Abstract
Protein misfolding disorders such as Alzheimer, Parkinson and transthyretin amyloidosis are characterized by the formation of protein amyloid deposits. Although the nature and location of the aggregated proteins varies between different diseases, they all share similar molecular pathways of protein unfolding, aggregation and amyloid deposition. Most effects of these proteins are likely to occur at the proteome level, a virtually unexplored reality. To investigate the effects of an amyloid protein expression on the cellular proteome, we created a yeast expression system using human transthyretin (TTR) as a model amyloidogenic protein. We used Saccharomyces cerevisiae, a living test tube, to express native TTR (non-amyloidogenic) and the amyloidogenic TTR variant L55P, the later forming aggregates when expressed in yeast. Differential proteome changes were quantitatively analyzed by 2D-differential in gel electrophoresis (2D-DIGE). We show that the expression of the amyloidogenic TTR-L55P causes a metabolic shift towards energy production, increased superoxide dismutase expression as well as of several molecular chaperones involved in protein refolding. Among these chaperones, members of the HSP70 family and the peptidyl-prolyl-cis-trans isomerase (PPIase) were identified. The latter is highly relevant considering that it was previously found to be a TTR interacting partner in the plasma of ATTR patients but not in healthy or asymptomatic subjects. The small ubiquitin-like modifier (SUMO) expression is also increased. Our findings suggest that refolding and degradation pathways are activated, causing an increased demand of energetic resources, thus the metabolic shift. Additionally, oxidative stress appears to be a consequence of the amyloidogenic process, posing an enhanced threat to cell survival.
Introduction
Protein misfolding and aggregation are common features in many neurodegenerative amyloid disorders, such as Alzheimer, Parkinson and transthyretin amyloidosis (ATTR) [1], [2]. In each case, a specific amyloidogenic protein misfold and follows a toxic aggregation pathway leading to a defined clinical outcome [1], [2]. A legion of factors that may trigger protein misfolding and aggregation has been implied in the development of this kind of pathologies, such as abnormal proteolysis, point mutations and post-translational modifications, namely phosphorylation, oxidation and glycation [2], [3]. Cell quality control mechanisms evolved to cope with protein misfolding and aggregation, including molecular chaperones and protein degradation pathways that prevent protein aggregation by either protein refolding or degradation [4]–[6]. However, in the disease process these mechanisms are not sufficient to prevent the accumulation of toxic protein aggregates and recent studies have implicated several components of the protein quality control system in neurodegenerative disorders of amyloid type (reviewed in [5], [6]). Thus, a decreased cell capacity to clear misfolded proteins may be directly involved in pathogenesis. In fact, one hypothesis to explain the late onset of several conformational disorders considers the loss of effectiveness of the protein quality control system with age, either arising from environmental insults, mutations or unidentified triggers [7]. Although the regulation of these pathways has been considered a therapeutic strategy [8] our knowledge of these processes is still very limited and therefore a deeper understanding of the cellular response mechanisms, at the proteome level, to misfolding and aggregation of amyloidogenic proteins is of great interest. Importantly, even in familial neuropathies, where genetic determinants play a key role, several observation points to the involvement of non-genetic factors. In ATTR, where the amyloid deposits are mainly composed of transthyretin (TTR), several TTR amyloidogenic point mutations with different degrees of amiloidogenicity (i.e., different tendency to misfold and aggregate) are associated to disease onset and progression [9], [10]. However, it was observed that many mutation carriers are asymptomatic throughout their lives and non-mutant TTR also forms amyloid, causing senile systemic amyloidosis [11]–[13]. Additionally, patients carrying the same mutation show a wide range of age at onset from 20 to 70 years [14]. We recently discovered that ATTR patients show increased protein glycation that decreases the chaperone activity of fibrinogen, a TTR protein binding partner, hence promoting transthyretin unfolding and aggregation in ATTR [15].
In this work we investigated the cellular responses to the misfolding and aggregation of an amyloidogenic protein using a high-throughput proteomics approach to screen differentially expressed proteins. For that purpose, we used yeast Saccharomyces cerevisiae as a host to express human TTR, used as a model amyloidogenic protein. As the general protein quality control systems are highly conserved, especially in eukaryotes, yeast is a valuable model to evaluate fundamental aspects of protein misfolding and the molecular mechanisms involved in the cellular response. Additionally, Saccharomyces cerevisiae was the first eukaryote to have its genome sequenced and, recently, its entire proteome has been mapped [16], [17]. Yeast has been successfully used to investigate fundamental aspects of protein misfolding involved in syndromes like amyotrophic lateral sclerosis, Huntington’s disease and Parkinson’s disease [18]–[20]. TTR was investigated as an amyloidogenic protein for two main reasons: first, it is one of the best structurally characterized amyloidogenic proteins and second, several known point mutations are associated to different degrees of amiloidogenicity and disease progression. This allows the discrimination between the cell response to the expression of a heterologous protein and the amyloidogenic version of the same protein. Thus, wild-type TTR and the highly amyloidogenic TTR variant L55P (leucine for proline at position 55) were selected for expression in yeast. Although these two variants only differ in one amino acid, TTR-L55P has a higher intrinsic propensity to misfold and aggregate and carriers of this amyloidogenic mutation have a very early age at disease onset [21].
Differential proteome changes between the control (cells carrying the empty plasmid), cells expressing TTR-wt (BTTR-wt) and cells expressing TTR-L55P (BTTR-L55P) were analyzed by 2D-DIGE coupled to protein identification by tandem MS. Expression of each TTR forms has no toxic effects on yeast and does not impair cell division, population growth and metabolism, despite the formation of TTR aggregates in yeast cells expressing the L55P variant. About 20 proteins were found to be differentially expressed in BTTR-wt while 70 were differentially expressed in BTTR-L55P. Changes in proteins involved in folding and degradation processes were detected, together with an increased expression of translation pathways. Major changes at the proteome level were associated with increased carbohydrate, energy and amino acid metabolism.
Results and Discussion
Protein Expression Analysis by 2D-DIGE
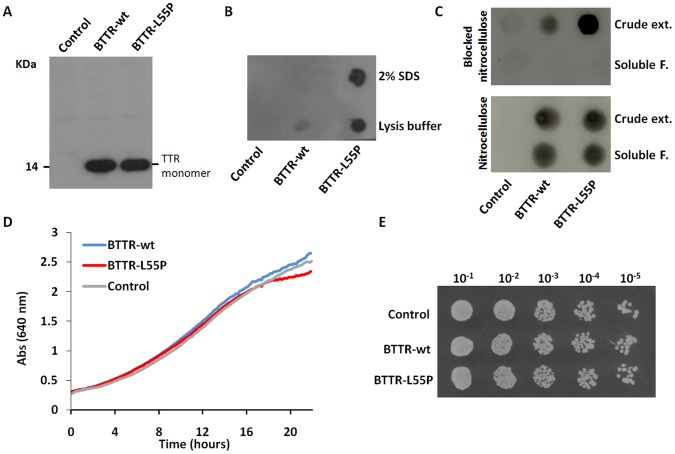
To quantitatively investigate proteome changes of yeast cells in response to TTR expression we analysed and compared the proteomes of yeast BY4741 cells carrying the plasmid without the insert (control), TTR-wt (BTTR-wt) and TTR-L55P variant (BTTR-L55P). TTR expression was confirmed by MS analysis (data not shown) and western blot, where similar expression levels of TTR-wt and TTR-L55P were detected (Figure 1A). To analyse the presence of TTR aggregates, a protein aggregate filtration assay was performed (Figure 1B). This microfiltration method is based on the finding that high-molecular mass amyloid-like aggregates are SDS-insoluble, being therefore retained in a 0.2 µm blocked membrane. In fact, if the nitrocellulose membrane is blocked previously to the filtration procedure, TTR-containing insoluble protein aggregates are retained while soluble TTR do not bind to the blocked membrane, in contrast to a regular dot-blot assay using non-blocked nitrocellulose membrane (Figure 1C). Thus this method is indeed a protein aggregation filter trap assay. Using blocked nitrocellulose, a positive signal was only obtained in the crude extract indicating the presence of TTR in the insoluble fraction. As shown in Figure 1B, TTR aggregates insoluble in 2% SDS were clearly observed in yeast cells expressing the amyloidogenic L55P variant (BTTR-L55P). SDS-insoluble aggregates were not detected in BTTR-wt and, as expected, in the control (Figure 1B). In addition, a substantially higher TTR amount was found in the insoluble protein fraction of BTTR-L55P in comparison to BTTR-wt (Figure 1B and C). Thus, even though no cell toxicity and growth defects were observed (Figure 1D and E), TTR-L55P variant, when expressed in yeast, forms high-molecular mass amyloid-like aggregates.
Figure 1. Characterization of TTR expression in yeast.
(A) Relative quantitation of TTR-wt and TTR-L55P expression by Western blot with anti-human TTR antibody. Since TTR-L55P aggregates, electrophoretic separation of proteins was performed in the presence of urea to compare the total protein amount. TTR monomer is observed with molecular masses of approximately 14 kDa only in cells expressing TTR (BTTR-wt and BTTR-L55P). Similar expression levels were observed for the two TTR variants. (B) Protein aggregate filtration assay of insoluble protein fraction. TTR aggregates insoluble in 2% SDS were observed only in yeast cells expressing the amyloidogenic L55P variant (BTTR-L55P). This indicates that TTR-L55P when expressed in yeast forms high molecular mass amyloid-like aggregates, in contrast to TTR-wt. A substantially higher TTR amount was also found in the insoluble protein fraction of BTTR-L55P in comparison with BTTR-wt. (C) Control assay. The crude extract and the soluble protein fraction were analysed through a nitrocellulose membrane (as a regular dot-blot assay) and a nitrocellulose membrane that was blocked prior to the filtration procedure. Growth curves (D) and dilution spot assay (E) of yeast cells expressing the TTR variants and the control shows that, although the highly amyloidogenic TTR variant L55P was expressed in yeast, no changes were detected in yeast cell growth and cell viability.
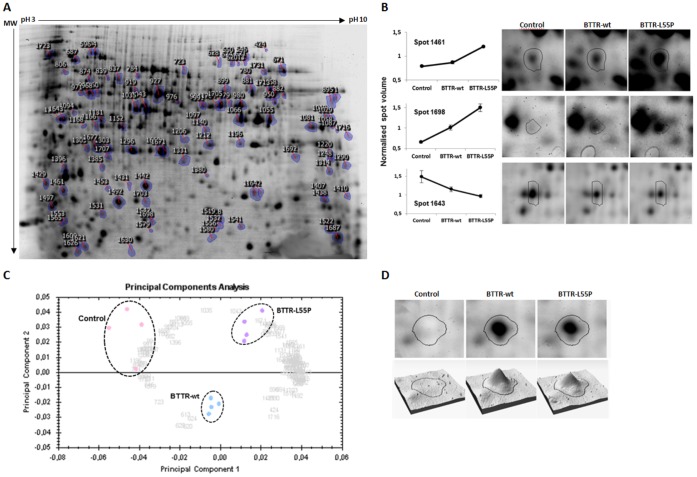
The 2D electrophoretic maps contained around 1800 spots (Figure 2A). From these, a total of 78 protein spots were detected with a statistically significant change in abundance (ANOVA p<0.05) with an absolute variation ≥1.3-fold from at least one experimental group. Examples of three spot patterns are shown in Figure 2B. Spot 1461 increase its abundance only in BTTR-L55P while spot 1643 decrease its abundance in both BTTR-wt and BTTR-L55P. The spot 1698 is differentially expressed in both experimental groups but shows a significantly higher abundance in BTTR-L55P compared to BTTR-wt. A principal component analysis (PCA) analysis shows that 2D gel images cluster into three well separated groups (Figure 2C), indicating a clear differentiation between the expression of the non-amyloidogenic TTR-wt and the amyloidogenic TTR-L55P form with significant changes in protein abundances. In fact, by directly comparing the 2D-DIGE maps of BTTR-wt and BTTR-L55P with the control, a total of 24 and 75 spots, respectively, were differentially expressed, highlighting a much higher induced change upon expression of the amyloidogenic TTR variant. Interestingly, of the 24 protein spots with changes in abundance between BTTR-wt/control, 21 spots were also differentially expressed in BTTR-L55P/control with a similar fold variation. These changes are likely to be due to the heterologous TTR expression and not because of TTR misfolding. However, 54 protein spots exhibited significant changes in abundance exclusively upon TTR-L55P expression, suggesting that relevant proteins involved in the cell response to TTR misfolding and aggregation are revealed with this approach.
Figure 2. 2D-DIGE differential protein expression analysis.
(A) Representative 2D-DIGE gel image showing the spot map corresponding to the internal standard (Cy2 labeling), which is common to all gels analyzed. Sample preparation and labeling were performed as described in the material and methods section. Indicated spots showed a statistically significant variation of spot volume with 95% confidence level (p<0.05) and a minimal fold variation of 1.3. These spots were removed for subsequent protein identification by mass spectrometry (see Table 1 for code assignment). (B) Example of one protein present in higher abundance only in BTTR-L55P (Ubiquitin-like protein SMT3, spot 1461), one protein with an increase abundance in BTTR-wt and BTTR-L55P (FK506-binding protein, spot 1698) and one protein present in lower abundance both in BTTR-wt and BTTR-L55P (60 S acidic ribosomal protein P0, spot 1643). The spots of interested are encircled and the graphs represent the normalized spot volume. The spot volumes are an average of the 4 biological replicates used for each experimental group. (C) PCA of the 2D-DIGE results. Each data point in the PCA plot represents the global expression values for all spots with a significant ANOVA value (p<0.05). The PCA clustered the 6 individual Cy3 and Cy5-labeled 2D-gel images into three discreet groups differentiated by two principle components PC1 and PC2, explaining a cumulative 82% of all the variation. A separation between the control (carrying the empty plasmid), cells expressing the non-amyloidogenic TTR-wt (BTTR-wt) and the amyloidogenic variant TTR-L55P (BTTR-L55P) is clearly observed. Replicate samples were encircled manually for illustration. (D) 2D-DIGE image analysis of the protein spot identified as human TTR. Figure shows the spot expression map and three-dimensional spot image for each experimental group.
Protein Identification and Gene Ontology Analysis
All spots highlighted in Figure 2A were picked and trypsin digested using the Ettan Spot Handling Workstation and the proteins were identified by MALDI-TOF-TOF MS. With this approach, we were able to identify the corresponding proteins in 73 spots, resulting in the identification of 70 unique proteins (Table 1). For the majority of the identified proteins, the molecular mass and isoelectric points determined on the 2D gel are consistent. In some cases, the same protein is identified in different spots across the 2D gel with different molecular mass and isoelectric point suggesting the presence of post-translational modifications and/or protein isoforms. All spots representing the same protein have a very similar regulation (see for example spot 991 and 994 both identified as alcohol dehydrogenase (ADH1) where a similar trend and fold variation was observed). In seven spots, more than one protein was identified (see Table 1). In some cases, MSMS data allowed the identification of a particular protein isoform (example, spot 613 identified as HSP75). In other cases, this was not possible and thus both protein isoforms are shown in table 1 (for example, spot 1542 identified as enolase 1 and/or enolase 2).
Table 1. Differentially expressed proteins identified by MALDI-TOF-TOF MSMS.
| Gene symbola) | Protein names | Control/BTTR-wt | Control/BTTR-L55P | Functionb) | Acces. codec) | Spotno.d) | MS datae) | ||||
| Fold Var. | P value | Fold Var. | P value | Prot. Score | Pep. score | #Matched MSMS peptides | |||||
| ACON | Aconitatehydratase, mitochondrial | +1.7 | 6.39E-03 | +1.4 | 5.64E-03 | CM | P19414 | 424 | 321 | 249 | 4 |
| ADH1 | Alcohol dehydrogenase 1 | −1.6 | 7.84E-03 | −1.5 | 1.24E-02 | CM | P00330 | 991 | 617 | 504 | 9 |
| ADH1 | Alcohol dehydrogenase 1 | −1.5 | 1.07E-03 | −1.4 | 1.19E-02 | CM | P00330 | 994 | 704 | 563 | 9 |
| ADH3 | Alcohol dehydrogenase 3, mitochondrial | +1.4 | 1.23E-02 | +1.5 | 2.75E-03 | CM | P07246 | 950 | 977 | 774 | 10 |
| ADK | Adenosine kinase | – | – | +1.4 | 3.62E-03 | NM | P47143 | 1094 | 537 | 429 | 4 |
| AIM29 | Altered inheritance rate of mitochondria protein 29 | −1.4 | 8.26E-03 | −1.4 | 1.29E-02 | U | P36154 | 1453 | 121 | 76 | 2 |
| ALF | Fructose-bisphosphatealdolase | – | – | +1.3 | 5.18E-03 | CM | P14540 | 1035 | 221 | 130 | 3 |
| ALF | Fructose-bisphosphatealdolase | – | – | +1.3 | 1.85E-02 | CM | P14540 | 1131 | 277 | 168 | 3 |
| ATPA | ATP synthase subunit alpha, mitochondrial | −1.4 | 1.29E-03 | – | – | EM | P07251 | 780 | 647 | 484 | 6 |
| ATPB | ATP synthase subunit beta, mitochondrial | – | – | +1.5 | 3.19E-04 | EM | P00830 | 971 | 639 | 519 | 8 |
| BCA1 | Branched-chain-amino-acid aminotransferase, mithocondrial | – | – | +1.6 | 2.60E-04 | AM | P38891 | 1565 | 130 | 110 | 2 |
| BCA2 | Branched-chain-amino-acid aminotransferase, cytosolic | – | – | +1.6 | 2.60E-04 | AM | P47176 | 1565 | 220 | 199 | 2 |
| BMH2 | Protein BMH2 | – | – | −1.4 | 8.98E-04 | PFD | P34730 | 1127 | 505 | 395 | 6 |
| CISY1 | Citrate synthase, mitochondrial | −1.4 | 1.11E-02 | +1.5 | 2.31E-01 | CM | P00890 | 881 | 167 | 118 | 2 |
| COX14 | Cytochrome c oxidase assembly protein COX14 | – | – | +1.5 | 1.62E-02 | U | P39103 | 1532 | 104 | 77 | 1 |
| CYPH | Peptidyl-prolylcis-trans isomerase | – | – | +1.7 | 7.20E-03 | PFD | P14832 | 1519 | 193 | 133 | 3 |
| DHE4 | NADP-specific glutamate dehydrogenase 1 | – | – | +1.6 | 3.17E-06 | EM | P07262 | 837 | 977 | 771 | 9 |
| DLD3 | D-lactate dehydrogenase [cytochrome] 3 | −1.4 | 1.29E-03 | – | – | CRH | P39976 | 780 | 1020 | 797 | 8 |
| DTD | D-tyrosyl-tRNA(Tyr) deacylase | – | – | +1.4 | 2.59E-02 | T | Q07648 | 1642 | 84 | 63 | 2 |
| DUG1 | Cys-Glymetallodipeptidase DUG1 | – | – | +1.6 | 3.17E-06 | PFD | P43616 | 837 | 343 | 303 | 4 |
| EF1A | Elongation factor 1-alpha | – | – | +1.4 | 2.82E-03 | T | P02994 | 891 | 802 | 692 | 8 |
| EF1A | Elongation factor 1-alpha | +1.3 | 1.82E-02 | +1.5 | 2.60E-03 | T | P02994 | 895 | 129 | 88 | 2 |
| EF1A | Elongation factor 1-alpha | +1.4 | 7.81E-03 | +1.6 | 7.65E-04 | T | P02994 | 1013 | 294 | 238 | 4 |
| EF1A | Elongation factor 1-alpha | – | – | +1.4 | 4.58E-03 | T | P02994 | 1029 | 846 | 714 | 8 |
| EF1B | Elongation factor 1-beta | – | – | +1.5 | 7.07E-05 | T | P32471 | 1429 | 236 | 222 | 3 |
| EF2 | Elongation factor 2 | – | – | +1.3 | 1.27E-03 | T | P32324 | 1621 | 86 | 78 | 2 |
| EIF3G | Eukaryotic translation initiation factor 3 subunit G | – | – | −1.3 | 1.85E-02 | T | Q04067 | 1097 | 712 | 570 | 8 |
| ENO1 | Enolase 1 | – | – | +1.5 | 3.07E-02 | CM | P00924 | 1542 | 618 | 567 | 6 |
| ENO2 | Enolase 2 | – | – | +1.5 | 3.07E-02 | CM | P00925 | 1542 | 532 | 501 | 6 |
| FHP | Flavohemoprotein | +1.6 | 1.75E-03 | +1.8 | 7.61E-06 | CRH | P39676 | 976 | 704 | 618 | 7 |
| FKBP | FK506-binding protein 1 | +1.4 | 1.91E-04 | +2.2 | 4.90E-04 | PFD | P20081 | 1698 | 163 | 110 | 3 |
| G3P2 | Glyceraldehyde-3-phosphate dehydrogenase 2 | – | – | +1.6 | 5.88E-03 | CM | P00358 | 1609 | 226 | 211 | 3 |
| G3P3 | Glyceraldehyde-3-phosphate dehydrogenase 3 | – | – | +1.6 | 5.88E-03 | CM | P00359 | 1609 | 226 | 211 | 3 |
| GBLP | Guanine nucleotide-binding protein subunit beta-like protein | – | – | +1.5 | 7.49E-03 | T | P38011 | 1541 | 315 | 223 | 3 |
| GBLP | Guanine nucleotide-binding protein subunit beta-like protein | – | – | +1.4 | 8.16E-04 | T | P38011 | 1556 | 108 | 92 | 1 |
| GPP1 | (DL)-glycerol-3-phosphatase 1 | – | – | −1.3 | 3.89E-03 | LM | P41277 | 1152 | 479 | 343 | 6 |
| HIS2 | Histidine biosynthesis trifunctional protein | +1.4 | 4.50E-03 | +1,5 | 3.63E-04 | AM | P00815 | 1692 | 419 | 367 | 6 |
| HSP10 | 10 kDa heat shock protein, mitochondrial | +1.7 | 9.55E-03 | +1.8 | 1.15E-03 | PFD | P38910 | 1522 | 465 | 347 | 4 |
| HSP60 | Heat shock protein 60, mitochondrial | +1.6 | 8.31E-03 | +1.8 | 2.12E-03 | PFD | P19882 | 687 | 383 | 204 | 4 |
| HSP60 | Heat shock protein 60, mitochondrial | – | – | +1.6 | 1.86E-02 | PFD | P19882 | 1438 | 369 | 307 | 4 |
| HSP71 | Heat shock protein SSA1 | – | – | +1.4 | 2.62E-03 | PFD | P10591 | 960 | 161 | 51 | 2 |
| HSP72 | Heat shock protein SSA2 | +1.3 | 1.40E-02 | +1.5 | 2.88E-03 | PFD | P10592 | 874 | 165 | 127 | 2 |
| HSP72 | Heat shock protein SSA2 | +1.4 | 8.91E-03 | +1.6 | 4.37E-04 | PFD | P10592 | 967 | 272 | 135 | 3 |
| HSP75 | Heat shock protein SSB1 | – | – | −1.5 | 4.53E-03 | PFD | P11484 | 613 | 177 | 138 | 4 |
| HSP77 | Heat shock protein SSC1, mitochondrial | +1.6 | 1.23E-02 | +1.4 | 1.28E-02 | PFD | P12398 | 594 | 310 | 228 | 6 |
| HSP77 | Heat shock protein SSC1, mitochondrial | +1.4 | 2.27E-02 | – | – | PFD | P12398 | 596 | 329 | 196 | 6 |
| IDHP | Isocitrate dehydrogenase [NADP], mitochondria | +2.5 | 7.38E-03 | +2.1 | 1.18E-02 | CM | P21954 | 1703 | 139 | 92 | 2 |
| IF4A | ATP-dependent RNA helicase eIF4A | −2.2 | 2.09E-02 | – | – | T | P10081 | 1396 | 170 | 111 | 3 |
| IF4B | Eukaryotic translation initiation factor 4B | – | – | +1.5 | 9.26E-05 | T | P34167 | 839 | 358 | 284 | 5 |
| ILV5 | Ketol-acid reductoisomerase, mitochondrial | – | – | −1.3 | 6.76E-03 | AM | P06168 | 980 | 565 | 414 | 7 |
| ILVB | Acetolactate synthase catalytic subunit, mitochondrial | – | – | −1.3 | 1.98E-02 | AM | P07342 | 571 | 152 | 87 | 4 |
| INO1 | Inositol-3-phosphate synthase | – | – | +1.5 | 8.65E-04 | CM | P11986 | 1168 | 282 | 255 | 3 |
| KPYK1 | Pyruvate kinase 1 | – | – | +1.5 | 4.33E-03 | CM | P00549 | 1140 | 345 | 197 | 4 |
| MPCP | Mitochondrial phosphate carrier protein | +2.4 | 5.97E-03 | +1.5 | 2.05E-02 | TP | P23641 | 1716 | 335 | 286 | 4 |
| NDK | Nucleoside diphosphate kinase | – | – | +1.3 | 3.39E-02 | NM | P36010 | 1407 | 294 | 288 | 5 |
| PDC1 | Pyruvate decarboxylase isozyme 1 | – | – | −1.3 | 1.30E-01 | CM | P06169 | 723 | 1390 | 1222 | 11 |
| PDI | Protein disulfide-isomerase | – | – | +1.3 | 1.05E-02 | PFD | P17967 | 1723 | 474 | 391 | 5 |
| PGK | Phosphoglycerate kinase | – | – | +1.4 | 2.67E-02 | CM | P00560 | 1220 | 457 | 348 | 5 |
| PGK | Phosphoglycerate kinase | +1.5 | 1.75E-02 | +1.6 | 1.62E-03 | CM | P00560 | 1248 | 511 | 431 | 6 |
| PUR92 | Bifunctional purine biosynthesis protein ADE17 | – | – | +1.3 | 1.44E-02 | NM | P38009 | 628 | 349 | 215 | 3 |
| PYRF | Orotidine 5'-phosphate decarboxylase | −1.4 | 1.97E-02 | −1.5 | 5.28E-03 | NM | P03962 | 1212 | 185 | 75 | 2 |
| PYRF | Orotidine 5'-phosphate decarboxylase | – | – | +1.5 | 2.64E-03 | NM | P03962 | 1385 | 86 | 63 | 2 |
| RL11A | 60S ribosomal protein L11-A | – | – | +1.5 | 7.07E-05 | T | P0C0W9 | 1429 | 565 | 482 | 6 |
| RL11B | 60S ribosomal protein L11-B | – | – | +1.5 | 7.07E-05 | T | Q3E757 | 1429 | 565 | 482 | 6 |
| RL12 | 60S ribosomal protein L12 | – | – | +1.3 | 5.32E-03 | T | P17079 | 1410 | 125 | 112 | 2 |
| RLA0 | 60S acidic ribosomal protein P0 | – | – | −1.5 | 2.02E-03 | T | P05317 | 1643 | 443 | 358 | 6 |
| RS12 | 40S ribosomal protein S12 | – | – | −1.4 | 3.05E-02 | T | P48589 | 1497 | 307 | 271 | 4 |
| RS3 | 40S ribosomal protein S3 | +1.7 | 9.55E-03 | +1.8 | 1.15E-03 | T | P05750 | 1522 | 212 | 199 | 2 |
| RS5 | 40S ribosomal protein S5 | – | – | −1.3 | 2.65E-05 | T | P26783 | 1196 | 261 | 153 | 3 |
| SAM2 | S-adenosylmethionine synthase 2 | – | – | +1.5 | 3.19E-04 | AM | P19358 | 971 | 253 | 191 | 3 |
| SMT3 | Ubiquitin-like protein SMT3 | – | – | +1.5 | 5.90E-06 | PFD | Q12306 | 1461 | 163 | 146 | 3 |
| SODC | Superoxide dismutase [Cu-Zn] | – | – | +1.4 | 9.64E-03 | CRH | P00445 | 1442 | 363 | 247 | 4 |
| SYDC | Aspartyl-tRNAsynthetase, cytoplasmic | – | – | +1.3 | 1.44E-02 | T | P04802 | 628 | 301 | 228 | 5 |
| SYRC | Arginyl-tRNAsynthetase, cytoplasmic | – | – | −1.3 | 5.76E-02 | T | Q05506 | 620 | 245 | 164 | 3 |
| SYRC | Arginyl-tRNAsynthetase, cytoplasmic | – | – | −1.4 | 4.33E-02 | T | Q05506 | 624 | 473 | 333 | 5 |
| TKT1 | Transketolase 1 | – | – | −1.3 | 1.73E-03 | CM | P23254 | 550 | 322 | 296 | 5 |
| TPIS | Triosephosphateisomerase | – | – | +1.4 | 1.61E-02 | CM | P00942 | 1296 | 432 | 342 | 5 |
| TPIS | Triosephosphate isomerase | – | – | +1.5 | 7.15E-03 | CM | P00942 | 1669 | 906 | 750 | 7 |
| TPIS | Triosephosphate isomerase | – | – | +1.4 | 6.10E-02 | CM | P00942 | 1671 | 513 | 443 | 4 |
| TTR | Human transthyretin | – | – | – | – | – | P02766 | 1492 | 592 | 534 | 6 |
| UBX1 | UBX domain-containing protein 1 | – | – | −1.4 | 1.65E-03 | PFD | P34223 | 806 | 682 | 562 | 7 |
| VDAC1 | Mitochondrial outer membrane protein porin 1 | – | – | +1.4 | 2.67E-02 | EM | P04840 | 1220 | 171 | 135 | 3 |
| YBD6 | UPF0001 protein YBL036C | – | – | −1.5 | 4.47E-04 | U | P38197 | 1206 | 156 | 109 | 3 |
| YJV7 | Uncharacterized protein YJL217W | – | – | +1.8 | 4.62E-03 | U | P40893 | 1380 | 173 | 117 | 3 |
All the listed proteins showed a statistical difference of spot volume ratio between the control/BTTR-wt and control/BTTR-L55P with an ANOVA p<0.05.
Gene code as in the yeast genome database (www.yeastgenome.org).
CM – carbohydrate metabolism; AM – amino acid metabolism; NM – nucleotide metabolism; EM – energy metabolism; LM – lipid metabolism; U – unknown; CRH – cell redox homeostasis; T – translation; TP – transport; PFD – protein folding and degradation.
Accession code of the uniprot database (www.uniprot.org).
Spot number on the master gel (see fig. 1A).
Summary of the protein identification results. The protein and the peptide score as given by the GPS Explorer software (Applied Biosystems). The number of peptides with MSMS data is also given.
A significant change in protein abundance was clearly detected for spot number 1492, absent from the control (Figure 2D). This spot was unequivocally identified as human TTR. This is a noteworthy observation for two main reasons: first, it shows that our experimental system leads to a high TTR expression level; second, the detection of this expected difference validates the approach we chose to detect quantitative differences in protein abundances.
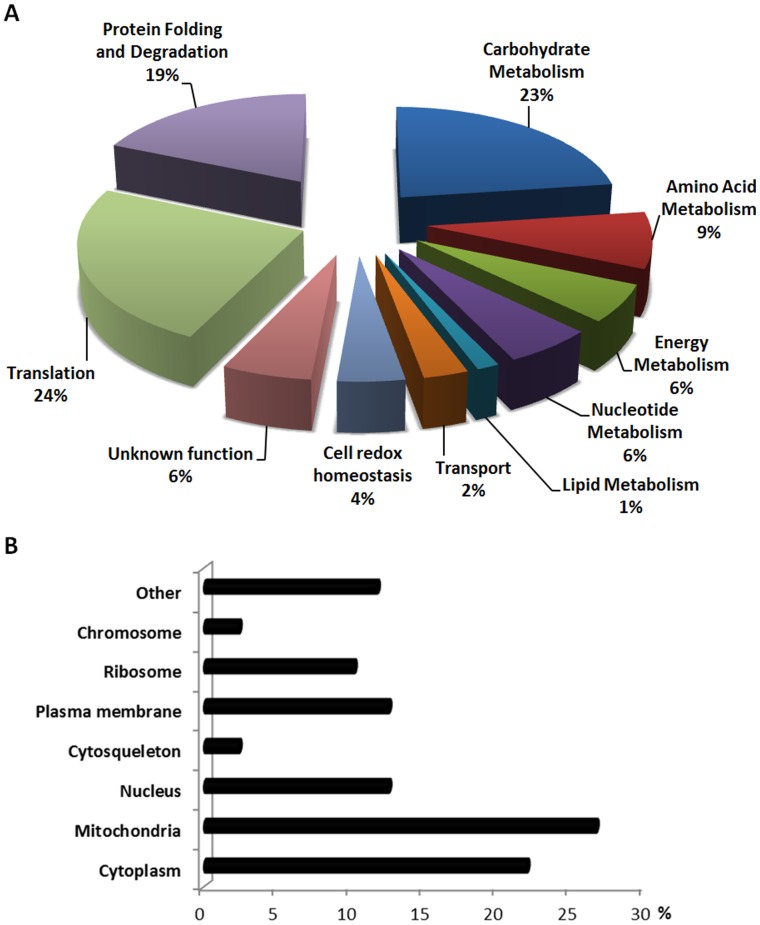
The identified proteins were categorised into functional groups and cellular location using Gene Ontology annotations. The 70 identified proteins fell into 10 functional categories (Figure 3A). About 45% are proteins related to cell metabolism, including carbohydrate (16 unique proteins, 23%), amino acid (6 proteins, 9%), energy (4 proteins, 6%), nucleotide (4 proteins, 6%) and lipid metabolism (1 protein, 1%). A significant number of the identified proteins (17 proteins; 24%) are involved in translation, including ribosomal proteins and translational factors. Noteworthy, a high number of the identified differentially expressed proteins are related to protein folding and degradation pathways (13 proteins, 19%). Several of these proteins have been described as stress response ones, involved in the response to an increase protein misfolding (as Hsp70 protein family). Proteins involved in transport (2 proteins, 3%), cell redox-homeostasis (3 proteins, 4%) and proteins with unknown or poorly characterized function (4 proteins, 6%) were also identified.
Figure 3. Gene ontology characterization of the identified differentially expressed proteins.
(A) Biological function. (B) Cellular location.
Concerning cellular location, a high number of identified proteins were from mitochondria (27%), while 22% were from cytoplasm, 12.5% from the plasma membrane and nucleus (Figure 3B).
Proteome Changes Induced by the Amyloidogenic TTR Variant
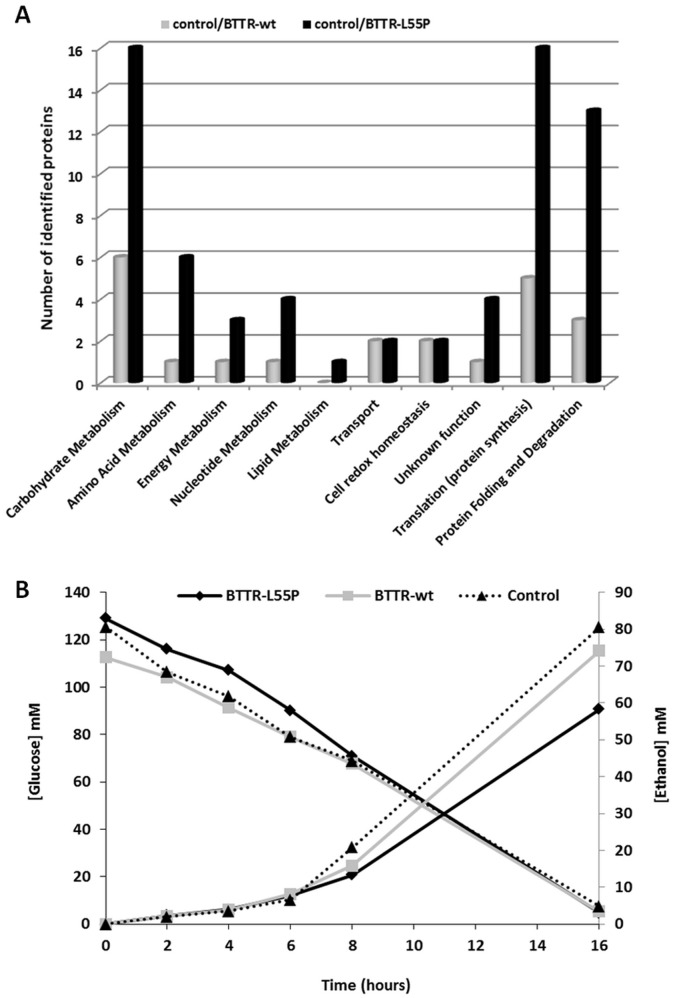
As described above, a clear differentiation was evident between the proteome of cells expressing the non-amyloidogenic TTR-wt and the highly amyloidogenic TTR-L55P variant. In BTTR-wt, 22 proteins were differentially expressed (Figure 4A), with 15 proteins up-regulated and 7 down-regulated (Figure 5, grey). By contrast, in cells expressing the amyloidogenic TTR-L55P, significant proteome changes were induced with 67 unique proteins differentially expressed, 49 being up-regulated and 18 down-regulated (Figure 4A, detailed in Figure 5, black). To further explore the involvement of the uncovered pathways involved in TTR misfolding, we performed a functional enrichment analysis using DAVID (Database for Annotation, Visualization and Integrated Discovery). This analysis revealed 8 functional clusters with a significantly enrichment score (Table 2). Significant changes in cell metabolism (namely glucose and amino acid metabolism) and also in the regulation of translation and protein synthesis was noticeable. Biological themes related to plasma membrane and mitochondria proteins and a functional enrichment in the molecular chaperones network and in protein refolding was observed. In contrast, the DAVID analysis using the proteins differentially expressed in BTTR-wt revealed three significant clusters only: mitochondrial matrix (FDR of 6.10E-07), tricarboxylic acid cycle (FDR of 3.60E-06) and plasma membrane enriched fraction (FDR of 5.40E-03).
Figure 4. Expression profile in BTTR-wt and BTTR-L55P versus the control.
(A) Number of the identified proteins according to the biological function, differentially expressed between the control and BTTR-wt (grey) and between the control and BTTR-L55P (black). Upon the expression of the amyloidogenic TTR variant L55P, a substantial increase in the number of differentially expressed proteins namely in proteins involved in cell metabolism, translation and protein folding and degradation is clearly detected. (B) D-Glucose consumption and ethanol production during cell growth in the control (dotted line), BTTR-wt (grey) and BTTR-L55P (black). A representative analysis is shown.
Figure 5. Detailed expression profiles for all the identified differentially expressed proteins, according to its functional categories: (A) cell metabolism; (B) unknown function; (C) Cell redox homeostasis; (D) protein folding and degradation; (E) translation.
Grey bars represent fold change in protein expression in BTTR-wt versus the control while black bars represent fold change in protein expression in BTTR-L55P versus the control. The vertical axis indicates the identified protein while the horizontal axis represents the fold variation in protein expression. Additional information for each protein, including full name, can be found in Table 1. For proteins identified in different spots (with slightly different fold variations) the average is represented in the graph.
Table 2. Functional annotation enrichment analysis of the identified proteins using the Database for Annotation, Visualization and Integrated Discovery (DAVID ) v6.7.
| AnnotationCluster | Annotation Terms | FDR |
| 1 | Glycolysis/Gluconeogenesis | 2.70E-07 |
| Pyruvate metabolic process | 2.00E-09 | |
| Alcohol catabolic process | 5.50E-09 | |
| 2 | Plasma membrane enriched fraction | 1.40E-10 |
| 3 | Regulation of translation | 1.90E-06 |
| Cytosolic ribosome | 1.10E-03 | |
| 4 | Cellular amino acid biosynthetic process | 4.10E-05 |
| Cellular amino acid catabolic process | 1.50E-04 | |
| Branched chain family amino acid metabolic process | 1.80E-04 | |
| Pantothenate and CoA biosynthesis | 3.00E-03 | |
| Glutamate biosynthetic process | 5.10E-04 | |
| 5 | Molecular chaperone | 2.60E-06 |
| Protein refolding | 1.20E-03 | |
| 6 | Mitochondrial matrix | 6.80E-05 |
| 7 | Metal-binding | 1.80E-04 |
| 8 | Nucleotide-binding | 5.20E-03 |
Annotation terms are representative of a particular cluster. FDR – False discovery rate.
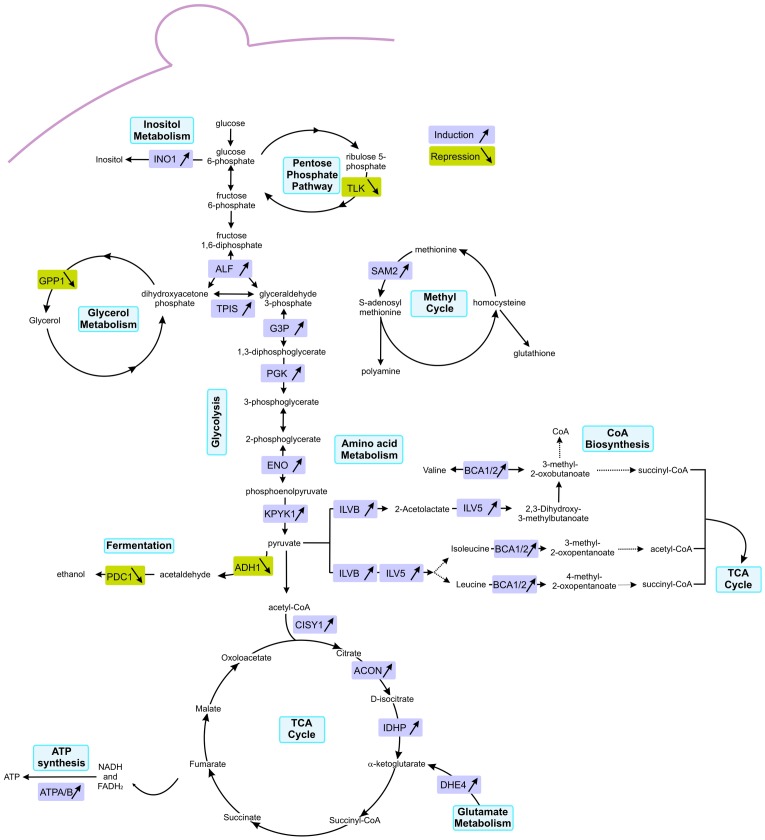
The high number of the differentially expressed proteins involved in metabolic processes hints that the cell response to protein misfolding stress is accompanied by active metabolic changes. The major metabolic pathways altered are illustrated in Figure 6. Following TTR-L55P expression, we detected an up-regulation of several glycolytic enzymes (Figures 5A and 6), pointing to an increase in glucose catabolism. In addition, the two enzymes that catalyse ethanol formation from pyruvate (PDC1, pyruvate decarboxylase 1 and ADH1, alcohol dehydrogenase) are down-regulated (Figures 5A and 6). This could reflect a shift in the pyruvate fate, from alcoholic fermentation to the TCA cycle and oxidative phosphorylation. Indeed, the TCA cycle enzymes citrate synthase (CISY1) aconitate hydratase (ACON) and isocitrate dehydrogenase (IDHP), which are responsible for the synthesis of α-ketoglutarate from acetyl-CoA, are up-regulated in BTTR-L55P in compassion to control cells. Consistently, an increased abundance of ATP synthase (ATPB) and a down-regulation of the pentose phosphate pathway enzyme transketolase (TKT1) and (DL)-glycerol-3-phosphatase 1 (GPP1), an enzyme involved in glycerol synthesis, was detected. Altogether, these results point to the channelling of glucose catabolism through the TCA cycle, leading to an increase ATP production via cell respiration (Figure 6). Notably, this metabolic change is not apparent in BTTR-wt (Figure 5A in grey). In these cells, D-glucose consumption and ethanol production are similar (Figure 4B).
Figure 6. Major metabolic pathways altered in BTTR-L55P.
Differentially expressed proteins are highlighted, those induced are framed in purple and those repressed are framed in green. Protein full names can be identified in Table 1.
The increased ATP demand is surely related with the higher energy needed to actively refold or degrade misfolded proteins. In fact, a similar metabolic change was apparent in the response to heat shock that is also characterized by an increased protein unfolding and aggregation [22]. An undesirable by-product of an increased ATP formation via cell respiration is the formation of reactive oxygen species such as superoxide anion which may activate the oxidative stress response. This explains the up-regulation found exclusively in BTTR-L55P of superoxide dismutase [Cu-Zn] (SODC). These data may suggest a link between protein misfolding and oxidative cellular stress derived from a higher ATP demand. It has been show that oxidative modifications may facilitate aggregation of amyloidogenic proteins [23]. Upon aging, cellular defences towards oxidative stress are compromised and oxidative protein modifications are also likely to accumulate, which may be synergistically linked to the disease onset [2].
Another major set of differentially expressed proteins are involved in translation, with a complex pattern of expression (6 proteins down-regulated and 10 up-regulated in BTTR-L55P) (Figure 5E). These changes may reflect an adaptive response to cellular stress. It was reported that elongation factors are up-regulated in response to stress conditions, such as oxidative stress [24]. In addition to its canonical role in translation, unique cellular activities, such as nuclear export, cytoskeleton organization and apoptosis, have been attributed to elongation factor protein family in eukaryotes [25]. Interestingly, a potential role in protein quality control and co-translational degradation has been suggested for these proteins [26]–[28]. Elongation factors interact with the 26S proteasome and this association increases when translation is inhibited [26]. Thus, the identified proteins may be important in the cell response to protein misfolding in a more complex way than a simple activation or inhibition of protein synthesis. Although further studies are needed to clarify this issue, our findings provide a good starting point by revealing potential protein targets.
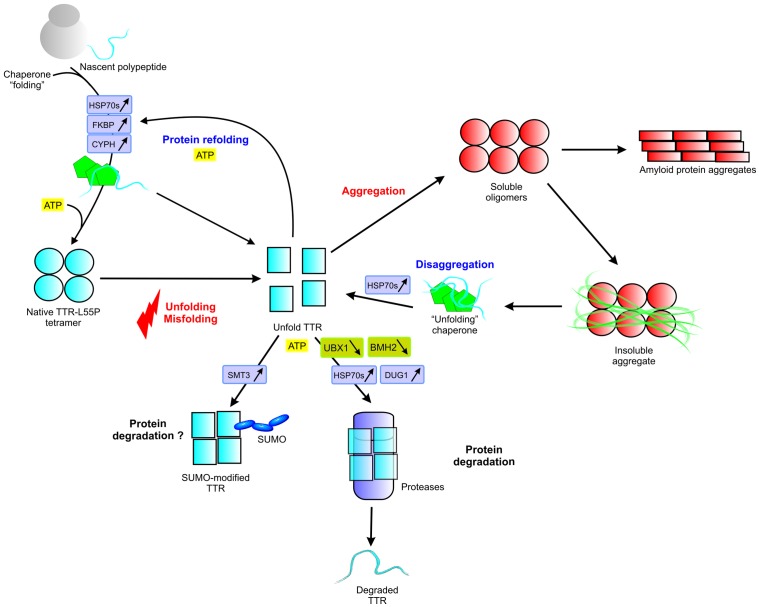
Several stress-response proteins involved in protein folding and/or degradation were also identified, with 13 proteins differentially expressed in BTTR-L55P (10 up-regulated and only 3 down-regulated, Figure 5D). Some heat shock proteins were also found up-regulated in BTTR-wt by a similar fold variation (HSP77, HSP60, HSP72 and HSP10; Figure 5D, in grey). Interestingly, HSP77, HSP60 and its co-chaperone HSP10 [29] are mitochondrial-resident chaperones, involved in folding of newly imported proteins to the mitochondria. The other identified proteins involved in protein folding and degradation changed their abundance only in BTTR-L55P. The potential role of these proteins in TTR misfolding and aggregation is illustrated in Figure 7.
Figure 7. TTR misfolding and protein quality control mechanisms.
Differentially expressed proteins are highlighted, those induced framed in purple and those repressed are framed in green. Protein full names are indicated in Table 1.
Two interesting protein revealed by our study are the cyclophilin FK506-binding protein 1 (FKBP) and cyclophilin A (CYPH). These proteins, that have a peptidyl-prolyl-cis-trans isomerase (PPIase) activity, were up-regulated in BTTR-L55P (Figure 5D). FKBP changed its abundance also in BTTR-wt but it increases significantly in BTTR-L55P (1.4 fold vs 2.2, Table 1). The involvement of PPIases in the cell response to protein misfolding and aggregation is still unclear. However, recent experiments implied this protein family in conformational neurodegenerative disorders. It was showed that FKBP52 overexpression reduced the Aβ peptide toxicity and increases the lifespan of flies expressing Aβ peptide, whereas loss of function of FKBP52 exacerbated these Aβ phenotypes [30]. Our previous results revealed that PPIase cyclophilin H is one of the major TTR interactuant in human plasma of ATTR patients [15]. In yeast cells, FKBP was shown to interact with the heat shock factor 1, a major regulator of the cell response to stress conditions such as heat shock and protein misfolding [31]. In addition to FKBP, CYPH was up-regulated by 1.7 fold exclusively in BTTR-L55P. The human homologue of yeast CYPH is a major Aβ-peptide interactuant found in the brain and elevated levels of this protein were reported in human Alzheimeŕs disease brains [32]. Moreover, it was showed that the Aβ oligomeric form has a greater affinity for CYPH, hinting for a relevant role of this protein in protein aggregation [32]. Interestingly, yeast CYPH interacts with several proteasome regulatory subunits and also with ubiquitin and SMT3 (yeast homologue of mammalian SUMO1) [33] suggesting a role in protein degradation associated processes. Altogether, these observations suggest an important role of PPIases in disease development and cellular responses to protein aggregation in the context of conformational disorders and are good protein targets for further studies.
DIGE Screen identified three additional molecular chaperones exclusively in BTTR-L55P: an increased abundance of HSP71 (SSA1 gene) and HSP72 (SSA2 gene) and a reduction in HSP75 (SSB1 gene). Using an animal model of ATTR, it was recently observed that TTR deposition leads to an increase in HSP70 expression [34], in agreement with our 2D-DIGE analysis. However, contrary to our study, the interplay between the different isoforms of the Hsp70 was not revealed, which is highly relevant considering that a functional difference between members of SSA and SSB is apparent [35], [36]. Both classes of chaperones affect de novo prion formation in yeast, although with opposite effects [37]. After heat shock, a similar trend was observed with the SSB isoform being down-regulated while the SSA isoforms is up-regulated [35].
Besides HSP70, the endoplasmic reticulum lumen resident protein disulfide isomerase (PDI) was up-regulated exclusively in BTTR-L55P, which appears to be in concordance with an up-regulation of the HSP70 resident endoplasmatic reticulum chaperone BiP in tissues affected with TTR aggregation [38]. Thus, even though a simple model organism was used in this study and TTR was used as a model amyloidogenic protein, it is likely that relevant protein targets, even for familial amyloidosis, was revealed by this screen.
The action of molecular chaperones is an essential first step to avoid protein aggregation. At this point, misfolded proteins are either refolded or degraded, avoiding their accumulation. It is now recognized that some molecular chaperones are also involved, either directly or indirectly, in protein disposal. HSP71, found up-regulated exclusively in BTTR-L55P, and its co-chaperone HSP40 specifically recognize misfolded protein domains and escort them for proteasome degradation [39]. Yeast HSP71 is in fact known to interact with proteasome regulatory subunits (like the 26S RPN 2) [40]. Noteworthy, the ubiquitin-like protein SMT3 was found up-regulated exclusively in BTTR-L55P (Figure 5). This protein displays 50% sequence identity with mammalian SUMO1 protein and is essential for yeast viability. Protein sumoylation is known to be involved in neurodegenerative disorders such as Alzheimer’s [41] and Huntington’s diseases [42] although its exact role is presently controversial. For the Huntington protein, sumoylation renders the protein more soluble and apparently more toxic by inhibiting its aggregation into inclusion bodies [42]. In Alzheimer’s disease, SUMO1 overexpression in tissue cultured cells co-transfected with the APP gene suppresses Aβ fragment accumulation [41]. Studies in human samples of ATTR patients showed that TTR aggregates lead to a significant increase in ubiquitin conjugates and an impairment of the ubiquitin-proteasome system was observed [43].
Two proteins involved in protein folding and degradation were down-regulated exclusively in BTTR-L55P: UBX domain-containing protein 1 (UBX1) and protein BMH2 (Brain Modulosignalin Homologue, member of the ubiquitous 14-3-3 gene family) (Figure 5D, in black). UBX1 is known to interact with proteasome regulatory subunits and with ubiquitylated proteins in vivo, being required for the degradation of an ubiquitylated model substrate [44]. Interestingly, UBX1 also interacts with FKBP [45], with several isoforms of the HSP70 family (HSP71 [40] and HSP77 [46]) and also with PDI [47], proteins differentially expressed in response to TTR-L55P expression. Concerning BMH2, it is likely to be related to carbohydrate metabolism and stress response [48], [49]. It was observed that the expression of BMH proteins is altered after exposure to several stress conditions such as heat shock or dithiotreitol [50], [51]. To our knowledge, no association between 14-3-3 proteins and TTR aggregation has been described until now.
Three proteins with unknown or poorly characterized functions were also identified (Figure 5B). In addition to a down-regulation of AIM29 (already detected in BTTR-wt), the UPF0001 protein YBL036C was also down-regulated, while the uncharacterized protein YJL217W and cytochrome c oxidase assembly protein COX14 increase its abundance. The biological function of these proteins is not yet known. Some data seem to relate the YJL217W protein in the regulation of enolase1 [52]. This could be related with the detected increase in glucose catabolism and, considering that yeast enolase1 also function as a heat shock protein [53], with the misfolding of TTR-L55P. This is an interesting hypothesis that requires further investigation, feasible in yeast considering that the ENO1 yeast null mutant is viable. COX14 Is an integral mitochondria membrane protein and a yeast null mutant for this protein displays a respiratory growth deficiency [54]. Thus, the increased expression of this protein may results in an improved mitochondria function. No significant human homology was found for this protein. Finally, UPF0001 protein YBL036C also increased significantly its abundance in BTTR-L55P. A BLAST homology search revealed that this yeast protein shares 43% identity with proline synthase co-transcribed bacterial homolog protein, whose function is nevertheless not yet known.
Concluding Remarks
The proteome response to protein misfolding and aggregation is a key factor in understanding onset and evolution of conformational disorders. By uncovering specific proteins and pathways effective therapeutic strategies may be devised. In this study, we performed a high-throughput proteomics analysis using the 2D-DIGE technology to screen changes in protein abundance upon the expression of an amyloidogenic protein (TTR-L55P variant) in comparison with the expression of the non-amyloidogenic version of the same protein (TTR-wt). With this approach, around 1800 individual protein spots were quantitatively analyzed. Our results showed a clear cut differentiation between the expression of TTR-wt and TTR-L55P, distinguishing the cell response to the expression of a heterologous protein from the cell response to protein misfolding and aggregation. The proteome changes observed by the expression of TTR-wt and TTR-L55P share about 20 proteins in common that may be attributed to the expression of a heterologous protein in yeast. Expression of the amyloidogenic TTR-L55P caused changes in about 50 exclusive proteins that may be specifically associated to amyloidogenic behavior and protein aggregation. Several heat shock protein (as HSP70 and HSP60) and members of PPIase family (cyclophilin A and FKBP) are up-regulated upon TTR-L55P expression. In addition, several other processes were influenced by the misfolding and aggregation of an amyloidogenic protein such as carbohydrate and amino acid metabolism, energy production, translation and oxidative stress response. The expression of the amyloidogenic TTR variant causes a metabolic shift towards energy production via mitochondrial cell respiration which is related to the high energy demand of refolding and degradation pathways. This in turn may promote oxidative stress. The up-regulation of mitochondrial chaperones (HSP60, HSP77, HSP10, protein disulfide-isomerase) is also consisted with increased oxidative stress and higher energy demands needed to face misfolding and aggregation.
Although in transthyretin amyloidosis TTR accumulates as amyloid deposits in the extracellular space, we believe that TTR is a good amyloidogenic protein model to investigate the cellular responses to the general misfolding and aggregation problem. TTR has a high number of amyloidogenic point mutations and thus the proteome response may be investigated with different amyloidogenic potential against a defined genetic background. It was recently discovered that intracellular signaling mechanisms related to extracellular TTR aggregates in ATTR was elicited [34], [38], [43]. Notably, similarly to our results, changes in the chaperone network (such as HSP70 protein family and endoplasmatic resident chaperones) and degradation processes were detected [38], [43]. Moreover, a striking parallel was found on the specific increased expression of PPIases in yeast expressing TTR-L55P, found to be a specific TTR interacting partner only in symptomatic ATTR subjects [15]. Yeast can thus be used as a model to investigate TTR effects on living cells that is highly relevant in the context of ATTR and other misfolding diseases. Moreover, more than 100 TTR point mutants are known to be associated to amyloid disease and the availability of a complete gene set collection of yeast single gene deletion mutants shows its potential for large scale screening.
Materials and Methods
Bacteria, Yeast Strains and Culture Conditions
Escherichia coli strain used (DH5α, F-; recA1; endA1; thi-1; gyrA96; hsdR17; supE44; relA1;φ89d; lacZ; DM15 λ-) was cultured in LB medium [1% (w/v) NaCl, 1% (w/v) tryptone, 0.5% (w/v) yeast extract] at 37°C. Solid LB medium contained 2% (w/v) agar. Transformed strains, carrying the plasmids, grew in LB medium supplemented with 0.1 mg.ml−1 ampicillin. Saccharomyces cerevisiae strain used was the BY4741 (genotype BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) from Euroscarf collection (Frankfurt, Germany). Strains were kept in YPGlu agar slopes [0.5% (w/v) yeast extract, 1% (w/v) peptone and 2% (w/v) D-glucose and 2% (w/v) agar] at 4°C and cultured in liquid YPGlu medium at 30°C. BY4741 strain carrying the TTR expression plasmids were cultured in YNB minimal medium without uracil [0.67% (w/v) yeast nitrogen base, 2% (w/v) D-glucose and 0.025% (w/v) L-methionine, L-histidine, L-leucine].
Plasmids and Yeast Transformation for TTR Expression
Plasmids p426GPD carrying the different TTR genes (TTR-wt and TTR-L55P) were a king gift of Dr. Tiago Outeiro (Cell and Molecular Neuroscience Unit, IMM, Portugal). Plasmid DNA extraction was performed using the Wizard Plus SV Minipreps DNA Purification System (Promega), following the manufacturer’s instructions. DNA concentration was evaluated spectrophotometrically at 260 nm and purity was assessed by standard procedures. BY4741 yeast strain was transformed by the lithium acetate method, and transformants were selected on minimal agar plates deficient in uracil, following standard procedures [55]. Yeast cells were also transformed with the p426GPD vector without the inserted gene as control. Yeast growth curves were monitored at 640 nm and phenotypic growth assays were carried out by spotting 3 µl of late-exponential-phase culture, sequentially diluted (approx. 2000–20 cells), in selective medium. Growth was recorded after 2 days at 30°C.
D-Glucose and ethanol were enzymatically assayed during cell growth using the D-Glucose assay kit and Ethanol assay kit (NZYTech), following the manufacturer instructions.
Western Blotting and Protein Aggregate Filtration Assay
TTR expression levels were relatively quantified by western blot using an anti-human TTR polyclonal antibody (SC 13098, Santa Cruz Biotechnology). Protein extraction was performed by glass bead lysis as previously described [56] and protein concentration was determined using the Bio-Rad Bradford assay reagent. Proteins (30 µg per lane) were separated using denaturant urea gel electrophoresis in a Mini-protean 3 system (Bio-Rad). Proteins were transferred to PVDF membranes (Immobilon-P PVDF, Millipore), using the Mini Trans-Blot system (Bio-Rad) and the membrane was blocked overnight at 4°C in TBS-T (50 mM Tris and 150 mM NaCl, pH 7.5 with 0.1% (v/v) Tween 20) containing 5% (w/v) skimmed milk. TTR polyclonal antibody was used at a dilution of 1∶5000. Ponceau S staining was used to monitor protein transfer and to confirm that equal amount of protein were loaded in each lane. Washes, secondary antibody and detection procedures were performed using the BM Chemiluminescence Western Blotting Kit (Roche), following the manufacturer’s instructions.
Protein aggregate filtration assay was performed essentially as described [57], [58], with slight modifications. Briefly, yeast cells expressing TTR variants were suspended in modified lysis buffer (50 mM Tris containing 5 mM MgCl2, 0.1 mM EDTA, 2 mM PMSF and proteases cocktail inhibitor) and lysed with glass beads as described [56]. Cell ghosts were removed by centrifugation at 2500 g for 5 min at 4°C. Supernatants were collected as the crude protein extract and protein concentrations in all samples were equalized. 150 µl Of the crude extract was centrifuged at 14000 g for 30 min at 4°C to separate the soluble from aggregated proteins. The pellet fractions, containing the insoluble aggregated proteins, were suspended in 2% (w/v) SDS or in lysis buffer. Samples were filtered on a dot-blot filtration unit (Bio-Rad) through a nitrocellulose membrane (0.2 µm pore size) that was previously blocked and pre-equilibrated with 2% (w/v) SDS. Filters were washed twice with 0.1% (w/v) SDS (except the samples suspended in lysis buffer, which were washed with the same buffer). TTR aggregates were imunodetected using the anti-human TTR polyclonal antibody at a dilution of 1∶5000. To assess the assay specify toward TTR protein aggregates and not soluble TTR, the crude extract and the soluble protein fraction were analysed through a nitrocellulose membrane (as a regular dot-blot assay) and a blocked nitrocellulose membrane.
Protein Sample Preparation and CyDye Protein Labeling
For 2D-DIGE analysis, cells were collected at mid-log phase of growth, harvested by centrifugation and suspended in 2D-DIGE labeling buffer [7 M urea, 2 M thiourea, 4% (w/v) CHAPS and 30 mM Tris] containing proteases inhibitors (PMSF and proteases cocktail inhibitor, Sigma). Yeast cells were lysed using glass beads as previously described [56]. All samples (four biological replicates) were processed in parallel. Protein extracts were clarified by centrifugation at 12000 g for 15 min at 4°C. The cell lysate pH was then carefully adjusted to 8.5 with NaOH and afterwards protein concentration was determined using the 2D Quant Kit (GE Healthcare) with BSA as standard. Prior to electrophoresis, protein extracts were labeled with CyDyes™ (GE Healthcare), following the manufacturer instructions. Briefly, proteins were labeled by mixing 240 pmol of fluorochromes (Cy3 or Cy5) with 30 µg of protein and incubated on ice for 30 min in the dark. Lysine (1 µl, 10 mM) was then added to quench the reaction and the samples were left on ice for 10 min in the dark. A pooled internal standard was performed by mixing 15 µg of each sample analyzed that was labeled with Cy2 dye and included in all gel runs. A dye swap was used between Cy3 and Cy5 to avoid problems associated with preferential labeling.
2D Gel Electrophoresis
For 2D gel electrophoresis, the two samples to be run on the same gel plus the internal standard were mixed before adding 2x lysis buffer [7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 6 µl.ml−1 DeStreak reagent (GE Healthcare)] and 2% (v/v) ampholytes immobilized pH gradient buffer (pH 3–10 NL, GE Healthcare) to a final volume of 125 µl. Isoelectric focusing was carried out on pH 3–10 IPG-strips (24 cm, non-linear gradient; GE Healthcare) using the IPGphor3 system from GE Healthcare. ImmobilineDryStrips were rehydrated overnight with 450 µl DeStreak Rehydration Solution (GE Healthcare), complemented with 1.5% ampholytes, before cup-loading of proteins and IEF on an EttanIPGphor Manifold (GE Healthcare). The migration was performed at 20°C (60 V for 2 h; gradient from 60 to 500 V for 5 h; hold 500 for 1 h, gradient from 500 to 1000 for 3 h; hold 1000 V for 1 h; gradient from 1000 V to 8000 V for 4 h, hold 8000 V until 64 000 Vh). After the IEF, IPGstrips were equilibrated twice for 15 min in equilibration buffer [50 mM Tris-HCl pH 8.8, 6 M urea, 30% (v/v) glycerol, 2% (w/v) SDS and 0.002% (w/v) bromophenol blue] supplemented with 1% (w/v) DTT and then with 2.5% (w/v) iodoacetamide. Second-dimension SDS-PAGE was performed using 1.0 mm large-format 12.5% polyacrilamide resolving gel and run at 20°C overnight with 1.5W per gel, using the EttanDALTtwelve system (GE Healthcare). Glass plates used for picking gels were treated with Bind-Silane solution [80% (v/v) ethanol, 2% (v/v) acetic acid and 0.1% (v/v) Bind-Silane (GE Healthcare)] before casting. The gels ran simultaneously, with a dye switching between repetitions, plus the internal standard. In the end, ninety micrograms of proteins were loaded on each 2D gel.
Scanning and Image Analysis
2D-DIGE gels were scanned at a pixel size of 100 µm using a Typhoon Imager 9400 (GE Healthcare) at three different wavelengths corresponding to the different CyDyes. Gel images were exported into Progenesis SameSpot V3 image analysis system (Nonlinear Dynamics), where quantitative analysis of protein spots was performed. Following automatic and subsequent manual editing, aligning and matching procedures as part of the Progenesis SameSpot workflow, ANOVA p-values between the samples were calculated within the Progenesis SameSpot software. Variation of protein expression was considered statistically significant if the absolute abundance variation was at least 1.3-fold between spots of any experimental group with a p<0.05 by ANOVA. Unsupervised PCA correlation analysis was performed using the statistical tool within the gel analysis software. Clustering of each sample was based on the expression pattern of each spot with a significant ANOVA p-value. The spots of interest were visually checked and selected for protein identification by mass spectrometry.
Spot Handling and Mass Spectrometry Analysis
Spots of interest were excised from gels using the EttanSpot Picker from the Ettan Spot Handling Workstation (GE Healthcare). After washing and desalting in 50 mM ammonium bicarbonate, 50% (v/v) methanol and 75% (v/v) acetonitrile, spots were then digested with Trypsin Gold for 6 h at 37°C (MS grade, Promega, 5 µg.mL–1 in 20 mM ammonium bicarbonate) using the Ettan Digester robot from the same workstation. Supernatants were collected, vacuum dried and peptides were thoroughly dissolved in 3 µl of 50% (v/v) acetonitrile containing 0.1% (v/v) trifluroacetic acid. Peptides were spotted on MALDI targets using a matrix consisting of 7 mg.ml−1 of α-cyano-hydroxycynamic in 50% (v/v) acetonitrile containing 0.1% (v/v) trifluroacetic acid. Monoisotopic peptide mass determinations were carried out using the MALDI-TOF/TOF 4800 Plus mass spectrometer (Applied Biosystems). For spots with a low signal, the peptide mixture was purified and concentrated using home-made chromatographic microcolumns using GELoader tips packed with POROS R2 as described [59]. MS experiments were performed in positive reflectron mode for monoisotopic peptide mass determination. The mass spectrometer was externally calibrated using the 4700 Calibration Mix (Applied Biosystems). MS spectra were collected in a result-independent acquisition mode, typically using 1000 laser shots per spectra and a fixed laser intensity of 3100 V. For tandem experiments, fifteen of the strongest precursors were selected for MS/MS, the weakest precursors being fragmented first. MS/MS analyses were performed using CID (Collision Induced Dissociation) with 1 kV collision energy at 1 × 106 torr air pressure. Spectra were collected using a fixed laser intensity of 4200 V and 2000 laser shots. Raw data were generated by the 4000 Series Explorer Software v3.0 RC1 (Applied Biosystems) and tryptic peptide contaminant m/z peaks resulting from trypsin auto-digestion were excluded when generating the peptide mass list used for comparison with the theoretical tryptic digest. Proteins were identified by the GPS explorer (Applied Biosystem) using the following search parameters: 1) carboxyiodomethylation of cysteine residues and methionine oxidation were taken as fixed and variable modifications, respectively; 2) tolerance of one missed cleavage; 3) maximum error tolerance of 50 ppm for MS data and 0.3 Da for the MS/MS data. Protein identifications were further confirmed using the ProteinPilot software (Applied Biosystem).
Gene ontology and pathway analysis
The identified proteins were categorized into functional groups using the gene ontologies annotations available at the Universal Protein Resource Protein Knowledge database (UniProtKB; http://www.uniprot.org/), Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) and Saccharomyces Genome Database (www.yeastgenome.org/). BLAST searches were performed using the UniProtKB tools. A functional annotation enrichment analysis was performed using DAVID (Database for Annotation, Visualization and Integrated Discovery) [60], [61], available at http://david.abcc.ncifcrf.gov/. In addition to gene ontology, the PIR (Protein Information resource), COG (Clusters of Orthologous Groups), Uniprot, KEEG, Interpro and SMART (Single Modular Architecture Research Tool) databases were used within DAVID to generate biological theme by grouping like terms, thereby creating functional annotation clusters. The results were manually checked for significant enriched terms.
Acknowledgments
The authors acknowledge Dr. Tiago Outeiro (Cell and Molecular Neuroscience Unit, IMM, Portugal) for the gift of TTR-expressing plasmids.
Funding Statement
Work was supported by grants SFRH/BPD/41037/2007 (R.A.G.), PEst-OE/QUI/UI0612/2011, PTDC/QUI/70610/2006 and REDE/1501/REM/2005 from the Fundação para a Ciência e a Tecnologia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ghiso J, Wisniewski T, Frangione B (1994) Unifying features of systemic and cerebral amyloidosis. Mol Neurobiol 8: 49–64. [DOI] [PubMed] [Google Scholar]
- 2. Ross CA, Poirier MA (2004) Protein aggregation and neurodegenerative disease. Nat Med 10 Suppl: S10–17 [DOI] [PubMed] [Google Scholar]
- 3. Gomes R, Sousa Silva M, Quintas A, Cordeiro C, Freire A, et al. (2005) Argpyrimidine, a methylglyoxal-derived advanced glycation end-product in familial amyloidotic polyneuropathy. Biochem J 385: 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bukau B, Weissman J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125: 443–451. [DOI] [PubMed] [Google Scholar]
- 5. Muchowski PJ, Wacker JL (2005) Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci 6: 11–22. [DOI] [PubMed] [Google Scholar]
- 6. Outeiro TF, Tetzlaff J (2007) Mechanisms of disease II: cellular protein quality control. Semin Pediatr Neurol 14: 15–25. [DOI] [PubMed] [Google Scholar]
- 7. Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, et al. (2007) Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science 317: 516–519. [DOI] [PubMed] [Google Scholar]
- 8. Chaudhuri TK, Paul S (2006) Protein-misfolding diseases and chaperone-based therapeutic approaches. FEBS J 273: 1331–1349. [DOI] [PubMed] [Google Scholar]
- 9. Connors LH, Lim A, Prokaeva T, Roskens VA, Costello CE (2003) Tabulation of human transthyretin (TTR) variants, 2003. Amyloid 10: 160–184. [DOI] [PubMed] [Google Scholar]
- 10. Saraiva MJ (2001) Transthyretin mutations in hyperthyroxinemia and amyloid diseases. Hum Mutat 17: 493–503. [DOI] [PubMed] [Google Scholar]
- 11. Olsson M, Norgren N, Obayashi K, Plante-Bordeneuve V, Suhr OB, et al. (2010) A possible role for miRNA silencing in disease phenotype variation in Swedish transthyretin V30M carriers. BMC Med Genet 11: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ando Y, Araki S, Ando M (1993) Transthyretin and familial amyloidotic polyneuropathy. Intern Med 32: 920–922. [DOI] [PubMed] [Google Scholar]
- 13. Westermark P, Sletten K, Johansson B, Cornwell GG, 3rd (1990) Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc Natl Acad Sci U S A 87: 2843–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Araki S (1995) Anticipation of age-of-onset in familial amyloidotic polyneuropathy and its pathogenesis. Intern Med 34: 703–704. [DOI] [PubMed] [Google Scholar]
- 15. da Costa G, Gomes RA, Guerreiro A, Mateus E, Monteiro E, et al. (2011) Beyond genetic factors in familial amyloidotic polyneuropathy: protein glycation and the loss of fibrinogen's chaperone activity. PLoS One 6: e24850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, et al. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455: 1251–1254. [DOI] [PubMed] [Google Scholar]
- 17.Nagaraj N, Kulak NA, Cox J, Neuhaus N, Mayr K, et al.. (2011) Systems-wide perturbation analysis with near complete coverage of the yeast proteome by single-shot UHPLC runs on a bench-top Orbitrap. Mol Cell Proteomics. [DOI] [PMC free article] [PubMed]
- 18. Krobitsch S, Lindquist S (2000) Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci U S A 97: 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Outeiro TF, Lindquist S (2003) Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science 302: 1772–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishida CR, Gralla EB, Valentine JS (1994) Characterization of three yeast copper-zinc superoxide dismutase mutants analogous to those coded for in familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A 91: 9906–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacobson DR, McFarlin DE, Kane I, Buxbaum JN (1992) Transthyretin Pro55, a variant associated with early-onset, aggressive, diffuse amyloidosis with cardiac and neurologic involvement. Hum Genet 89: 353–356. [DOI] [PubMed] [Google Scholar]
- 22. Albrecht D, Guthke R, Brakhage AA, Kniemeyer O (2010) Integrative analysis of the heat shock response in Aspergillus fumigatus. BMC Genomics 11: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conway KA, Rochet JC, Bieganski RM, Lansbury PT Jr (2001) Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science 294: 1346–1349. [DOI] [PubMed] [Google Scholar]
- 24. Bektas M, Akcakaya H, Aroymak A, Nurten R, Bermek E (2005) Effect of oxidative stress on in vivo ADP-ribosylation of eukaryotic elongation factor 2. Int J Biochem Cell Biol 37: 91–99. [DOI] [PubMed] [Google Scholar]
- 25. Mateyak MK, Kinzy TG (2010) eEF1A: thinking outside the ribosome. J Biol Chem 285: 21209–21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, et al. (2005) Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol 25: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gonen H, Smith CE, Siegel NR, Kahana C, Merrick WC, et al. (1994) Protein synthesis elongation factor EF-1 alpha is essential for ubiquitin-dependent degradation of certain N alpha-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc Natl Acad Sci U S A 91: 7648–7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hotokezaka Y, Tobben U, Hotokezaka H, Van Leyen K, Beatrix B, et al. (2002) Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J Biol Chem 277: 18545–18551. [DOI] [PubMed] [Google Scholar]
- 29. Haslbeck M (2002) sHsps and their role in the chaperone network. Cell Mol Life Sci 59: 1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanokawa-Akakura R, Cao W, Allan K, Patel K, Ganesh A, et al. (2010) Control of Alzheimer's amyloid beta toxicity by the high molecular weight immunophilin FKBP52 and copper homeostasis in Drosophila. PLoS One 5: e8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bandhakavi S, Xie H, O'Callaghan B, Sakurai H, Kim DH, et al. (2008) Hsf1 activation inhibits rapamycin resistance and TOR signaling in yeast revealed by combined proteomic and genetic analysis. PLoS One 3: e1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Du H, Guo L, Fang F, Chen D, Sosunov AA, et al. (2008) Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer's disease. Nat Med 14: 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, et al. (2005) Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 280: 4102–4110. [DOI] [PubMed] [Google Scholar]
- 34. Santos SD, Fernandes R, Saraiva MJ (2010) The heat shock response modulates transthyretin deposition in the peripheral and autonomic nervous systems. Neurobiol Aging 31: 280–289. [DOI] [PubMed] [Google Scholar]
- 35. Peisker K, Chiabudini M, Rospert S (2010) The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim Biophys Acta 1803: 662–672. [DOI] [PubMed] [Google Scholar]
- 36. James P, Pfund C, Craig EA (1997) Functional specificity among Hsp70 molecular chaperones. Science 275: 387–389. [DOI] [PubMed] [Google Scholar]
- 37. Shorter J, Lindquist S (2008) Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J 27: 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Teixeira PF, Cerca F, Santos SD, Saraiva MJ (2006) Endoplasmic reticulum stress associated with extracellular aggregates. Evidence from transthyretin deposition in familial amyloid polyneuropathy. J Biol Chem 281: 21998–22003. [DOI] [PubMed] [Google Scholar]
- 39. Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, et al. (2007) The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell 18: 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, et al. (2009) An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol 5: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y, Wang H, Wang S, Quon D, Liu YW, et al. (2003) Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc Natl Acad Sci U S A 100: 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, et al. (2004) SUMO modification of Huntingtin and Huntington's disease pathology. Science 304: 100–104. [DOI] [PubMed] [Google Scholar]
- 43. Santos SD, Cardoso I, Magalhaes J, Saraiva MJ (2007) Impairment of the ubiquitin-proteasome system associated with extracellular transthyretin aggregates in familial amyloidotic polyneuropathy. J Pathol 213: 200–209. [DOI] [PubMed] [Google Scholar]
- 44. Schuberth C, Richly H, Rumpf S, Buchberger A (2004) Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep 5: 818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, et al. (2010) The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643. [DOI] [PubMed] [Google Scholar]
- 47. Kim JH, Zhao Y, Pan X, He X, Gilbert HF (2009) The unfolded protein response is necessary but not sufficient to compensate for defects in disulfide isomerization. J Biol Chem 284: 10400–10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bruckmann A, Steensma HY, Teixeira De Mattos MJ, Van Heusden GP (2004) Regulation of transcription by Saccharomyces cerevisiae 14-3-3 proteins. Biochem J 382: 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ichimura T, Kubota H, Goma T, Mizushima N, Ohsumi Y, et al. (2004) Transcriptomic and proteomic analysis of a 14–3-3 gene-deficient yeast. Biochemistry 43: 6149–6158. [DOI] [PubMed] [Google Scholar]
- 50. Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, et al. (2002) Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem 277: 31079–31088. [DOI] [PubMed] [Google Scholar]
- 52. Choi ID, Jeong MY, Ham MS, Sung HC, Yun CW (2008) Novel Ree1 regulates the expression of ENO1 via the Snf1 complex pathway in Saccharomyces cerevisiae. Biochem Biophys Res Commun 377: 395–399. [DOI] [PubMed] [Google Scholar]
- 53. Iida H, Yahara I (1985) Yeast heat-shock protein of Mr 48,000 is an isoprotein of enolase. Nature 315: 688–690. [Google Scholar]
- 54. Glerum DM, Koerner TJ, Tzagoloff A (1995) Cloning and characterization of COX14, whose product is required for assembly of yeast cytochrome oxidase. J Biol Chem 270: 15585–15590. [DOI] [PubMed] [Google Scholar]
- 55.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al.. (1999) Current protocols in molecular biology. New York, NY: Wiley-Inter-science.
- 56. Gomes RA, Vicente Miranda H, Silva MS, Graca G, Coelho AV, et al. (2006) Yeast protein glycation in vivo by methylglyoxal. Molecular modification of glycolytic enzymes and heat shock proteins. FEBS J 273: 5273–5287. [DOI] [PubMed] [Google Scholar]
- 57. Kramer ML, Schulz-Schaeffer WJ (2007) Presynaptic alpha-synuclein aggregates, not Lewy bodies, cause neurodegeneration in dementia with Lewy bodies. J Neurosci 27: 1405–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kramer ML, Behrens B, Schulz-Schaeffer WJ (2008) Selective detection, quantification, and subcellular location of α-synuclein aggregates with a protein aggregate filtration assay. BioTechniques 44: 403–411. [DOI] [PubMed] [Google Scholar]
- 59. Gobom J, Nordhoff E, Mirgorodskaya E, Ekman R, Roepstorff P (1999) Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom 34: 105–116. [DOI] [PubMed] [Google Scholar]
- 60. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 61. Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]