Abstract
Background and Aims
The actual number of domestications of a crop is one of the key questions in domestication studies. Answers to this question have generally been based on relationships between wild progenitors and domesticated descendants determined with anonymous molecular markers. In this study, this question was investigated by determining the number of instances a domestication phenotype had been selected in a crop species. One of the traits that appeared during domestication of common bean (Phaseolus vulgaris) is determinacy, in which stems end with a terminal inflorescence. It has been shown earlier that a homologue of the arabidopsis TFL1 gene – PvTFL1y – controls determinacy in a naturally occurring variation of common bean.
Methods
Sequence variation was analysed for PvTFL1y in a sample of 46 wild and domesticated accessions that included determinate and indeterminate accessions.
Key Results
Indeterminate types – wild and domesticated – showed only synonymous nucleotide substitutions. Determinate types – observed only among domesticated accessions – showed, in addition to synonymous substitutions, non-synonymous substitutions, indels, a putative intron-splicing failure, a retrotransposon insertion and a deletion of the entire locus. The retrotransposon insertion was observed in 70 % of determinate cultivars, in the Americas and elsewhere. Other determinate mutants had a more restricted distribution in the Americas only, either in the Andean or in the Mesoamerican gene pool of common bean.
Conclusions
Although each of the determinacy haplotypes probably does not represent distinct domestication events, they are consistent with the multiple (seven) domestication pattern in the genus Phaseolus. The predominance of determinacy in the Andean gene pool may reflect domestication of common bean prior to maize introduction in the Andes.
Keywords: Determinate growth habit, mutability, TFL1y, centroradialis, domestication, common bean, Phaseolus vulgaris
INTRODUCTION
Domestication is a selection process conducted by humans – either consciously or unconsciously – among wild plants for adaptation to human cultivation and consumption. This selection process has brought about marked changes in the morphology and physiology of crop plants (Evans, 1993). Indeed one of the hallmarks of full domestication among crop plants is their inability to survive without human intervention (Darwin, 1859). One of the traits selected during crop domestication is a more compact growth habit, manifested, for example in the legumes, by a series of traits such as reduced branching, shorter internodes, fewer nodes, reduced twining and, in some cases, a determinate stem ending (Smartt, 1990). The wild relatives of food legumes are generally viny, herbaceous plants, with a high level of branching, many nodes, long and twining internodes, and diageotropic branch growth. The vininess allows plants to compete with surrounding plants for light in the shrubby or arboreal vegetation in which these wild plants grow naturally (e.g. Debouck et al., 1993; Freyre et al., 1996). Under these conditions, a non-viny, determinate plant is presumably at a severe disadvantage and is unlikely to survive. Determinacy is, therefore, a trait selected during or after domestication.
The growth habit of legumes, such as common bean, is constructed in modular fashion (e.g. Tanaka and Fujita, 1979). Individual modules consist of a subtending internode, a leaf and, in later stages of growth, an inflorescence in the axil of the leaf. In plants with an indeterminate growth habit, the production of new modules continues until senescence. Wild common beans are all indeterminate, as is a sizable fraction of domesticated beans. In determinate domesticated types, the production of modules is interrupted by the appearance of a terminal inflorescence in the early stages of plant development, e.g. around the time of production of the fifth trifoliolate node (Debouck, 1991). Thus, stems have a finite length, and flowering occurs earlier than in indeterminate types. In Phaseolus, a determinate growth habit exists not only in domesticated common bean (P. vulgaris), but also in domesticated runner bean (P. coccineus) and lima bean (P. lunatus). The determinate growth habit has been exploited for crop breeding to decrease plant biomass and to optimize allocation between vegetative and reproductive growth (Cober and Tanner, 1995). Since the determinate growth habit allows mechanical harvesting with a shorter growing cycle, cultivars with the determinacy trait have been preferred in several crop species, such as tomato, soybean and common bean (Pnueli et al., 1998; Kelly, 2001; Boote et al., 2003). In common bean, determinacy has been adopted at higher latitudes or in cooler climates to select earlier varieties adapted to shorter growing seasons. Many bush snap bean varieties are determinate because the trait shortens the period of pod production and leads to a more homogeneous harvest.
Two phenotypic loci have been reported that control determinacy in common bean. The fin locus was first identified in 1915 (Norton, 1915) and has since been mapped to chromosome Pv01 (Koinange et al., 1996). This locus appears to be responsible for determinacy in most varieties with a determinate growth habit, most of whom have an origin in the Andean gene pool of common bean (Singh et al., 1991). A second, as yet unnamed locus, was mapped on chromosome Pv07 (Kolkman and Kelly, 2003) and may be responsible for determinacy observed in some Michigan navy bean cultivars, which arose from an artificial mutagenesis programme in the 1950s (Kelly, 2001).
We have reported that one of the arabidopsis Terminal Flower 1 (TFL1) homologues in common bean, PvTFL1y, co-segregated with the fin locus on Pv01 in a recombinant inbred line population resulting from a cross between an indeterminate, wild and a determinate, domesticated genotype (Kwak et al., 2008). The arabidopsis TFL1 gene is a member of the CETS family (TFL1, Flowering Locus T, Mother of FT and TFL1, Arabidopsis thaliana centroradialis homologue, Twin sister of ft and brother of ft and tfl; Kobayashi et al., 1999). Centroradialis (CEN) from Antirrhinum and Self-pruning (sp) from tomato are also homologous to TFL1. This family has an important role in controlling the switch from a vegetative to a reproductive phase and inflorescence morphology (Pnueli et al., 1998; Ratcliffe and Riechmann, 2002; Carmel-Goren et al., 2003). TFL1 homologues have been identified in some legume species (Bennloch et al., 2007). In pea, two homologous loci were identified: PsTFL1a as the Determinate (DET) gene and PsTFL1c as the Late Flowering (LF) gene (Foucher et al., 2003). Ljcen1, the Lotus japonicus TFL1 homologue, is expressed in the inflorescence meristem; its ectopic expression in arabidopsis delays flowering (Guo et al., 2006). In soybean, a homologue of TFL1 is responsible for the determinate growth habit controlled by the Dt1 locus (Tian et al., 2010) and the region including the Dt1 locus on soybean chromosome 19 is collinear to the part of common bean chromosome Pv01 harbouring the fin locus (S. Repinski and P. Gepts, unpubl. results). In addition, co-segregation between the fin locus and determinacy is maintained in a much larger (n approx. 1470) population, expression of PvTFL1y determinate mutants is sharply reduced, and complementation in A. thaliana of the mutant tfl1 phenotype with the wild-type PvTFL1 gene has been demonstrated (Repinski et al., 2012). Thus, PvTFL1y is the gene that underlies the determinacy locus fin at the phenotypic level on chromosome Pv01 (Kwak et al., 2008).
Here we address the origin and diversity of the mutation(s) in the PvTFL1y gene leading to the determinacy phenotype in the domesticated gene pool by analysing the geographic and gene pool distribution of mutations and haplotypes in a sample of 349 wild and domesticated common bean accessions. Wild common bean consists of two geographic gene pools, Mesoamerican and Andean, which had already diverged prior to domestication (Gepts and Debouck, 1991; Kwak and Gepts, 2009). Domesticated common bean originated from those two gene pools through independent domestication events (Gepts and Debouck, 1991; Kwak and Gepts, 2009; Kwak et al., 2009). Thus, we examine whether this essential feature of bean domestication originated in one of the gene pools or both of them, and whether it originated from a single mutation or multiple mutations at the PvTFL1y locus.
MATERIALS AND METHODS
Plant materials
The sequence of PvTFL1y was determined in a sub-sample of 47 accessions of common bean (Phaseolus vulgaris; Supplementary Data Table S1) taken from a larger sample of 349 accessions that represented the entire P. vulgaris germplasm and whose genetic structure has been characterized with microsatellites (Kwak and Gepts, 2009). The 47 accessions included a sample of wild and domesticated entries from the Andean and Mesoamerican gene pools. All determinate accessions belonged to the type I growth habit as defined by Singh (1982); the number of nodes bearing trifololiate leaves on the main stem varies from four to seven (D.G. Debouck and O. Toro, pers. obs.).
High-throughput genomic DNA extraction
The plant tissue samples were freeze-dried in a lyophilizer for 2 d. Genomic DNA was extracted using a modified high-throughput cetyltrimethylammonium bromide (CTAB) method with stainless steel beads (Chen and Ronald, 1999). Two stainless steel beads were put into each well and the block was sealed with a sealing mat. The block was placed under the grinder and the dried plant tissue samples were powdered for 6 min. Then, 800 µL of lysis buffer [2 % CTAB, 1·4 m NaCl, 20 mm EDTA (pH 8·0), 100 mm Tris base, 1 % β-mercaptoethanol, 5 % polyvinylpolypyrrolidone (PVPP), and RNase A] was added. Samples were sealed with adhesive foil sealer tape and then incubated at 65 °C in a hybridization oven for 20 min. After incubation, 800 µL of sample solutions were transferred to a new 96 deep-well block and 800 µL of chloroform:isoamylalcohol (24:1) was added. After mixing by inverting five times, samples were centrifuged for 10 min at 4000 g at room temperature. The upper aqueous layers were transferred into another 96-well block and DNA was precipitated by the addition of the same volume of isopropanol. After centrifugation for 10 min at 4000 g at room temperature, DNA was washed twice with 70 % ethanol and dissolved in 100 µL of TE (10 mm Tris–HCl, 0·1 mm EDTA, pH 8·0).
Association study
To test the association of the retrotransposon insertion in PvTFL1y and determinacy, a sample of 349 accessions comprising 100 wild and 249 domesticated accessions; 194 Andean and 155 Mesoamerican accessions) was genotyped using the TFL1-la and TFL1-F4 primers (Repinski et al., 2012). PCR mixtures contained approx. 45 ng of total genomic DNA, 200 mm dNTP, 0·2 µm TFL1y-la and TFL1-F4 primers, Expand long template buffer with 1·75 mm MgCl2, and 0·5 U of expand long template PCR Taq polymerase (Roche Applied Science) in a 30 µL total reaction volume. The PCR cycle consisted of 2 min at 94 °C and ten cycles of 10 s at 94 °C, 30 s at 56 °C and then 4 min at 68 °C, followed by 25 cycles of 15 s at 94 °C, 30 s at 56 °C and then 4 min at 68 °C with a 20 s increase every cycle with a PTC-220 thermocycler (MJ Research). Accessions whose DNA was not amplified with the TFL1-la and TFL1-F4 primer pair were tested with the TFL1-R7 (5′-GAGCTCACACTCCTTTTCTC-3′) and TFL1-F12(5′-CAAACCAACAGTAAAAACCA-3′) primer pair, with the same PCR cycles. A 5 µL aliquot of amplified product with 1 µL of loading buffer were loaded in a 1·5 % agarose gel in 0·5× TBE and genotyped for their size.
Individual sequences have been deposited in GenBank (accession nos JN418219–418266)
Analysis of genetic and geographic relationships of PvTFL1y
Using primers designed based on G12873 and Midas sequences, the sequences of PvTFL1y from 47 accessions were determined; haplotypes were identified based on the presence or absence of specific polymorphisms (Supplementary Data Table S2). A haplotype network of PvTFL1y coding sequences including three introns was generated using TCS ver. 1·21 (Clement et al., 2000) after gap coding (Simmons and Ochoterena, 2000). Since the A4d haplotype (observed in accessions G02686, G04647, G24705 and G24800) does not contain coding sequences of PvTFL1y, it was excluded from the haplotype network. The presence/absence of the retrotransposon insertion haplotype among domesticated types was displayed in the previously published Neighbor–Joining tree and STRUCTURE bar plot (Kwak and Gepts, 2009).
RESULTS
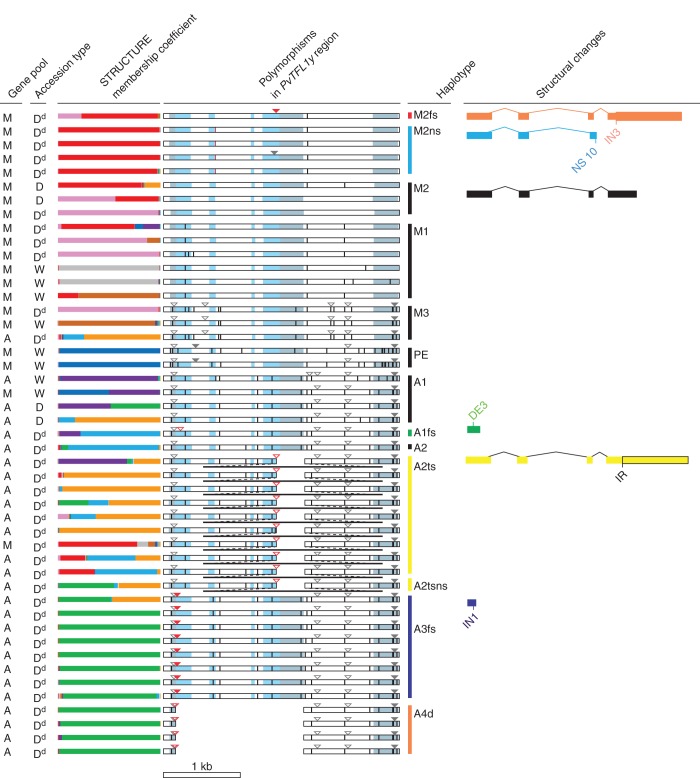
The polymorphic sites in PvTFL1y sequences from 47 common bean accessions are shown in Fig. 1. At this locus, we identified 47 nucleotide substitutions and 14 insertions or deletions (Supplementary Data Tables S1 and S2). Among them, eight mutations are expected to lead to a marked change in gene function. Most strikingly, a 4171 bp insertion was observed in the fourth exon (mutation IR; Fig. 1 and Supplementary Data Table S2; Repinski et al., 2012). This mutation is expected to lead to a frameshift, with a change in amino acid sequence and a lengthening of the protein coded at the 3′ end of the fourth exon. The other mutations (Fig. 1 and Supplementary Table S2) included four frameshift mutations: two with a 2 bp insertion in exons (IN1 and IN3), one with a 2 bp deletion (DE3) and one exon–intron boundary mutation (NS10) leading to a putative splicing failure and altered gene structure. In addition, there was a deletion of the entire coding region (DE2), and two non-synonymous mutations (NS9 and NS18; Supplementary Data Table S2). Interestingly, all mutations expected to lead to a marked change in gene function were found only among determinate (domesticated) accessions. The marked loss of expression expected from these mutations has been confirmed by quantitative PCR for two of the mutations, the retrotransposon insertion (in an Andean background; mutation IR, Supplementary Data Table S2) and the putative splicing mutant (in a Mesoamerican background; mutation NS10, Supplementary Data Table S2).
Fig. 1.
Haplotype diversity in the PvTFL1y region. Each row is a different accession, presented in the same order as in Supplementary Tables S1 and S3. Gene pool: assignment for each accession – M, Mesoamerican; A, Andean. Accession type – domestication status of accessions: W, wild (all indeterminate); D, domesticated indeterminate; Dd, domesticated determinate. STRUCTURE membership coefficient – ancestry of accessions in nine STRUCTURE clusters (Kwak et al., 2009) as indicated by the length of each coloured segments: pink, races Jalisco and Durango; red, race Mesoamerica; grey, Mesoamerican wild; brown, Mesoamerican and Colombian wild; dark blue, ancestral Peru and Ecuador wild; purple, Andean wild; green, race Peru; light blue, race Chile; orange, race Nueva Granada. Polymorphisms in the PvTFL1y region – light blue, coding region; grey, untranslated region; horizontal bar, nucleotide substitution; open triangle, deletion; filled triangle, insertion; black vertical bar, 4171 bp retrotransposon insertion, not to scale. Polymorphisms expected to affect gene expression are indicated as red-coloured symbols. Information about each polymorphism is given in Supplementary Data Table S2. Haplotype – haplotype of PvTFL1y. Black bar, wild type; coloured bar, mutations with severe structural changes expected: d, deletion of the entire coding region; fs, frameshift; ns, non-synonymous substitution; ts, transposon insertion. Structural changes – the expected structural changes are depicted and the mutations leading to changes are shown. DE, deletion; IN, insertion; IR, insertion of retrotransposon; NS, non-synonymous substitution.
A haplotype network illustrates the relationships among the different haplotypes at the PvTFL1y locus (Fig. 2). The genetic background (Andean or Mesoamerican) of individual accessions was determined by microsatellite fingerprinting (Kwak and Gepts, 2009). Two haplotypes in PvTFL1y were correlated with determinate phenotypes in the Mesoamerican gene pool (M2ns and M2fs) and five haplotypes in the Andean gene pool (A3fs, A2, A2ts, A2tsns and A1fs). Initially, the A4d haplotype could not be assigned to the Andean or Mesoamerican gene pool since the entire coding sequence was deleted. However, the adjacent 5′ upstream and 3′ downstream sequences showed that the A4d haplotype could clearly be assigned to the Andean gene pool (Fig. 1). Some determinate accessions shared their haplotype with indeterminate accessions (M1, M2 and M3), implying the existence of unidentified mutations in PvTFL1y or other determinacy genes.
Fig. 2.
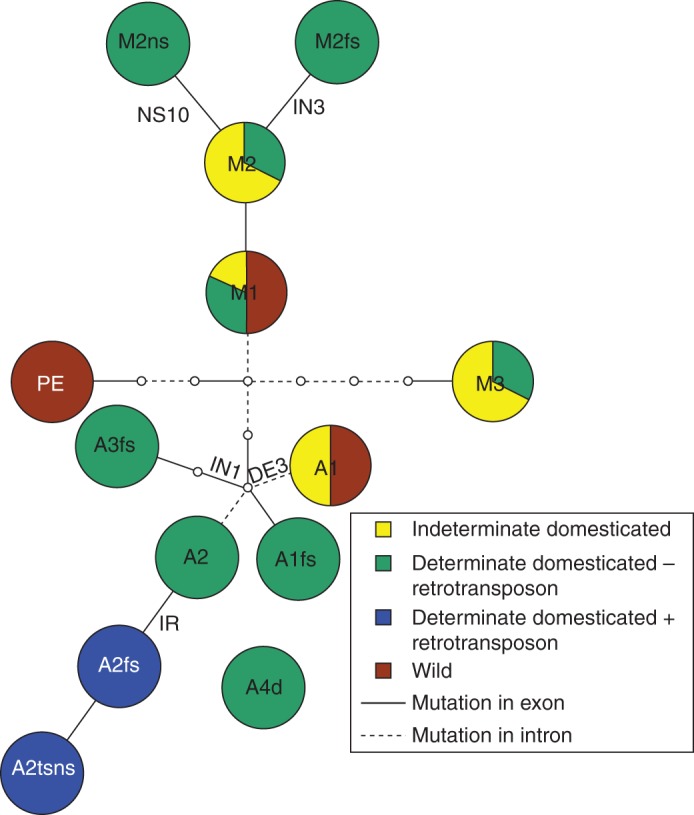
Haplotype network of PvTFL1y. The frequency of determinate, retrotransposon insertion and domesticated accessions for each haplotype is shown as a proportion of colour in the node circle. Yellow, indeterminate domesticated; green, determinate domesticated without retrotransposon insertion; blue, determinate domesticated with retrotransposon insertion; brown, wild. For haplotype abbreviations, see text. Mutations in exon and intron are as indicated in the key. Nodes without circles were not sampled in this study.
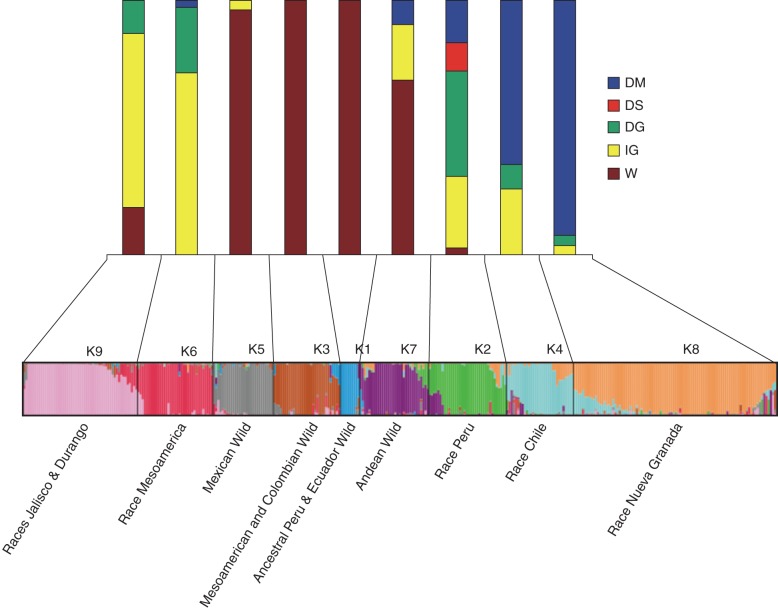
The association test revealed that determinate types were distributed mainly in Andean subpopulations, as determined by a STRUCTURE analysis: K2, K4, K7 and K8 (Fig. 3; Kwak and Gepts, 2009). Among the 160 determinate accessions included in this study, 73 % showed the retrotransposon insertion in PvTFL1y. Conversely, all accessions with the retrotransposon insertion in PvTFLly were determinate and Andean, except accession G03807 (Brasil2), which has a Mesoamerican background (Fig. 3, Group K6, and Supplementary Table S3). Although the determinate accessions with retrotransposon insertion were closely related genetically, they were distributed broadly in South and North America, Europe, Africa and Asia. All determinate types originating outside the American continent (23 accessions from Europe, seven from Africa and four from Asia) showed the haplotype with the retrotransposon insertion (A2ts and A2tsns). Determinate accessions without retrotransposon insertion in PvTFL1y were distributed in both Andean (K2, K4 and K8) and Mesoamerican (K6 and K9) subpopulations (Fig. 3), further underscoring the diversity of mutations leading to the determinacy phenotype. Also, four accessions with haplotype A4d, characterized by a deletion of the entire PvTFL1y sequence, were found only in the Andean region, specifically on the eastern slope of the Andes in Colombia and Peru, in sub-population K2 (Fig. 3, Supplementary Data Table S3).
Fig. 3.
The distribution of determinacy in each sub-population (K1–K9) as identified by STRUCTURE (Kwak and Gepts, 2009). The lower part of the figure represents a STRUCTURE bar graph of wild and domesticated bean simple sequence repeat diversity according to Kwak and Gepts (2009). The upper part of the figure represents the proportion of four classes of bean accessions based on status (wild vs. domesticated), growth habit (determinate vs. indeterminate) and presence or absence of a retrotransposon insertion in the PvTFL1y locus: DM, domesticated, determinate, retrotransposon insertion into PvTFL1y region; DS, domesticated, determinate, deletion of whole PvTFL1y region; DG, domesticated, determinate, wild-type PvTFL1y region; IG, domesticated, indeterminate, wild-type PvTFL1y region; W, wild, indeterminate, wild-type PvTFL1y region.
DISCUSSION
Independent data based on co-segregation (Kwak et al., 2009; Repinski et al., 2012), gene expression and complementation (Repinski et al., 2012) show that PvTFL1y is the molecular locus underlying the phenotypic locus fin (Repinski et al., 2012), the recessive allele of which controls determinacy. Our present results provide additional information supporting the role of PvTFL1y. All the variation observed among indeterminate entries involved synonymous substitutions (Fig. 1) that probably did not affect the expression of PvTFL1y in these entries. In contrast, among determinate entries, several mutation types (Fig. 1 and Supplementary Data Table S2) were observed that potentially cause reduction or elimination of gene expression. These types include deletion of the entire locus (haplotype A4d), non-synonymous mutation (haplotype M2ns), indels (haplotype M2fs, A1fs and A3fs), a potential splicing site alteration (haplotype M2ns) and a retrotransposon insertion in the fourth exon (haplotypes A2ts and A2tsns). The predicted elimination of gene expression in two of these haplotypes (M2ns and A2ts) was verified experimentally (Repinski et al., 2012). These expression-disrupting mutations were only observed among determinate types and are consistent with the recessiveness of the trait.
Three haplotypes (M1, M2 and M3) included both determinate and indeterminate types (Fig. 2), suggesting that sequences responsible for determinacy in these accessions are not situated within the sequence covered by the current analysis. These sequences could be located immediately adjacent to the PvTFL1y sequence, perhaps in a regulatory sequence 5′ upstream of PvTFL1y, or at other determinacy loci in the genome, such as the PvTFL1x and PvTFL1z loci.
Haplotypes involving the retrotransposon insertion deserve more scrutiny. First, this type of mutation confirms the importance of transposons in general as a factor shaping evolution (Barabaschi et al., 2012; Kejnovsky et al., 2012; Leitch and Leitch, 2012; Slotkin et al., 2012). Evidence is growing that transposons have influenced the crop domestication process. For example, they have increased the expression of genes involved in morphological development, such as the Tb1 locus in maize (Studer et al., 2011), have been involved in the re-structuring of certain domesticated Gossypium species (Palmer et al., 2012), and render certain genes more inducible by external stresses such as cold (Naito et al., 2009). Secondly, it may be more than a coincidence that the insertion occurred in the fourth exon of the PvTFL1y gene. In A. thaliana, experiments show that the fourth exon is crucial for the full inhibitory effect of the TFL1 gene to be expressed (Ahn et al., 2006). Assuming that the mechanism of action is similar in P. vulgaris, natural insertional mutagenesis in the fourth exon may ensure strong inactivation of PvTFL1y and a stable and highly heritable expression of the determinacy phenotype. Thirdly, of all the PvTFL1y haplotypes, the retrotransposon insertion is the most frequent one. It occurs in 73 % of the determinate accessions. Outside of the Americas, it is the only PvTFL1y haplotype among determinates. The predominance of this haplotype among determinate domesticated types could be attributed to several, non-mutually exclusive reasons. The A2ts haplotype may have achieved its predominance by virtue of being the earliest haplotype to appear in the Andean domesticated gene pool. Other haplotypes would have appeared later and achieved a more limited geographic distribution. Alternatively, the A2ts haplotype is linked to a factor promoting domestication in addition to determinacy. Selection for this additional factor combined with hitchhiking could speed up the spread of the A2ts haplotype. A quantitative trait locus (QTL) analysis of the domestication syndrome of common bean has shown that the fin locus is linked to both the Ppd (photoperiod sensitivity) locus and a QTL for internode length (L5) on chromosome Pv01 (Koinange et al., 1996). Further research is needed to isolate the genes involved and determine the genotypes at these linked loci in the domesticated sample involved. Further research is needed to determine the specific phenotypic effects (in addition to determinacy) especially on growth habit of the different PvTFL1y haplotypes.
The haplotype tree of PvTFL1y sequence variation (Fig. 2) is consistent with the overall genealogy of common bean, as determined through numerous studies (e.g. Kwak and Gepts, 2009; Kwak et al., 2009). Wild types (all of which are indeterminate) as well as indeterminate domesticated types are located in the centre of the tree, whereas determinate domesticated types were located at the end of branches. This topology confirms the derived nature of determinacy. Furthermore, there are two major branches, one including Andean accessions and the other Mesoamerican accessions (as determined by independent, neutral microsatellite markers), confirming that determinacy has been selected independently in the two major geographic gene pools. With two exceptions (Suppelementary Data Fig. S1 and Table S1), there is congruity between haplotypes and gene pool assignments, i.e. all the haplotypes were observed in a single gene pool. The first exception was haplotype A2ts, the major Andean PvTFL1y haplotype, which was also represented in a single determinate Mesoamerican accession, G03807 or Brasil2. Microsatellite diversity analyses show that Brasil2 has the characteristic microsatellite complement of a Mesoamerican accession (Kwak and Gepts, 2009). Our observations therefore suggest that Brasil2 acquired the determinacy trait from an Andean accession by an initial hybridization followed by multiple backcrosses to one or more Mesoamerican parents. This somewhat unlikely scenario would account for the existence of an Andean PvTFL1y allele in a predominantly Mesoamerican background. The second exception is G24999, which carries a Mesoamerican PvTFL1y haplotype (M3) in an Andean microsatellite background and probably arose through a similar introgression procedure to that of Brasil2. The existence of these two exceptions is important because it shows the high level of congruence between haplotypes, and gene pool membership is not due to reproductive isolation between the two gene pools, but presumably through the limited opportunities for hybridization between these geographically isolated gene pools. Genes causing F1 Andean × Mesoamerican dwarf lethality – located on the linkage groups Pv02 and Pv11 (Hannah et al., 2007) – are also unlinked to the fin-PvTFL1 loci (located on linkage group Pv01).
The higher frequency of determinacy in the Andean domesticated gene pool can be traced back to the early stages of agriculture in the Andean domestication centre. Whereas in the Mesoamerican domestication centre, farmers could use maize (domesticated in Mesoamerica, presumably in the Balsas River basin) as a physical support for viny common bean varieties, a plant that could fulfil a similar function was not domesticated in the Andes as most crop plants in the Andes are of lesser stature or stem strength compared with maize (e.g. root crops, tarwi or lupine, and quinoa). Common bean remnants in the Andes have been retrieved in contexts associated with charcoal, the latter dated to 7700 calibrated years BP (Kaplan and Lynch, 1999). The dissemination of maize from its centre of domestication in Mexico into Panama and Ecuador has been dated to 7800–7000 calibrated years BP (Dickau et al., 2007) and 5300–4950 calibrated years BP (Zarrillo et al., 2008), respectively. In Peru, the oldest maize remains documented so far were dated to 3600–4000 calibrated years BP (Perry et al., 2006). Plant starch grains embedded in tooth calculus have been retrieved from an early archaeological site in northern Peru. These starch grains, dated to 5500–9000 BP, originated from three legume species (common bean, peanut and Inga sp.) and squash (Piperno et al., 2008). No maize was described at this site. Thus, there may have been an incipient agricultural period in which maize had not yet been integrated into the Andean cropping and human nutritional systems. In the absence of maize as a physical support, selection for a bush growth habit would have taken place.
The PvTFL1y results show a different pattern of distribution among domesticated gene pools from that of genes selected during domestication in other crop species. The information available so far suggests that the tb1 gene in maize, which increases apical dominance and reduces branching, has a unique origin (Clark et al., 2006) and that it became fixed early in maize domesticated history (Jaenicke-Després et al., 2003). In the case of rice, a species with two major geographic gene pools like common bean (indica and japonica), several domestication alleles have a unique origin in one or the other gene pool based on haplotype information in spite of the existence of the two gene pools. Two alleles (the Wxb-derived wx, conditioning grain stickiness, Yamanaka et al., 2004; and rc, controlling red pericarp, Sweeney et al., 2007) were widely distributed across japonica and indica types. Both Wxb-derived wx and rc originated in the japonica gene pool, implying that there was a wholesale introgression of these domestication alleles from one gene pool into the other. The other three alleles (Wxa-derived wx, Yamanaka et al., 2004; rc-s, Sweeney et al., 2007; and qsh1, Konishi et al., 2006) originated in the japonica or indica gene pools but acquired a more limited distribution across the rice domesticated gene pool. These rice examples differ from the situation in common bean where PvTFL1y-associated determinacy alleles originated in both the Andean and Mesoamerican gene pools. Taken together, the pattern of domestication in Phaseolus is one of multiple occurrences, involving several species (five domesticated species), different gene pools within species (Andean and Mesoamerican gene pools) and repeated selection of the same trait within gene pools. This situation may be due to the relatively young age of the genus amounting to some 4 million years (Delgado-Salinas et al., 2006). This time frame has allowed species to diverge enough to develop contrasting adaptations such as those shown by the five domesticated species, but has been insufficient for the different species to lose a putative predisposition to domestication.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank M. Welsh (Western Regional Plant Introduction Station, USDA, Prosser, WA, USA) for supplying seed samples. This work was supported by the USDA CSREES NRI Plant Genome program to P.G. and a graduate student scholarship from the UC Davis Department of Plant Science to M.K.
LITERATURE CITED
- Ahn JH, Miller D, Winter VJ, et al. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO Journal. 2006;25:605–614. doi: 10.1038/sj.emboj.7600950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabaschi D, Guerra D, Lacrima K, et al. Emerging knowledge from genome sequencing of crop species. Molecular Biotechnology. 2012;50:250–256. doi: 10.1007/s12033-011-9443-1. [DOI] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Serrano-Mislata A, Madueno F. Floral initiation and inflorescence architecture: a comparative view. Annals of Botany. 2007;100:659–676. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boote KJ, Jones JW, Batchelor WD, Nafziger ED, Myers O. Genetic coefficients in the CROPGRO-soybean model: links to field performance and genomics. Agronomy Journal. 2003;95:32–51. [Google Scholar]
- Carmel-Goren L, Liu YS, Lifschitz E, Zamir D. The SELF-PRUNING gene family in tomato. Plant Molecular Biology. 2003;52:1215–1222. doi: 10.1023/b:plan.0000004333.96451.11. [DOI] [PubMed] [Google Scholar]
- Chen DH, Ronald PC. A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Molecular Biology Reporter. 1999;17:53–57. [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nature Genetics. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cober E, Tanner J. Performance of related indeterminate and tall determinate soybean lines in short-season areas. Crop Science. 1995;35:361–364. [Google Scholar]
- Darwin C. On the origin of species by means of natural selection. London: J. Murray; 1859. [Google Scholar]
- Debouck DG. Systematics and morphology. In: Van Schoonhoven A, Voysest O, editors. Common beans; research for crop improvement. Wallingford, UK: CAB Publishing; 1991. [Google Scholar]
- Debouck DG, Toro O, Paredes OM, Johnson WC, Gepts P. Genetic diversity and ecological distribution of Phaseolus vulgaris in northwestern South America. Economic Botany. 1993;47:408–423. [Google Scholar]
- Delgado-Salinas A, Bibler R, Lavin M. Phylogeny of the genus Phaseolus (Leguminosae): a recent diversification in an ancient landscape. Systematic Botany. 2006;31:779–791. [Google Scholar]
- Dickau R, Ranere AJ, Cooke RG. Starch grain evidence for the preceramic dispersals of maize and root crops into tropical dry and humid forests of Panama. Proceedings of the National Academy of Sciences, USA. 2007;104:3651–3656. doi: 10.1073/pnas.0611605104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LT. Crop evolution, adaptation, and yield. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- Foucher F, Morin J, Courtiade J, et al. DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. The Plant Cell. 2003;15:2742–2754. doi: 10.1105/tpc.015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyre R, Ríos R, Guzmán L, Debouck D, Gepts P. Ecogeographic distribution of Phaseolus spp. (Fabaceae) in Bolivia. Economic Botany. 1996;50:195–215. [Google Scholar]
- Gepts P, Debouck DG. Origin, domestication, and evolution of the common bean, Phaseolus vulgaris. In: Voysest O, Van Schoonhoven A, editors. Common beans: research for crop improvement. Wallingfor, UK: CAB Publishing; 1991. [Google Scholar]
- Guo XZ, Zhao Z, Chen JH, Hu XH, Luo D. A putative CENTRORADIALIS/TERMINAL FLOWER 1-like gene, Ljcen1, plays a role in phase transition in Lotus japonicus. Journal of Plant Physiology. 2006;163:436–444. doi: 10.1016/j.jplph.2005.04.037. [DOI] [PubMed] [Google Scholar]
- Hannah MA, Kramer KM, Geffroy V, et al. Hybrid weakness controlled by the dosage-dependent lethal (DL) gene system in common bean (Phaseolus vulgaris) is caused by a shoot-derived inhibitory signal leading to salicylic acid-associated root death. New Phytologist. 2007;176:537–549. doi: 10.1111/j.1469-8137.2007.02215.x. [DOI] [PubMed] [Google Scholar]
- Jaenicke-Despres V, Buckler ES, Smith BD, Gilbert MTP, Cooper A, Doebley J, Paabo S. Early allelic selection in maize as revealed by ancient DNA. Science. 2003;302:1206–1208. doi: 10.1126/science.1089056. [DOI] [PubMed] [Google Scholar]
- Kaplan L, Lynch T. Phaseolus (Fabaceae) in archaeology: AMS radiocarbon dates and their significance for pre-Columbian agriculture. Economic Botany. 1999;53:261–272. [Google Scholar]
- Kejnovsky E, Hawkins JS, Feschotte C. Plant transposable elements: biology and evolution. In: Wendel JF, Greilhuber J, Dolezel J, Leitch IJ, editors. Plant genome diversity. Vol. 1. Vienna: Springer; 2012. pp. 17–34. [Google Scholar]
- Kelly JD. Remaking bean plant architecture for efficient production. Advances in Agronomy. 2001;71:109–143. [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Koinange EMK, Singh SP, Gepts P. Genetic control of the domestication syndrome in common-bean. Crop Science. 1996;36:1037–1045. [Google Scholar]
- Kolkman JM, Kelly JD. QTL conferring resistance and avoidance to white mold in common bean. Crop Science. 2003;43:539–548. [Google Scholar]
- Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- Kwak M, Gepts P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae) Theoretical and Applied Genetics. 2009;118:979–992. doi: 10.1007/s00122-008-0955-4. [DOI] [PubMed] [Google Scholar]
- Kwak M, Velasco DM, Gepts P. Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris) Journal of Heredity. 2008;99:283–291. doi: 10.1093/jhered/esn005. [DOI] [PubMed] [Google Scholar]
- Kwak M, Kami JA, Gepts P. The putative Mesoamerican domestication center of Phaseolus vulgaris is located in the Lerma-Santiago basin of Mexico. Crop Science. 2009;49:554–563. [Google Scholar]
- Leitch AR, Leitch IJ. Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytologist, 2012;194:629–646. doi: 10.1111/j.1469-8137.2012.04105.x. [DOI] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- Norton JB. Inheritance of habit in the common bean. American Naturalist. 1915;49:547–561. [Google Scholar]
- Palmer SA, Clapham AJ, Rose P, et al. Archaeogenomic evidence of punctuated genome evolution in Gossypium. Molecular Biology and Evolution. 2012;29:2031–2038. doi: 10.1093/molbev/mss070. [DOI] [PubMed] [Google Scholar]
- Perry L, Sandweiss DH, Piperno DR, et al. Early maize agriculture and interzonal interaction in southern Peru. Nature. 2006;440:76–79. doi: 10.1038/nature04294. [DOI] [PubMed] [Google Scholar]
- Piperno DR, Dillehay TD. Starch grains on human teeth reveal early broad crop diet in northern Peru. Proceedings of the National Academy of Sciences, USA. 2008;105:19622–19627. doi: 10.1073/pnas.0808752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Carmel-Goren L, et al. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125:1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Riechmann JL. Arabidopsis transcription factors and the regulation of flowering time: a genomic perspective. Current Issues in Molecular Biology. 2002;4:77–91. [PubMed] [Google Scholar]
- Repinski SL, Kwak M, Gepts P. The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theoretical and Applied Genetics. 2012;124:1539–1547. doi: 10.1007/s00122-012-1808-8. [DOI] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology. 2000;49:369–381. [PubMed] [Google Scholar]
- Singh SP. A key for identification of different growth habits of Phaseolus vulgaris L. Annual Report of the Bean Improvement Cooperative. 1982;25:92–95. [Google Scholar]
- Singh SP, Gepts P, Debouck DG. Races of common bean (Phaseolus vulgaris L., Fabaceae) Economic Botany. 1991;45:379–396. [Google Scholar]
- Slotkin RK, Nuthikattu S, Jiang N. The impact of transposable elements on gene and genome evolution. In: Wendel JF, Greilhuber J, Dolezel J, Leitch IJ, editors. Plant genome diversity. Vol. 1. Vienna: Springer; 2012. pp. 35–58. [Google Scholar]
- Smartt J. Grain legumes: evolution and genetic resources. Cambridge: Cambridge University Press; 1990. [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nature Genetics. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Cho YG, Park YJ, Williamson SH, Bustamante CD, McCouch SR. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genetics. 2007;3 doi: 10.1371/journal.pgen.0030133. e133. http://dx.doi.org/10.1371/journal.pgen.0030133 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Fujita K. Photosynthesis and yield components in relation to grain yield of the field beans. Journal of the Faculty of Agriculture, Hokkaido University. 1979;59:145–238. [Google Scholar]
- Tian Z, Wang X, Lee R, et al. Artificial selection for determinate growth habit in soybean. Proceedings of the National Academy of Sciences, USA. 2010;107:8563–8568. doi: 10.1073/pnas.1000088107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Nakamura I, Watanabe KN, Sato YI. Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theoretical and Applied Genetics. 2004;108:1200–1204. doi: 10.1007/s00122-003-1564-x. [DOI] [PubMed] [Google Scholar]
- Zarrillo S, Pearsall DM, Raymond JS, Tisdale MA, Quon DJ. Directly dated starch residues document early formative maize (Zea mays L.) in tropical Ecuador. Proceedings of the National Academy of Sciences, USA. 2008;105:5006–5011. doi: 10.1073/pnas.0800894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.