distal to the root (e.g. Fig. 4-1A). These nodules contained each of the four regions typically observed in young indeterminate nodules including meristematic cells (I) as well as the zones of pre-fixation (II), inter-fixation (II–III) and fixation (III). A senescent zone (IV) was not observed in these nodules but is instead, more often observed in older clover nodules. The following were also observed: infection threads, bacteria in zones I and II and bacteroids in inter-zone II–III and zone III (Fig. 4-1B) with commonly one (Fig. 4-1A) but sometimes two bacteroids per symbiosome. These effective bacteroids were present as elongated rod shapes and in Y-, L- (Fig. 4-1C) and V-shapes. The variation of these Y-, L- and V- shapes may be related to the plane of view.
Fig. 4.
Comparison of symbiotic effectiveness resulting from inoculation of T. subterraneum ‘Campeda’ with R. leguminosarum bv. trifolii strains (1) WSM1325 (Effective), (2) SRDI565 (Sub-optimal), (3). SRDI943 (Sub-optimal) and (4). WSM2304 (Ineffective-differentiated). Images: (A) segments of the root system at 5 weeks post-inoculation; (B) stained sections of nodules imaged by light microscope; (C) electron micrographs of bacteroids. Scale bars: (1C) and (3C) = 5 µm; (2C) and (4C) = 2 µm. In (B) the boxes indicate the approximate region from which the section was taken for electron microscopy (C) and the nodule zones are indicated (Z1, nodule meristem; ZII, pre-fixation; ZII–ZIII, inter-fixation; ZIII, nitrogen-fixation; ZIV, senescent zone). Additional abbreviations in (B): UC, uninfected plant cell; BC, plant cells containing bacteroids; IT, infection thread. Abbreviations in (C): PBU, symbiosome; PBM, symbiosome membrane; Y, Y-shaped bacteroid; L, L-shaped bacteroid; S, starch; PCW, plant cell wall; SB; senescing bacteroid; GM, ghost membranes.
Sub-optimal nodules (S)
Nodule Function of the four Sub-optimal symbiotic combinations varied from 24 % to 57 % (Table 3). A defining feature of the Sub-optimal nodules of T. subterraneum ‘Campeda’–SRDI565 and T. purpureum–SRDI565 was the presence of a zone of senescence (IV) where plant cell wall breakage and loss of peribacteroid membrane integrity was observed. This occupied at least one-third of the total nodule volume (Fig. 4-2B). Nodules from the sub-optimal (24 % NF) combination of T. subterraneum ‘Campeda’–SRDI943 were variably shaped along the root system (Fig. 4-3A) and lacked nodule zones; however, senescing plant cells containing senescing bacteroids were observed adjacent to bacteroid-dense plant cells (Fig. 4-3B). Like-wise T. subterraneum ‘Clare’–SRDI943 nodules lacked a senescent zone but senescing bacteroids and remnants of plant and bacterial membranes were observed.
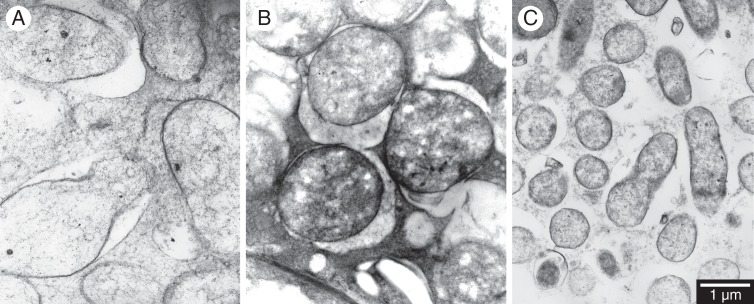
Y-shaped bacteroids, common in effective nodules, were absent in the Sub-optimal nodules of T. purpureum–SRDI565 but were present in low abundance in the Sub-optimal nodules of T. subterraneum ‘Campeda’–SRDI565. In addition, swollen and elongated V-shaped and L-shaped bacteroids visualized in effective nodules were common to T. subterraneum ‘Campeda’–SRDI565 but less common to T. purpureum–SRDI565. Furthermore nodules of both of these sub-optimal combinations contained more irregularly shaped bacteroids than their effective counterparts. Both the sub-optimal symbiosomes of T. subterraneum ‘Campeda’–SRDI565 and SRDI943 were on average shorter in length (2–3 µm and 0·5–1·6 µm, respectively) than the symbiosomes of the effective T. subterraneum ‘Campeda’–WSM1325 (2–4·5 µm; e.g. Fig. 5).
Fig. 5.
Comparison of bacteroid dimensions within nodules of T. subterraneum ‘Campeda’ carrying R. leguminosarum bv. trifolii (R. l. trifolii strains). Nodule samples are (A) WSM1325 (Effective); (B) SRDI565 (Sub-optimal) and (C) WSM2304 (Ineffective-differentiated). Note that bacteroid length and width was determined at a lower magnification (×2000–4000) than the higher magnification displayed here (×11 000). The scale in (C) also applies to (A) and (B).
Ineffective-differentiated nodules (ID)
The Ineffective-differentiated category includes T. subterraneum ‘Campeda’, T. subterraneum ‘Clare’ and T. purpureum with WSM2304 and T. polymorphum with SRDI565. These nodules were significantly smaller (Table 2 and, for example, T. subterraneum ‘Campeda’–WSM2304 in Fig. 4-4A) than the Effective nodules and they lacked distinct nodulation zones with the exception of T. purpureum–WSM2304 nodules which had semi-defined zones I, II and IV. Infection threads, bacteria and bacteroids were present in all Ineffective-differentiated nodules (e.g. Fig. 4-4B). Electron micrographs of T. subterraneum ‘Campeda’–WSM2304 showed the presence of intact symbiosomes with either immature-rod shaped bacteria (symbiosome length 1–1·5 µm) or elongating bacteroids (symbiosome length up to 3·5 µm) which were less swollen (symbiosome width range of 1–1·4 µm) than effective bacteroids (symbiosome width of 2·1–2·6 µm) (e.g. Fig. 5). The majority of the WSM2304 bacteroids extracted from T. subterraneum ‘Campeda’, T. subterraneum ‘Clare’ and T. purpureum were elongated rod-shaped and oval-shaped and infrequently Y-, L- and V-shaped. Senescing bacteroids were observed with partial to complete loss of peribacteroid membranes and loss of the symbiosome intracellular content as indicated by a less electron-dense symbiosome and a more electro-dense plant cytoplasmic area (external to the symbiosome; Fig. 4-4C). The presence of plant cells carrying senescent bacteroids adjacent to those carrying intact bacteroids and the lack of defined nodulation zones were similarly observed in the sub-optimal T. subterraneum ‘Campeda’–SRDI943 combination. It is pertinent to mention that the presence and density of bacteroids in these ID nodules and in T. subterraneum ‘Campeda’–SRDI943 nodules varied but is the more frequent phenotype observed.
Ineffective-vegetative nodules (IV)
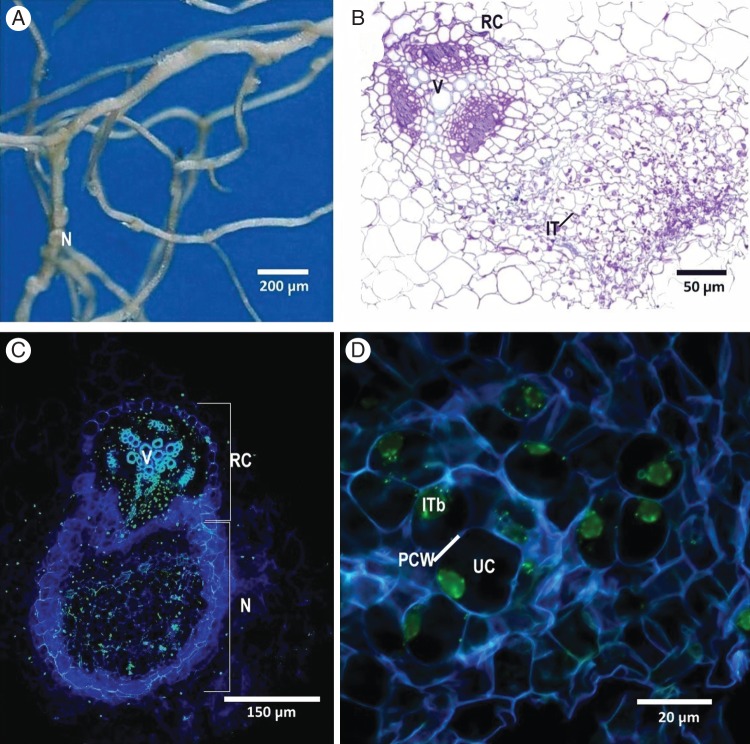
Nodules of the Ineffective-vegetative combinations of T. subterraneum ‘Clare’–SARDI565, T. polymorphum–SRDI943 and T. polymorphum–WSM1325 lacked bacteroids but contained infection threads and bacteria. The confocal sections of T. subterraneum ‘Clare’–SRDI565 nodules stained for nucleic acids (SYTO 13 stains green) confirmed that the infection threads were filled with bacterial cells (Fig. 6) and was compared with T. subterraneum ‘Campeda’–SRDI565 where stained bacteroids were visualized within the cytosol of the plant cells (data not shown).
Fig. 6.
Ineffective-vegetative phenotype arising from the interactions between T. subterraneum ‘Clare’ and R. leguminosarum bv. trifolii strain SRDI565: (A) root system with nodules (N) at 32 d post-inoculation; (B) nodule section stained with methylene blue and azur II and viewed by light microscopy; (C) and (D) CLSM images of a nodule stained with Syto 13 (green) and Calcofluor White (blue). Abbreviateions: UC, uninfected plant cell; ITb, infection threads filled with bacteria; PCW, plant cell wall; RC, root cortex; V, vascular tissue; IT, infection thread.
Bacteroid function
To determine if the reduced nodule function of Sub-optimal plant–Rhizobium combinations was caused by a limitation in the number of bacteroids, two clover genotypes with the greatest variability in nodule effectiveness with each of the four Rhizobium strains were selected for analysis of bacteroid abundance (Table 4). The Effective nodules of T. subterraneum ‘Campeda’ (with WSM1325) and T. purpureum (with WSM1325) contained a significantly higher number of bacteroids than those carried by the Sub-optimal or Ineffective nodules on that same host (Table 4). To determine if the bacteroids of the ineffective-differentiated nodules of T. purpureum and T. subterraneum ‘Campeda’ had an active nitrogenase we assessed their acetylene reduction (AR) activity. There was no detectable AR activity in nodules of T. purpureum–WSM2304 and only low levels in T. subterraneum ‘Campeda’–WSM2304. In contrast, AR activity was detected in each of the Sub-optimal and Effective combinations (Table 4).
Table 4.
The bacteroid pool of nodules collected from T. subterraneum ‘Campeda’ and T. purpureum plants 32 d after inoculation with R. leguminosarum bv. trifolii strains WSM1325, SRDI565, SRDI943 and WSM2304
| Plant | Strain | Nodule number plant−1 | Bacteroid number plant1 (×106) | Bacteroid number nodule−1 (×104) | AR detected |
|---|---|---|---|---|---|
| T. subterraneum ‘Campeda’ | WSM1325 | 14a | 1·00a | 7·88b | + |
| SRDI565 | 25b | 1·05a | 4·32a | + | |
| SRDI943 | 22b | 0·77a | 3·83a | + | |
| WSM2304 | 27b | 0·54a | 2·08a | + | |
| T. purpureum | WSM1325 | 5a | 0·61a | 13·44b | + |
| SRDI565 | 11b | 0·42a | 3·35a | + | |
| SRDI943 | 4a | 0·43a | 10·02ab | + | |
| WSM2304 | 9b | 0·18a | 2·04a | – |
AR, Acetylene reduction.
Different letters indicate treatments are significantly different within a plant species (P < 0·05, Duncan's multiple range test). Each plant species was analysed separately due to large differences in plant size.
This assessment of each combination of four Trifolium spp. with four R. leguminosarum bv. trifolii strains shows that abnormalities at several levels in nodule development and function contribute to the development of sub-optimal clover symbioses. Five Effective, four Sub-optimal, four Ineffective-differentiated and three Ineffective-vegetative nodulation phenotypes were identified in this study. The grouping of phenotypes in this way has allowed us to identify common developmental abnormalities and to hypothesize the underlying genetic causes.
DISCUSSION
In the Sub-optimal and Ineffective nodule phenotypes, abnormalities occurred after initiation of nodule organogenesis and infection thread initiation, suggesting that recognition in the early stages of symbiosis exists. A closer examination of the nodule structure, bacteroid ultrastructure and bacteroid abundance demonstrated that the development of Sub-optimal clover nodules was associated with the early senescence of functional bacteroids whilst Ineffective-vegetative nodules were associated with the failed endocytosis of bacteria from infection threads. The Ineffective-differentiated phenotype was the result of a low quantity of bacteroids, which had failed to differentiate beyond initial stages of elongation resulting in reduced nodule function.
The Sub-optimal nodules were less effective in N2-fixation outputs (biomass, shoot nitrogen and nodule function) than the effective nodules. Sub-optimal combinations varied in their nodule function relative to the effective strain (24–57 %), but commonly possessed fully differentiated bacteroids which have an active nitrogenase. The presence of an enlarged senescent zone in Sub-optimal T. subterraneum ‘Campeda’ and T. purpureum nodules infected with R. l. bv. trifolii strain SRDI565 and the dominance of irregularly shaped bacteroids indicates that bacteroid senescence had accelerated. This senescence should be considered premature given that it was not observed in other 5-week-old Effective nodules in this study.
Senescence in mature nodules involves a switch from a carbon sink to a nutrient source and up-regulation of various defence and oxidative stress responses (Van de Velde et al., 2006). Senescence has been characterized in pea nodules as a reactive oxygen species (ROS)-induced senescence (Matamoros et al., 2006). Redox buffers such as ascorbic acid and glutathione which scavenge ROSs have been identified in pea nodules (Matamoros et al., 1999) and may be required to minimize ROS accumulation and reduce the negative effect on nodule enzymes such as nitrogenase and leghaemoglobin reductase (Matamoros et al., 2006). These redox buffers are also important in stress perception and signalling, and may therefore be involved in perceiving nitrogen limitations and inducing nodule senescence.
Inoculation of four hosts with one strain (SRDI565) induced a diverse range of nodule phenotypes (S, ID and IV). The Sub-optimal nodules of T. subterraneum ‘Campeda’ and T. purpureum formed in response to SRDI565 had an equivalent bacteroid number per nodule. In contrast, the nodule function (Fixed Nshoot/g nodule) for T. purpureum–SRDI565 was almost half that of T. subterraneum ‘Campeda’–SRDI565. These findings suggest that the bacteroids of T. purpureum–SRDI565 are less active, which may be the result of a number of factors; e.g. insufficient synthesis of the nitrogenase protein, reduced availability of plant metabolites such as pyruvate required for electron transfer to nitrogenase (Riedel et al., 1995), branched-chain amino acids (Prell et al., 2009) or homocitrate, a component of the iron molybdenum cofactor of nitrogenase (Hakoyama et al., 2009).
Observations of Ineffective-differentiated nodules in this study were similar to that described by Bergersen (1955). In addition to Bergersen's findings, we observed that senescing bacteroids were more common than in the Effective nodules and that differentiated bacteroids were also observed in some plant cells. These differentiated bacteroids appear to have been disrupted in their development either during or post-elongation and either did not have an active nitrogenase or displayed low nitrogenase activity. Vasse et al. (1990) observed a similar disruption to bacteroid differentiation at either stage 2 or 3 in ineffective alfalfa nodules induced by fix::Tn5 mutant derivatives of Ensifer meliloti, which supports our conclusion of the stage of abortion of Ineffective-differentiated bacteroids.
In contrast to the Ineffective-differentiated nodules, the Ineffective-vegetative nodules did not contain bacteroids but did contain a small number of bacteria; the origin and identity of these is unknown, but we speculate that they are endophytic bacteria normally associated with clover roots. The presence of bacteria within infection threads was confirmed by CLSM in T. subterraneum ‘Clare’–SRDI565 nodule cells. Infection thread complications may be caused by deficiencies in secretion of acidic exopolysaccharide (EPS) by the infecting rhizobial strain (Skorupska et al., 1995) or by the type of EPS produced (Broughton et al., 2006). Since SRDI565 is not visibly deficient in EPS in comparison to the other strains investigated here (data not shown), we propose that the type of EPS produced by SRDI565 should be investigated.
The host identity is clearly a determinant of N2-fixation compatibility (see Table 2 and Fig. 3). Not surprisingly T. polymorphum, which is genetically isolated from the other species (subgenus Involucrarium; Table 1), had a narrow compatibility for symbiosis, forming Effective nodules solely with WSM2304. Specific host genes have been identified that regulate N2-fixation. For example, a Fix− mutant of Lotus japonicus (fen1) forms morphologically normal but functionally ineffective nodules due to its inability to synthesize homocitrate, which is essential for nitrogenase synthesis (Hakoyama et al., 2009). The Ineffective-differentiated nodules in this study likewise carry structurally stable symbiosomes; however those present were less swollen. There is evidence to suggest that bacteroid maturation events including swelling, membrane permeabilization, endoreduplication and loss of viability are associated with the availability of plant (nodule-specific cysteine-rich) peptides (Terpolilli et al., 2012).
In this comparison of nodule phenotypes, we have shown that there are three major responses to N2-fixation incompatibility between Trifolium spp. and R. l. trifolii strains: (1) failure of bacterial endocytosis from infection threads into plant cortical cells; (2) cessation of bacteroid differentiation post developmental stage 2; and (3) premature senescence of functional bacteroids. Future steps to target genes involved in the incompatibility response will be assisted by the genome sequences of R. l. trifolii strains used in this study. Sequencing of 13 additional R. l. trifolii strains is currently underway at the Joint Genome Institute (11 strains from CSP-231 and two from GEBA-RNB) and will further assist with comparative and functional studies of members of the R. leguminosarum clade. The development of a rapid genetic screen could enable researchers to assess the N2-fixation compatibility of soil-residing strains of R. l. trifolii with specific clover cultivars prior to their recommendation for agricultural use.
ACKNOWLEDGEMENTS
This work was financially supported by the Grains Research and Development Corporation via the National Rhizobium Program UMU00032 and the Crop and Plant Research Institute (Murdoch University). We thank Tian Rui and Liza Parkinson for their technical assistance with experiments. We gratefully acknowledge Prof. Janet Sprent and Prof. Sally Smith for their critical reading of the manuscript, and Prof. Craig Atkins for the use of his GC equipment.
LITERATURE CITED
- Anon Register of Australian Herbage Plant Cultivars. B. Legumes. 1. Clover. Trifolium purpureum Lois. (purple clover) cv. Paratta. 1972 http://www.pi.csiro.au/ahpc/legumes/pdf/paratta.pdf. (accessed 16 May 2012) [Google Scholar]
- Ballard R, Craig A, Charman N. Nodulation and growth of pasture legumes with naturalised soil rhizobia. 2. Balansa clover (Trifolium michelianum Savi) Journal of Experimental Agriculture. 2002;40:939–948. [Google Scholar]
- Bergersen FJ. The cytology of bacteroids from root nodules of subterranean clover (Trifolium subterraneum L.) Journal of General Microbiology. 1955;13:411–419. doi: 10.1099/00221287-13-3-411. [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Hanin M, Relic B, et al. Flavonoid-inducible modifications to rhamnan O antigens are necessary for Rhizobium sp. strain NGR234-legume symbioses. Journal of Bacteriology. 2006;188:3654–3663. doi: 10.1128/JB.188.10.3654-3663.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock DG, Anderson DS. Evaluation of the Minolta SPAD-502 chlorophyll meter for nitrogen management in corn. Journal of Plant Nutrition. 1998;21:741–755. [Google Scholar]
- Denton MD, Coventry DR, Bellotti WD, Howieson JG. Distribution, abundance and symbiotic effectiveness of Rhizobium leguminosarum bv. trifolii from alkaline pasture soils in South Australia. Australian Journal of Experimental Agriculture. 2000;40:25–35. [Google Scholar]
- Drew E, Ballard R. Improving N2 fixation from the plant down: compatability of Trifolium subterraneum L. cultivars with soil rhizobia can influence symbiotic performance. Plant and Soil. 2010;327:261–277. [Google Scholar]
- Drew EA, Charman N, Dingemanse R, Hall E, Ballard RA. Symbiotic performance of Mediterranean Trifolium spp. with naturalised soil rhizobia. Crop and Pasture Science. 2011;62:903–913. [Google Scholar]
- Ellison NW, Liston A, Steiner JJ, Williams WM, Taylor NL. Molecular phylogenetics of the clover genus (Trifolium – Leguminosae) Molecular Phylogenetics and Evolution. 2006;39:688–705. doi: 10.1016/j.ympev.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Gladstones J, Collins W. Subterranean clover as a naturalized plant in Australia. Journal of the Australian Institute of Agricultural Science. 1983;49:191–202. [Google Scholar]
- Hakoyama T, Niimi K, Watanabe H, et al. Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation. Nature. 2009;462:514–517. doi: 10.1038/nature08594. [DOI] [PubMed] [Google Scholar]
- Haynes JG, Czymmek KJ, Carlson CA, Veereshlingam H, Dickstein R, Sherrier DJ. Rapid analysis of legume root nodule development using confocal microscopy. New Phytologist. 2004;163:661–668. doi: 10.1111/j.1469-8137.2004.01138.x. [DOI] [PubMed] [Google Scholar]
- Herridge DF, Peoples MB, Boddey RM. Global inputs of biological nitrogen fixation in agricultural systems. Plant and Soil. 2008;311:1–18. [Google Scholar]
- Hirsch AM. Developmental biology of legume nodulation. New Phytologist. 1992;122:211–237. doi: 10.1111/j.1469-8137.1992.tb04227.x. [DOI] [PubMed] [Google Scholar]
- Howieson JG, Yates R, O'Hara GW, Ryder M, Real D. The interaction of Rhizobium leguminosarum biovar trifolii in nodulation of annual and perennial Trifolium spp. from diverse centres of origin. Australian Journal of Experimental Agriculture. 2005;45:199–207. [Google Scholar]
- Italy ISplCF. Trifolium subterraneum subsp. Subterranean Clover brachycalycinum ‘Campeda. Plant Variety Journal. 2000;13:74. [Google Scholar]
- Labandera CA, Vincent JM. Competition between an introduced strain and native Uruguayan strains of Rhizobium trifolii. Plant and Soil. 1975;42:327–347. [Google Scholar]
- Laguerre G, Mavingui P, Allard MR, et al. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Applied and Environmental Microbiology. 1996;62:2029–2036. doi: 10.1128/aem.62.6.2029-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight T. Efficiency of isolates of Rhizobium in the cowpea (Vigna unguiculata) group, with proposed additions to this group. Queensland Journal of Agricultural Science. 1949;6:61–76. [Google Scholar]
- Matamoros MA, Moran JF, Iturbe-Ormaetxe I, Rubio MC, Becana M. Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiology 121. 1999:879–888. doi: 10.1104/pp.121.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros MA, Loscos J, Coronado MJ, et al. Biosynthesis of ascorbic acid in legume root nodules. Plant Physiology. 2006;141:1068–1077. doi: 10.1104/pp.106.081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SH, Elliot RM, Sullivan JT, Ronson CW. Host-specific regulation of symbiotic nitrogen fixation in Rhizobium leguminosarum biovar trifolii. Microbiology. 2007;153:3184–3195. doi: 10.1099/mic.0.2007/006924-0. [DOI] [PubMed] [Google Scholar]
- Morris AC, Djordjevic MA. The Rhizobium leguminosarum biovar trifolii ANU794 induces novel developmental responses on the subterranean clover cultivar Woogenellup. Molecular Plant–Microbe Interactions. 2006;19:471–479. doi: 10.1094/MPMI-19-0471. [DOI] [PubMed] [Google Scholar]
- Navarro E, Simonet P, Normand P, Bardin R. Characterization of natural populations of Nitrobacter spp. using PCR/RFLP analysis of the ribosomal intergenic spacer. Archives of Microbiology. 1992;157:107–115. doi: 10.1007/BF00245277. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annual Review of Plant Biology. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiology and Molecular Biology Reviews. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prell J, White JP, Bourdes A, Bunnewell S, Bongaerts RJ, Poole PS. Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proceedings of the National Academy of Sciences of the USA. 2009;106:12477–12482. doi: 10.1073/pnas.0903653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prell J, Bourdes A, Kumar S, et al. Role of symbiotic auxotrophy in the Rhizobium–legume symbioses. PLoS One. 2010;5:pe13933. doi: 10.1371/journal.pone.0013933. http://dx.doi.org/10.1371/journal.pone.0013933 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve WG, O'Hara G, Chain P, et al. Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM1325, an effective microsymbiont of annual Mediterranean clovers. Standards in Genomic Sciences. 2010a;2:347–356. doi: 10.4056/sigs.852027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve WG, O'Hara G, Chain P, et al. Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM2304, an effective microsymbiont of the South American clover Trifolium polymorphum. Standards in Genomic Sciences. 2010b;2:66–76. doi: 10.4056/sigs.44642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson K, Jarret L, Finke E. Embedding in epoxy resins for ultrathin sectioning in electron microscopy. Stain Technology. 1960;35:313–323. doi: 10.3109/10520296009114754. [DOI] [PubMed] [Google Scholar]
- Riedel KU, Jouanneau Y, Masepohl B, Puhler A, Klipp W. A Rhizobium meliloti ferredoxin (FdxN) purified from Escherichia coli donates electrons to Rhodobacter capsulatus nitrogenase. European Journal of Biochemistry. 1995;231:742–746. doi: 10.1111/j.1432-1033.1995.tb20756.x. [DOI] [PubMed] [Google Scholar]
- Riley IT, Dilworth MJ. Cobalt requirement for nodule development and function in Lupinus angustifolius L. New Phytologist. 1985;100:347–359. [Google Scholar]
- Rys GJ, Bonish PM. Effectiveness of Rhizobium trifolii populations associated with Trifolium species in Taranaki, New Zealand. New Zealand Journal of Experimental Agriculture. 1981;9:327–335. [Google Scholar]
- Skorupska A, Bialek U, Urbaniksypniewska T, Vanlammeren A. Two types of nodules induced on Trifolium pratense by mutants of Rhizobium leguminosarum bv trifolii deficient in exopolysaccharide production. Journal of Plant Physiology. 1995;147:93–100. [Google Scholar]
- Slattery JF, Coventry DR. Acid-tolerance and symbiotic effectiveness of Rhizobium leguminosarum bv. trifoliii isolated from subterranean clover growing in permanent pastures. Soil Biology and Biochemistry. 1995;27:111–115. [Google Scholar]
- Sprent JI. Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytologist. 2007;174:11–25. doi: 10.1111/j.1469-8137.2007.02015.x. [DOI] [PubMed] [Google Scholar]
- Spurr A. A low viscosity epoxy resin embedding medium for electron microscopy. Journal of Ultrastructure Research. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Terpolilli JJ, O'Hara GW, Tiwari RP, Dilworth MJ, Howieson JG. The model legume Medicago truncatula A17 is poorly matched for N2 fixation with the sequenced microsymbiont Sinorhizobium meliloti 1021. New Phytologist. 2008;179:62–66. doi: 10.1111/j.1469-8137.2008.02464.x. [DOI] [PubMed] [Google Scholar]
- Terpolilli JJ, Hood GA, Poole PS. What determines the efficiency of N2-fixing Rhizobium–legume symbioses? Advances in Microbial Physiology. 2012;60:325–389. doi: 10.1016/B978-0-12-398264-3.00005-X. [DOI] [PubMed] [Google Scholar]
- Timmers AC, Soupene E, Auriac MC, et al. Saprophytic intracellular rhizobia in alfalfa nodules. Molecular Plant–Microbe Interactions. 2000;13:1204–1213. doi: 10.1094/MPMI.2000.13.11.1204. [DOI] [PubMed] [Google Scholar]
- Unkovich MJ, Baldock J, Peoples MB. Prospects and problems of simple linear models for estimating symbiotic N2 fixation by crop and pasture legumes. Plant and Soil. 2009;329:75–89. [Google Scholar]
- Van den Bos RC, Broughton WJ. Viability of Rhizobium leguminosarum endosymbionts in different stages of development. Archives of Microbiology. 1981;129:349–352. [Google Scholar]
- Van de Velde W, Guerra JC, De Keyser A, et al. Aging in legume symbiosis: a molecular view on nodule senescence in Medicago truncatula. Plant Physiology. 2006;141:711–720. doi: 10.1104/pp.106.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Camut S, Truchet G. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. Journal of Bacteriology. 1990;172:4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venable J, Coggeshall R. A simplified lead citrate stain for use in electron microscopy. The Journal of Cell Biology. 1965;25:407–408. doi: 10.1083/jcb.25.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JM. A manual for the practical study of the root-nodule bacteria. IBP handbook. Oxford: Blackwell Scientific Publications; 1970. [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CW, Reeves DW, Himelrick DG. Relationships between chlorophyll meter readings and leaf chlorophyll concentration, N status, and crop yield: a review. Proceedings of the Agronomy Society of New Zealand. 1993;23:1–9. [Google Scholar]
- Yates RJ, Howieson JG, Reeve WG, et al. Host-strain mediated selection for an effective nitrogen-fixing symbiosis between Trifolium spp. and Rhizobium leguminosarum biovar trifolii. Soil Biology and Biochemistry. 2008;40:822–833. [Google Scholar]
- Yates RJ, Howieson JG, Reeve WG, O'Hara GW. A re-appraisal of the biology and terminology describing rhizobial strain success in nodule occupancy of legumes in agriculture. Plant and Soil. 2011;348:255–267. [Google Scholar]