Abstract
Two important signaling systems involved in the growth and development of plants, those triggered by the photoreceptor phytochrome and the hormone abscisic acid (ABA), are involved in the regulation of expression of the NPR1 gene of Lemna gibba. We previously demonstrated that phytochrome action mediates changes in ABA levels in L. gibba, correlating with changes in gene expression evoked by stimulation of the phytochrome system. We have now further characterized phytochrome- and ABA-mediated regulation of L. gibba NPR1 gene expression using a transient particle bombardment assay, demonstrating that regulatory elements controlling responses to both stimuli reside within 156 nucleotides upstream of the transcription start. Linker scan (LS) analysis of the region from −156 to −70 was used to identify two specific requisite and nonredundant cis-acting promoter elements between −143 to −135 (LS2) and −113 to −101 (LS5). Mutation of either of these elements resulted in a coordinate loss of regulation by phytochrome and ABA. This suggests that, unlike the L. gibba Lhcb2*1 promoter, in which phytochrome and ABA regulatory elements are separable, the phytochrome response of the L. gibba NPR1 gene can be attributed to alterations in ABA levels.
We previously showed that D treatments of light-grown plants of both Lemna gibba and Arabidopsis thaliana resulted in significant increases in endogenous ABA concentrations (Weatherwax et al., 1996). More importantly, the phytochrome-signaling pathway was implicated in mediating these internal changes in ABA levels. Because these ABA changes occur rather gradually in response to changing light regimes, we chose to investigate whether a specific gene that shows a response to light over a similar time scale might, in fact, be regulated by the effects of phytochrome on endogenous ABA levels.
The NPR1 gene of L. gibba is a member of a class of genes isolated based on their increased level of expression in D-treated plants and decreased level in response to brief R illumination. The increase in transcription of the L. gibba NPR1 gene in response to D is fairly slow, with only a 30% increase detectable 6 h after initial D treatment of intermittent-R-grown plants (Okubara and Tobin, 1991). The transcription of the NPR1 gene can be negatively regulated by phytochrome action, with detectable decreases in transcription occurring within 2 to 4 h in response to a brief R illumination (Okubara et al., 1993). This time frame is in sharp contrast to the more rapid R-induced transcriptional changes observed within 15 to 30 min for other phytochrome-responsive genes (for review, see Terzaghi and Cashmore, 1995), including the Lhcb genes (Tobin, 1981). The predicted NPR1 protein bears strong resemblance to the late embryogenesis abundant (LEA) proteins, the genes of which are induced by ABA during seed maturation (Okubara and Tobin, 1991); we found that the expression of NPR1 could be regulated in a dose-dependent fashion by changes in ABA levels (Williams et al., 1994).
Therefore, we sought to determine the extent of NPR1 transcriptional regulation that might be occurring solely because of phytochrome-mediated changes in ABA levels and what sequences in the promoter might govern these responses. The promoter of the NPR1 gene does not contain the motifs found to be necessary for phytochrome repression of the well-characterized oat phyA gene (Bruce et al., 1991). There is also no similarity to the REα and REβ motifs that mediate phytochrome regulation of the L. gibba Lhcb2*1 gene (Degenhardt and Tobin, 1996). These 10-bp regulatory element motifs, which include highly conserved sequences found in promoters of other Lhcb genes, are likely to function as cis-acting elements that repress promoter activity in D. However, mutations within these Lhcb regulatory elements, which are sufficient to abolish the R-induced increase in Lhcb transcription, continue to allow ABA regulation. This finding demonstrates that the promoter regulatory elements for ABA repression and phytochrome induction of the Lhcb2*1 gene must be separable (Weatherwax et al., 1996). We now address the issue of separable phytochrome and ABA promoter regulatory elements in the L. gibba NPR1 promoter.
Previous work using the L. gibba particle bombardment transient assay demonstrated phytochrome repression and ABA induction in a 5′ deletion construct containing −354 bp from the transcriptional start (Williams et al., 1994). In this study we used a targeted LS mutagenic approach to define more precisely specific promoter elements that could mediate the NPR1 responses to phytochrome and ABA. Our results demonstrate that there are at least two separate cis-acting elements that are necessary to mediate ABA inducibility. Significantly, unlike the situation with the L. gibba Lhcb2*1 gene, mutation of either of these elements resulted in not only a loss of ABA induction but also of phytochrome repression.
MATERIALS AND METHODS
Growth and Treatment of Plants
Lemna gibba L. G-3 was grown aseptically on liquid E medium in continuous white light at 27°C. Etiolated plants were supplemented with 3 μm kinetin and grown in intermittent (2 min/8 h) R (Tobin, 1981). Plants were treated for 10 min with far red light to convert Pfr to Pr before being placed in D.
Promoter Constructs
The NlaIII-XhoI fragment from NR11 (Williams et al., 1994) was inserted into the SphI and SalI sites on pDR101 (Riggs and Chrispeels, 1987). The resulting construct, designated NPR1–156, contained −156 to +430 of the L. gibba NPR1 promoter relative to the transcription start in a translational fusion to the LUC reporter gene.
Eight pairs of annealed oligonucleotides were synthesized, substituting different lengths of the sequence CTTGCTAGCATCC, containing an NheI site, for equal-length segments of the entire 87-bp region from −156. LS oligonucleotides were first cloned into the HindIII and BsaAI sites of p11XH. The mutant oligonucleotides synthesized for replacement were cloned directly into the HindIII site in the NPR1–156 deletion construct.
LS1: AGCTTTTGCTAGCATCCTCGGCAATTTTAGATAAAGACGTCCATTTTTTCGACGCGTGTCGTTAC; GTAACGACACGCGTCGAAAAAATGGACGTCTTTATCTAAAATTGCCGAGGATGCTAGCAAA
LS2: AGCTTCATGCAAAGAGGTTGCTAGCATTAGATAAAGACGTCCATTTTTTCGACGCGTGTCGTTAC; GTAACGACACGCGTCGAAAAAATGGACGTCTTTATCTAATGCTAGCAACCTCTTTGCATGA
LS3: AGCTTCATGCAAAGAGGTCGGCAATTTGCTAGCAAGACGTCCATTTTTTCGACGCGTGTCGTTAC; GTAACGACACGCGTCGAAAAAATGGACGTCTTGCTAGCAAATTGCCGACCTCTTTGCATGA
LS4: AGCTTCATGCAAAGAGGTCGGCAATTTTAGATAACTTGCTAGCATCCTTCGACGCGTGTCGTTAC; GTAACGACACGCGTCGAAGGATGCTAGCAAGTTATCTAAAATTGCCGACCTCTTTGCATGA
LS5: AGCTTCATGCAAAGAGGTCGGCAATTTTAGATAAAGACGTCCATTTTTCTTGCTAGCATCCTTAC; GTAAGGATGCTAGCAAGAAAAATGGACGTCTTTATCTAAAATTGCCGACCTCTTTGCATGA
LS6: AGCTTCATGCAAAGAGGTCGGCAATTTTAGATAAAGACGTCCATTTTTTCGACGCGTGTCGTTGCTAGCATAAACGTCGTGGAAGGACGAGTCTTTGAGGGCA; CGCGTGCCCTCAAAGACTCGTCCTTCCACGACGTTTATGCTAGCAACGACACGCGTCGAAAAAATGGACGTCTTTATCTAAATTGCCGACCTCTTTGCATGA
LS7:AGCTTCATGCAAAGAGGTCGGCAATTTTAGATAAAGACGTCCATTTTTTCGACGCGTGTCGTTACGTGGCGTTGCTAGCATCAAGGACGAGTCTTTGAGGGCA; CGCGTGCCCTCAAAGACTCGTCCTTGATGCTAGCAACGCCACGTAACGACACGCGTCGAAAAAATGGACGTCTTTATCTAAAATTGCCGACCTCTTTGCATGA
LS8:AGCTTCATGCAAAGAGGTCGGCAATTTTAGATAAAGACGTCCATTTTTTCGACGCGTGTCGTTACGTGGCGAAACGTCGTGGTTGCTAGCATCTTTGAGGGCA; CGCGTGCCCTCAAAGATGCTAGCAACCACGACGTTTCGCCACGTAACGACACGCGTCGAAAAAATGGACGTCTTTATCTAAAATTGCCGACCTCTTTGCATGA
Standard techniques were used for all DNA manipulations (Sambrook et al., 1989). All constructs were sequenced using the Sequenase dideoxy sequencing kit (United States Biochemical). Analysis of transcription factor-binding sites on the NPR1 promoter was performed using the TESS transcription element search software of J. Schug and G.C. Overton (University of Pennsylvania, Philadelphia; http://agave.humgen.upenn.edu/tess/index.html).
Transient Assays
Microprojectile bombardments were performed as previously described (Williams et al., 1994). Plants were bombarded with wild-type NPR1–156 or mutant promoter constructs containing a series of CTTGCTAGCATCC LS substitutions fused to the firefly LUC reporter. A minimal rice (Oryza sativa L.) Act promoter fused to the uidA (Act) reporter (McElroy et al., 1990) was included as an internal control for transformation efficiency. Three different amounts of the internal control, giving different ratios of reporter:internal control DNA were used for bombardments: 5.0:1.5, 5.0:1.0, and 5.0:0.5 μg. Following bombardment, plants either remained in D or received 2 min of R. Plants were then returned to D for 16 to 18 h before being assayed for LUC and GUS activities (Okubara et al., 1993). Where indicated, either 10 μm ABA or water was added to the plants 4 h before bombardment. Background LUC and GUS activities from plants bombarded with gold particles only were subtracted from all raw experimental LUC and GUS values. The reporter activity for each treatment (five or more independent transformations) was determined by analysis of covariance using the internal standard activity as the dependent variable. Differences between the average ratio of reporter to internal standard activity were tested for significance by the Student's t test. Values for the NPR1–156 (wild type) R treatment group were used to normalize data from multiple experiments.
RESULTS
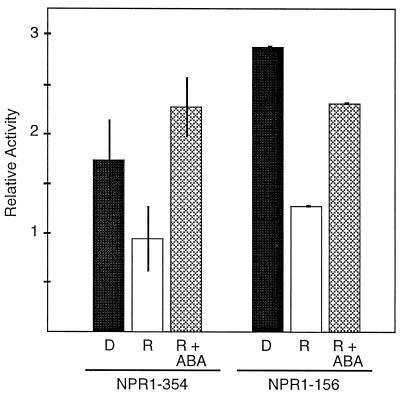
We previously showed that phytochrome and ABA regulation of the NPR1 promoter can be mediated by a sequence containing 354 bp upstream of the transcription start site. Figure 1 shows a comparison of the phytochrome and ABA responsiveness of this 354-bp promoter construct (NPR-354) to a shorter promoter construct containing 156 bp upstream of the transcription start site (NPR-156). Deletion of the additional 200 bp had no qualitative effect on overall expression levels. The shorter NPR-156 construct continued to display both the phytochrome-mediated reduction in relative activity from the D level in response to R and ABA-mediated induction. Furthermore, the magnitude of the responses to both phytochrome action and ABA application was comparable to what was observed in the longer (NPR-354) promoter construct. Thus, sequences downstream of 156 from the transcription start contain sufficient information for both phytochrome repression and ABA induction of the NPR1 gene.
Figure 1.
Sequences downstream of −156 from the transcription start confer a response to both ABA and phytochrome action. Following bombardment with the NPR1–354 or NPR1–156 constructs, intermittent R-grown L. gibba were treated with D (black bars), 2 min of R (white bars), or 2 min of R plus 10 μm ABA (cross-hatched bars); D and R samples received water as a control. Bombarded plants were returned to D for 16 to 18 h before reporter gene activity was assayed. The normalized ratios of reporter LUC to internal standard GUS activities are reported as relative activities; se values are shown.
Design and Rationale of LS Mutations
A series of LS substitutions was made in the context of the NPR1–156 promoter construct; the eight mutant constructs scanned the region between −156 and −70 from the start of transcription. The range of each LS mutation was designed to comprise specific regions of the promoter, which bear similarity to previously identified regulatory cis-elements in other phytochrome-regulated or ABA-inducible genes. Thus, the range of nucleotides altered was not equivalent for all LS constructs. The individual LS mutations and the corresponding promoter elements are summarized in Figure 2. LS4, 6, and 7 each mutate a region containing an ACGT core sequence, which has been shown to mediate binding to various plant bZIP transcription factors (Foster et al., 1994). These bZIPs include the G-box binding factor family members, which bind to sequences present in a diverse array of promoters, including many regulated by light (Donald and Cashmore, 1990; Weisshaar et al., 1991), and EmBP-1, which can mediate ABA regulation (Guiltinan et al., 1990; Niu and Guiltinan, 1994). LS3 disrupts a GATA sequence, a motif that has been implicated in the regulation of many light-responsive promoters (for review, see Terzaghi and Cashmore, 1995; see also Anderson and Kay, 1995; Degenhardt and Tobin, 1996; Puente et al., 1996). GATA sequences have also been studied within the extended context of the I-box motif (Buzby et al., 1990; Donald and Cashmore, 1990; Borello et al., 1993), which has also been implicated as important in light regulation. LS5 alters a region that bears substantial homology to the CE3 motif in the barley HVA1 gene (Shen et al., 1996) and to motif III in the rice rab16B gene (Ono et al., 1996); these motifs were shown to be required for ABA responsiveness. Finally, LS8 disrupts a sequence with homology to the Em2 motif in the wheat Em gene (Marcotte et al., 1989).
Figure 2.
LS constructs in the L. gibba NPR1 promoter. The wild-type sequence of the L. gibba NPR1 promoter between −156 and −70 from the transcriptional start site is given. The range of each LS construct is shown directly below the corresponding sequence, with mutated residues given in lowercase. Black boxes indicate the positions of ACGT motifs; the gray box indicates the position of a CE3-like motif. WT, Wild type.
Determination of Sequences Necessary for Phytochrome Responsiveness
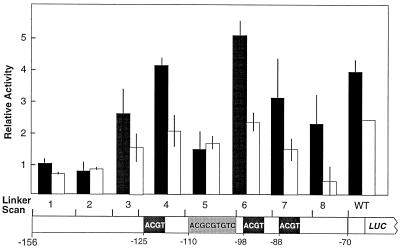
Each of the LS constructs was tested for phytochrome regulation in etiolated L. gibba by the particle bombardment transient assay. Figure 3 shows the results of these experiments. There were varying levels of expression of these constructs, suggesting that many of the mutations affected quantitative elements. For example, LS1 and 2 reduced the overall expression level by about 4-fold, and LS5 reduced the expression level by about 2-fold, relative to the wild-type NPR-156 construct. LS1 showed a slight but significant (P < 0.05) decrease in response to R. Only LS2 and 5 showed a lack of response to R; all other LS constructs retained the response to phytochrome.
Figure 3.
Phytochrome responsiveness of L. gibba NPR1 LS constructs. The range of each LS construct is diagrammed on a schematic of the NPR1 promoter between −156 and −70 from the transcriptional start. Black boxes indicate the positions of ACGT motifs; the gray box indicates the position of a CE3-like motif. Following bombardment with the designated LS mutant or wild-type NPR1–156 construct, plants were treated with D (black bars) or 2 min of R (white bars) and returned to D for 16 to 18 h before reporter gene activity was assayed. The normalized ratios of reporter LUC to internal standard GUS activities are reported as relative activities; se values are shown. WT, Wild type.
Determination of Sequences Necessary for ABA Responsiveness
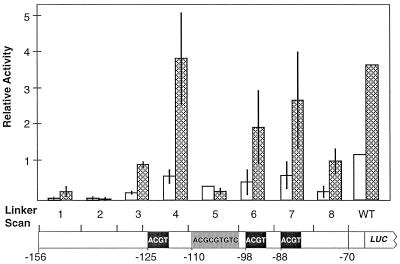
Each of the LS constructs was tested for ABA inducibility in etiolated L. gibba by the transient assay. Figure 4 shows the results of these experiments. Significant induction of ABA was retained by LS1, 3, 4, 6, 7, and 8 constructs, whereas LS2 and 5 demonstrated a loss of ABA induction. Again, LS1 gave the smallest response (2-fold). LS3 and 8 showed lower overall expression, but the 3-fold increase in response to ABA was similar to the wild-type NPR-156 construct.
Figure 4.
ABA responsiveness of L. gibba NPR1 LS constructs. The range of each LS construct is diagrammed on a schematic of the NPR1 promoter between −156 and −70 from the transcriptional start. Black boxes indicate the positions of ACGT motifs; the gray box indicates the position of a CE3-like motif. Following bombardment with the designated LS mutant or wild-type NPR1–156 construct, plants were treated with 2 min of R plus water (white bars) or 10 μm ABA (cross-hatched bars) and returned to D for 16 to 18 h before reporter gene activity was assayed. The normalized ratios of reporter LUC to internal standard GUS activities are reported as relative activities; se values are shown. WT, Wild type.
We note that the relative levels of expression of the wild-type NPR-156 construct differ in the data presented in Figures 3 and 4. Since plants that had only been treated with the different light regimes (D versus R) were used in Figure 3, whereas plants that had been given water or 10 μm ABA 4 h prior to bombardment and light treatments were used in Figure 4, the resulting data are consistent but not precisely equivalent. In general, we have observed that the addition of water to plants in the transient assay reduced overall expression levels (data not shown). However, in Figure 3, the highest expression in the D-treated plants was observed with the LS4, 6, 7, and wild-type constructs; the same constructs yielded the highest expression in response to ABA addition in Figure 4.
Reanalysis of LS2 and 5 Constructs
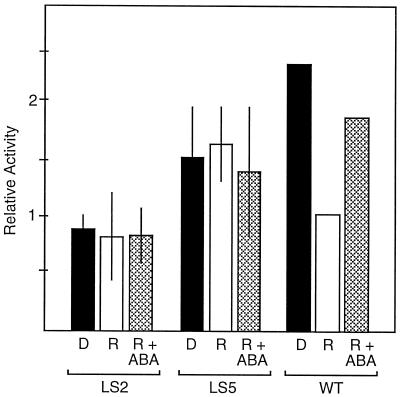
Since LS2 and 5 both showed quantitative and qualitative differences in comparison with the wild-type NPR1–156 construct, we reexamined the response of these promoter mutations to both phytochrome and ABA action. The relative ratio of the NPR LS::LUC construct to the internal standard Act::GUS DNA during the previous bombardments was 5.0 μg of NPR:1.0 μg of Act (Figs. 1 and 3) and 5.0 μg of NPR:1.5 μg of Act (Fig. 4). Varying this ratio over a 3-fold range did not affect the expression levels of most of the LS constructs (data not shown); however, LS2 and 5 relative activities increased upon increasing the bombardment ratio to 5 μg of NPR:0.5 μg of Act. This finding is consistent with other reports that coexpression of two promoters can be dependent on relative promoter strength (Rolfe and Tobin, 1991). Although the wild-type NPR1–156 promoter construct is insensitive to varying the ratio to Act::GUS, the LS2 and 5 constructs exhibit much weaker transcriptional activity and thus are more sensitive to higher levels of coexpression from the (presumably stronger) Act promoter. Therefore, we used the higher relative ratio of NPR:Act DNA (5.0:0.5 μg) to retest the LS2 and 5 constructs for phytochrome and ABA responsiveness. Figure 5 shows the results from a single representative experiment, in which the wild-type NPR-156 construct displayed a 2-fold reduction in activity with R treatment and a 2-fold induction by ABA. LS2 showed neither a decrease in expression with R treatment nor an induction by application of ABA. LS5 exhibited 50% higher activity than the LS2 construct; however, it also displayed no response to R or ABA treatments. The experiments under these conditions confirmed that both the LS2 and 5 mutations led to a loss of ABA induction, accompanied by an abolishment of the phytochrome response.
Figure 5.
Phytochrome and ABA regulatory elements converge in the L. gibba NPR1 promoter. Following bombardment with NPR1-LS2 (LS2), NPR1-LS5 (LS5), or the wild-type NPR1–156 (WT) construct, plants were treated with D (black bars), 2 min of R (white bars), or 2 min of R plus 10 μm ABA (cross-hatched bars); D and R samples received water as a control. Bombarded plants were returned to D for 16 to 18 h before reporter gene activity was assayed. The normalized ratios of reporter LUC to internal standard GUS activities are reported as relative activities; se values are shown.
DISCUSSION
We have shown that mutation of either of the two limited segments of the NPR1 promoter can abolish both phytochrome- and ABA-responsive gene expression. A comparison of these NPR1 regulatory sequences with other sequence motifs that have been characterized for their response to phytochrome or ABA action suggests that the NPR1 promoter elements are not equivalent to any previously established phytochrome- or ABA-responsive motifs.
The NPR1 gene was originally isolated as an example of a gene negatively regulated by phytochrome (Okubara and Tobin, 1991). Further work showed that sequences downstream of the −198 nucleotide relative to the start of transcription were necessary for this regulation (Williams et al., 1994). At present, RE1 motifs (CATGGGCGCGG) in the oat phyA gene promoter (Bruce et al., 1991) remain the only identified sequence element involved in negative regulation by phytochrome; a comparison with the NPR1 sequence yielded a similar match only at +235, within the presumptive coding region of the NPR1 gene. A closely related motif, RE3 (GATCTGGTGGGAGCTAG), has recently been defined in the pea AS1 gene (Neuhaus et al., 1997); a tetramer of the RE3 motif was able to confer negative regulation by white light to a reporter construct in a microinjection transient assay. However, there is no significant homology to the RE3 motif in the L. gibba NPR1 promoter. The RE1 and RE3 motifs share a core element containing a TGGG sequence; although there are many occurrences of this core sequence within the NPR1 promoter (at −681, −600, −421, −387, −352, −218, and −194), these all fall outside of the −156 region, which we have shown contains sufficient information to confer both phytochrome- and ABA-responsive gene expression.
LS analysis of a region 156 bp upstream of the start of transcription of the L. gibba NPR1 promoter revealed two independent regions of the promoter, each of which was necessary for ABA inducibility. Loss of ABA induction was accompanied by a loss of negative regulation by phytochrome action, suggesting that these promoter sequences act to control responses to both stimuli. This suggests that phytochrome regulation of NPR1 gene expression is acting primarily through alterations in endogenous ABA levels in the plant. The NPR1 promoter does contain three ACGT core sequence elements homologous to Em1a and Em1b ABA-responsive elements in the wheat Em promoter (Marcotte et al., 1992) at −125, −98, and −88 bp, respectively, from the transcriptional start. These Em1a and Em1b sequences were found to be necessary and sufficient to confer ABA regulation to a minimal 35S CaMV core promoter (Marcotte et al., 1989); however, these ABA-responsive elements displayed nonredundancy in their relative contribution to transactivation by the Viviparous1 (VP1) factor (Vasil et al., 1995). None of the three ACGT core elements in the NPR1 promoter (LS4, 6, and 7) appeared to be essential for ABA induction. However, these ACGT elements may have a redundant function; this question remains to be addressed in the future.
In addition to these ACGT core motifs, another class of response elements has been characterized in ABA-induced genes from monocots. The NPR1 promoter contains a sequence at −110 (ACGCGTGTCGT) that bears a strong similarity to both the synthetic hex3 (Lam and Chua, 1991) element (and the related rab16B motif III, Ono et al., 1996) and the coupling element (CE3; (ACGCGTGTCCTC) in the barley HVA1 promoter (Shen et al., 1996). The hex3 element (GACGCGTGGC) was sufficient to confer ABA inducibility when fused to the −90 35S CaMV promoter; similarly, the closely related rab16B motif III sequence (GCCGCGTGGC) also behaved as an ABA-responsive element when fused to the −46 35S CaMV promoter. In the HVA1 promoter, the CE3 element “couples” with an adjacent ACGT motif to provide a synergistic response to ABA relative to the response mediated by either element alone; thus, mutation of both CE3 and ACGT elements was required to abolish the response to ABA. A recent report also shows that this HVA1 promoter fused to green fluorescent protein exhibited both ABA- and D-induced expression in maize leaf protoplasts (Sheen, 1996). In fact, the NPR1 promoter displays a similar organization of CE3-like and ACGT elements (at −110 and −98 bp, respectively); however, we did not observe any evidence of coupling behavior in the NPR1 promoter, because mutation of CE3-like sequences in LS5 alone was sufficient to negate ABA inducibility of the NPR1 promoter. Thus, the functioning of the sequence in LS5 most closely resembles the hex3 (motif III) in its lack of coupling to an adjacent element.
LS1 mutates a region containing an oct-1 motif (Rosales et al., 1987); this disruption of an SV-40-like enhancer element may partially explain the overall reduction in expression from this mutation. As expected, the LS1 construct also showed considerable transcriptional interference from the internal standard; however, because it did display ABA induction (P < 0.05), it apparently does not alter an ABA regulatory motif. The sequences altered in the LS2 construct do not bear any significant homology with any known transcription factor-binding site; therefore, the decrease in relative activity of this promoter construct cannot be ascribed to any known function. In addition, there is no significant homology of the LS2 region with any characterized ABA-inducible motif; therefore, these sequences represent a novel ABA regulatory motif.
Significantly, mutations within either the LS2 or 5 sequences were sufficient to abolish the response to ABA. Thus, there do not appear to be any additive or synergistic effects of these promoter motifs within the NPR1 gene, unlike the rice Osem gene, which contains multiple interacting motifs (Hattori et al., 1995). The consequences of two nonredundant regulatory motifs are not clear; however, similar dual requisite and nonredundant ABA-responsive elements have been demonstrated in the rab16B promoter (Ono et al., 1996) and the wheat Em promoter (Vasil et al., 1995). We also did not examine whether subtle quantitative differences in sensitivity to or threshold of ABA induction exist between the LS2 and 5 elements; this may be precluded by the level of resolution of our transient assay.
In summary, we have shown that 156 nucleotides upstream of the transcription start site of the L. gibba NPR1 promoter are sufficient to confer both phytochrome repression and ABA induction in a transient assay system. Using an LS mutagenesis scheme, we have identified two cis-acting elements that behave as convergent phytochrome- and ABA-response control elements, respectively, in the L. gibba NPR1 promoter. Taking into account the role of phytochrome action in altering endogenous ABA levels in L. gibba, the phytochrome response of the NPR1 gene can be attributed to alterations in ABA levels.
ACKNOWLEDGMENT
We thank Kiet Lam for maintaining the cultures of L. gibba.
Abbreviations:
- Act
actin
- CaMV
cauliflower mosaic virus
- D
dark
- LS
linker scan
- LUC
luciferase
- R
red light
Footnotes
This research was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 95-37304-2324 to E.M.T.).
LITERATURE CITED
- Anderson SL, Kay SA. Functional dissection of circadian clock- and phytochrome-regulated transcription of the Arabidopsis CAB2 gene. Proc Natl Acad Sci USA. 1995;92:1500–1504. doi: 10.1073/pnas.92.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borello U, Ceccarelli E, Giuliano G. Constitutive, light-responsive and circadian clock-responsive factors compete for the different I-box elements in plant light-regulated promoters. Plant J. 1993;4:611–619. doi: 10.1046/j.1365-313x.1993.04040611.x. [DOI] [PubMed] [Google Scholar]
- Bruce WB, Deng X-W, Quail PH. A negatively acting DNA sequence element mediates phytochrome-directed repression of phyA gene transcription. EMBO J. 1991;10:3015–3024. doi: 10.1002/j.1460-2075.1991.tb07852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzby JS, Yamada T, Tobin EM. A light-regulated DNA binding activity interacts with a conserved region of a Lemna gibba rbcS promoter. Plant Cell. 1990;2:805–814. doi: 10.1105/tpc.2.8.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Tobin EM. A DNA binding activity for one of two closely defined phytochrome regulatory elements in an Lhcb promoter is more abundant in etiolated than in green plants. Plant Cell. 1996;8:31–41. doi: 10.1105/tpc.8.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RGK, Cashmore AR. Mutation of either G-box or I-box sequences profoundly affects expression from the Arabidopsis rbcs-1A promoter. EMBO J. 1990;9:1717–1726. doi: 10.1002/j.1460-2075.1990.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua N-H. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994;8:192–200. doi: 10.1096/fasebj.8.2.8119490. [DOI] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WM, Quatrano RS. A leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;150:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Hattori T, Terada T, Hamasuna S. Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J. 1995;7:913–925. doi: 10.1046/j.1365-313x.1995.07060913.x. [DOI] [PubMed] [Google Scholar]
- Lam E, Chua N-H. Tetramer of a 21 base pair synthetic element confers seed expression and transcriptional enhancement in response to water stress and abscisic acid. J Biol Chem. 1991;266:17131–17135. [PubMed] [Google Scholar]
- Marcotte WR, Jr, Guiltinan MJ, Quatrano RS. ABA-regulated gene expression: cis-acting sequences and trans-acting factors. Biochem Soc Trans. 1992;20:93–97. doi: 10.1042/bst0200093. [DOI] [PubMed] [Google Scholar]
- Marcotte WR, Jr, Russell SH, Quatrano RS. Abscisic acid-responsive sequences from the Em gene of wheat. Plant Cell. 1989;1:969–976. doi: 10.1105/tpc.1.10.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Hiratsuka K, Yamagata H, Chua N-H. Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 1997;16:2554–2564. doi: 10.1093/emboj/16.10.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Guiltinan MJ. DNA binding specificity of the wheat bZIP protein EmBP-1. Nucleic Acids Res. 1994;22:4969–4978. doi: 10.1093/nar/22.23.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeda K, Salinas J, Chua N-H. A tobacco bZIP transcription activator (TAF-1) binds to a G-box motif conserved in plant genes. EMBO J. 1991;10:1793–1802. doi: 10.1002/j.1460-2075.1991.tb07704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubara PA, Tobin EM. Isolation and characterization of three genes negatively regulated by phytochrome action in Lemna gibba. Plant Physiol. 1991;96:1237–1245. doi: 10.1104/pp.96.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubara PA, Williams SA, Doxsee RA, Tobin EM. Analysis of genes negatively regulated by phytochrome action in Lemna gibba and identification of a promoter region required for phytochrome responsiveness. Plant Physiol. 1993;101:915–924. doi: 10.1104/pp.101.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Izawa T, Chua N-H, Shimamoto K. The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol. 1996;112:483–491. doi: 10.1104/pp.112.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 1996;15:3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Riggs CD, Chrispeels M. Luciferase reporter gene cassettes for plant gene expression studies. Nucleic Acids Res. 1987;15:8115. doi: 10.1093/nar/15.19.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe SA, Tobin EM. Deletion analysis of a phytochrome-regulated monocot rbcS promoter in a transient assay system. Proc Natl Acad Sci USA. 1991;88:2683–2686. doi: 10.1073/pnas.88.7.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales R, Vigneron M, Macchi M, Davidson I, Xiao JH, Chambon P. In vitro binding of cell-specific and ubiquitous nuclear proteins to the octamer motif of the SV40 enhancer and related motifs present in other promoters and enhancers. EMBO J. 1987;6:3015–3025. doi: 10.1002/j.1460-2075.1987.tb02607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sheen J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science. 1996;274:1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- Zhang QP, Ho T-H, Shen D. Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Tobin EM. Phytochrome-mediated regulation of messenger RNAs for the small subunit of ribulose 1,5-bisphosphate carboxylase and the light-harvesting chlorophyll a/b protein in Lemna gibba. Plant Mol Biol. 1981;1:35–51. doi: 10.1007/BF00023012. [DOI] [PubMed] [Google Scholar]
- Vasil V, Marcotte WR, Jr, Rosenkrans L, Cocciolone SM, Vasil IK, Quatrano RS, McCarty DR. Overlap of Viviparous1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell. 1995;7:1511–1518. doi: 10.1105/tpc.7.9.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherwax SC, Ong MS, Degenhardt J, Bray EA, Tobin EM. The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol. 1996;111:363–370. doi: 10.1104/pp.111.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar B, Armstrong GA, Block A, da Costa e Silva O, Halbrock K. Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promoter element with functional relevance in light responsiveness. EMBO J. 1991;10:1777–1786. doi: 10.1002/j.1460-2075.1991.tb07702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Weatherwax SC, Bray EA, Tobin EM. NPR genes, which are negatively regulated by phytochrome action in Lemna gibba L. G-3, can also be positively regulated by abscisic acid. Plant Physiol. 1994;105:949–954. doi: 10.1104/pp.105.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]