Abstract
Background
Pyruvate-decarboxylase negative (Pdc-) strains of Saccharomyces cerevisiae combine the robustness and high glycolytic capacity of this yeast with the absence of alcoholic fermentation. This makes Pdc-S. cerevisiae an interesting platform for efficient conversion of glucose towards pyruvate-derived products without formation of ethanol as a by-product. However, Pdc- strains cannot grow on high glucose concentrations and require C2-compounds (ethanol or acetate) for growth under conditions with low glucose concentrations, which hitherto has limited application in industry.
Results
Genetic analysis of a Pdc- strain previously evolved to overcome these deficiencies revealed a 225bp in-frame internal deletion in MTH1, encoding a transcriptional regulator involved in glucose sensing. This internal deletion contains a phosphorylation site required for degradation, thereby hypothetically resulting in increased stability of the protein. Reverse engineering of this alternative MTH1 allele into a non-evolved Pdc- strain enabled growth on 20 g l-1 glucose and 0.3% (v/v) ethanol at a maximum specific growth rate (0.24 h-1) similar to that of the evolved Pdc- strain (0.23 h-1). Furthermore, the reverse engineered Pdc- strain grew on glucose as sole carbon source, albeit at a lower specific growth rate (0.10 h-1) than the evolved strain (0.20 h-1). The observation that overexpression of the wild-type MTH1 allele also restored growth of Pdc-S. cerevisiae on glucose is consistent with the hypothesis that the internal deletion results in decreased degradation of Mth1. Reduced degradation of Mth1 has been shown to result in deregulation of hexose transport. In Pdc- strains, reduced glucose uptake may prevent intracellular accumulation of pyruvate and/or redox problems, while release of glucose repression due to the MTH1 internal deletion may contribute to alleviation of the C2-compound auxotrophy.
Conclusions
In this study we have discovered and characterised a mutation in MTH1 enabling Pdc- strains to grow on glucose as the sole carbon source. This successful example of reverse engineering not only increases the understanding of the glucose tolerance of evolved Pdc-S. cerevisiae, but also allows introduction of this portable genetic element into various industrial yeast strains, thereby simplifying metabolic engineering strategies.
Keywords: Inverse metabolic engineering, Reverse metabolic engineering, Whole genome sequencing, Glucose tolerance, by-product reduction, MTH1 allele
Background
Saccharomyces cerevisiae continues to prove its potential as an excellent microbial production platform of many bulk chemicals [1-4]. While traditionally S. cerevisiae has mainly been used for its high speed and capacity to convert sugars into ethanol and CO2, presently its robustness and genetic accessibility are also much appreciated in many metabolic engineering efforts for production of bio-based fuels [5-7] and chemicals [8-11]. In fact, in several industrial processes, including those centered around pyruvate-derived products such as malate [12,13] or lactate [14-16], ethanol is now considered an undesired by-product.
Even under fully aerobic conditions, S. cerevisiae converts part of its sugar substrate to ethanol when confronted with high sugar concentrations [17]. Conversion of glucose to ethanol yields much less ATP than complete conversion to CO2 and H2O via respiratory dissimilation, which is a drawback in ATP-requiring production processes [18]. The strong tendency of S. cerevisiae towards alcoholic fermentation is thought to have evolved as a mechanism to outcompete other organisms by the resulting fast glucose uptake and build-up of growth-inhibiting ethanol concentrations [19,20]. Although beneficial in natural environments, in many applied contexts this phenomenon lowers product yields. Therefore, several metabolic engineering studies have sought to disrupt aerobic fermentation of sugars by S. cerevisiae[21-26].
A powerful approach to prevent alcoholic fermentation in S. cerevisiae is elimination of pyruvate decarboxylase, which catalyzes the first step in the conversion of pyruvate to ethanol. S. cerevisiae strains in which all three structural genes encoding pyruvate decarboxylase (PDC1PDC5 and PDC6[27]) were deleted, did not produce ethanol, but were unable to grow in the presence of high glucose concentrations and, when grown in glucose-limited cultures, required the addition of ethanol or acetate to growth media, due to their inability to synthesize cytosolic acetyl-CoA from pyruvate [23-25]. To overcome these deficiencies, a Pdc- yeast was selected for growth on glucose as the sole carbon source in an evolutionary engineering experiment [25]. First, C2-carbon source prototrophic mutants were selected by prolonged cultivation in glucose-limited chemostat cultures, in which the acetate concentration in the medium gradually decreased to zero. Subsequently, a mutant able to grow at high glucose concentrations was selected by cultivation in serial shake flask cultures. The resulting evolved mutant could grow at a growth rate of 0.20 h-1 on synthetic medium with glucose as the sole carbon source and proved to be an efficient pyruvate producer [25].
Elucidation of the genetic background of glucose tolerance in Pdc-S. cerevisiae is not only of fundamental interest, but is also required to enable its fast introduction in metabolic engineering strategies. The process of elucidating and subsequent reconstruction of a desired phenotypic trait is known as reverse metabolic engineering [28,29]. Reverse engineering of phenotypes obtained by laboratory evolution has the added benefit that potential detrimental effects of random mutations obtained during evolution can be eliminated. Identification of relevant mutations is an essential step in reverse metabolic engineering. Transcriptional profiling of the evolved Pdc- mutant during growth in nitrogen-limited chemostat cultures revealed the altered expression of many hexose transporters (Hxt) in this evolved strain compared to a wild type strain [25]. It was found that the summed transcript abundance of all HXT genes represented on the arrays (HXT1 to HXT10HXT12HXT14, and HXT16) was four-fold lower in the evolved Pdc- strain than in a Pdc+ reference strain [25].
Transcription of HXT genes in S. cerevisiae is predominantly regulated via the transcriptional regulator Rgt1 [30-33], which also regulates MIG2 and STD1 expression [34-36]. MIG2 and STD1 are both down-regulated in the evolved Pdc- strain [25]. Rgt1 is regulated by the concerted action of the glucose sensors Rgt2 and Snf3, which relay the extracellular glucose signal via the paralogous repressors Mth1 and Std1 to Rgt1 [33,36-40]. In the absence of extracellular glucose, Mth1 and Std1 are in a complex with Rgt1, Ssn6 and Tup1 resulting in the transcriptional repression or activation of Rgt1 targets [41-43]. In the presence of glucose, the conformation of the glucose sensors Rgt2 and Snf3 is thought to change, which facilitates the phosphorylation of Mth1 and Std1 by Yck1 [38]. When phosphorylated, Mth1 and Std1 are targeted for degradation [38]. The absence of Mth1 or Std1 enables phosphorylation of Rgt1 [30,42,44], which is subsequently released from the promoters of, amongst others, the Hxt transporters [30-33]. The altered transcript profiles of HXT genes in the evolved, glucose-tolerant Pdc-S. cerevisiae strain might therefore be explained by mutations in this regulatory network. For a comprehensive review and graphical representation of the regulation of the HXT transporters see Gancedo et al. 2008 [36].
The goal of the present study was to identify the mutation(s) responsible for the ability of the evolved Pdc- strain isolated by Van Maris et al. (2004) to grow on high concentrations of glucose as sole carbon source. Our results identified a mutation in MTH1, whose impact on growth on glucose in the absence of added C2-compounds was investigated after reintroduction in an ancestral Pdc-S. cerevisiae.
Results
An evolved Pdc-S. cerevisiae strain has an internal deletion within MTH1
To investigate the genetic basis of the ability of the evolved Pdc-S. cerevisiae strain (TAM) to grow at high glucose concentrations, the strain was crossed with a pdc1 pdc5 strain of the opposite mating type (pdc1 pdc5 strains are unable to grow on glucose despite the presence of the weakly expressed PDC6 gene [23]). The resulting diploid was able to grow on 20 g l-1 glucose, indicating that the glucose tolerance of the TAM strain is a dominant trait. Upon sporulation of this diploid and dissection on YP medium supplemented with 2% ethanol (v/v), only one of 23 tetrads yielded four viable spores, twelve yielded three, six yielded two, and four yielded one. Fifty-two spores from the nineteen tetrads with more than one viable spore were tested for growth on YPD, and it was found that 28 were strongly glucose-tolerant, six were weakly so, and eighteen were glucose-sensitive. These results, coupled with the observation that no tetrad yielded more than two glucose sensitive segregants, strongly suggested that the trait is monogenic. PCR analysis of the segregants showed that PDC6 segregated independently from the glucose tolerant phenotype.
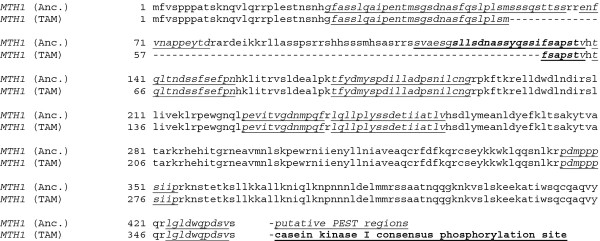
Several alleles of MTH1, which encodes a transcriptional regulator involved in glucose sensing, are known to dominantly suppress the glucose sensitivity of several other glucose-intolerant mutants [32,45-49]. A hypothesis to explain the glucose tolerance of the evolved Pdc- strain TAM could be that it is caused by a mutation in the MTH1 gene. This would be consistent with the previously observed transcriptional changes of HXT genes in nitrogen-limited, glucose-grown chemostat cultures [25]. Sequencing of the 1302 basepairs of the MTH1 ORF from the evolved Pdc- strain revealed a 225 bp internal deletion spanning from position 169 to 393 of MTH1 (Figure 1). This new allele was named MTH1-ΔT. The deletion found in MTH1-ΔT does not disrupt the reading frame but it affects two important characteristics of the protein. Firstly, it eliminates a sequence phosphorylated by the yeast casein kinase Yck1, required for degradation of Mth1 [38] (Figure 1). Secondly, it removes amino acid stretches rich in serine and other amino acids which may form PEST regions that are associated with proteins that have short half-lives [50].
Figure 1 .
Alignment of the Mth1 amino acid sequences from TAM and RWB837. TAM is a Pdc- strain evolved for growth on glucose and RWB837 is the ancestral Pdc- strain from which the TAM strain was derived. The casein kinase I consensus phosphorylation site is underlined and bold. The deleted region is rich in amino acid stretches in serine and other amino acids which may form PEST regions that are associated with proteins that have short half-lives [50,53].
Introduction of MTH1-ΔT in the ancestral Pdc- strain restores growth on glucose
If the mutation in MTH1 found in the evolved Pdc- strain is responsible for its glucose tolerant phenotype, substitution of the chromosomal wild-type allele of MTH1 in a non-evolved Pdc- strain should render it glucose tolerant. This was experimentally tested by integration of a cassette containing MTH1-ΔT in the MTH1 locus of RWB837 and subsequent selection of uracil-auxotrophic revertants with only the MTH1-ΔT allele. After confirmation of correct integration, this yielded strain IMI076 (Pdc-MTH1-ΔTura3). Since quantitative growth studies are best performed using prototrophic strains [51], the URA3 gene was subsequently repaired, resulting in strains IMI078 (Pdc-MTH1-ΔT), IMI082 (Pdc-MTH1) and IMI083 (evolved Pdc-).
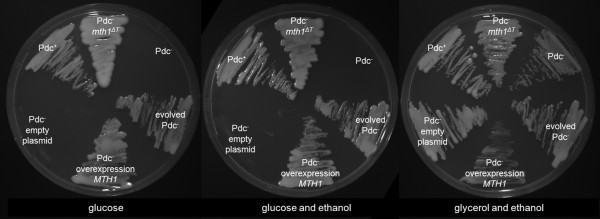
As expected for Pdc-S. cerevisiae, all strains grew on the positive-control plates containing glycerol and ethanol as the carbon source (Figure 2). As previously reported, the parental Pdc- strain (IMI082 (Pdc-MTH1)) did not grow in the presence of 20 g l-1 glucose, whereas the evolved Pdc- strain (IMI083 (evolved Pdc-)) did grow. The unevolved Pdc- strain carrying the MTH1-ΔT allele (IMI078 (Pdc-MTH1-ΔT)), grew equally well as the evolved Pdc- strain on plates with 20 g l-1 glucose supplemented with 0.3% (v/v) ethanol as C2-source, thereby demonstrating that the internal deletion in MTH1 is sufficient to confer glucose tolerance to Pdc-S. cerevisiae.
Figure 2 .
Growth ofS. cerevisiaestrains with differentMTH1alleles on synthetic medium agarose plates with 20 g l-1glucose as the sole carbon source (left plate), 20 g l-1glucose supplemented with 0.3% (v/v) ethanol (middle plate) or 2% (v/v) glycerol and 0.3% (v/v) ethanol (right plate). The strains used were: IMI078 (Pdc-MTH1-ΔT), IMI082 (Pdc-), IMI083 (evolved Pdc-), IMZ104 (Pdc- overexpression MTH1), IMZ103 (Pdc- empty plasmid), CEN.PK113-7D (Pdc+ reference). Plates were incubated at least 3 days at 30°C.
If the deletion of the phosphorylation site in MTH1-ΔT resulted in decreased degradation of the Mth1 protein and thereby in an increased abundance of Mth1 in the cell, direct overexpression of the native MTH1 might also confer glucose tolerance to a Pdc- strain. To challenge this hypothesis, the native MTH1 gene was expressed from the strong PGK1 promoter on a multicopy plasmid in the ancestral Pdc- strain RWB837 (yielding strain IMZ104). The transformed strain grew on agarose plates with 20 g l-1 glucose and 0.3% (v/v) ethanol, whereas the empty plasmid control (strain IMZ103) did not grow (Figure 2).
Characterization of evolved Pdc- and reverse engineered MTH1-ΔT Pdc-S. cerevisiae in bioreactors
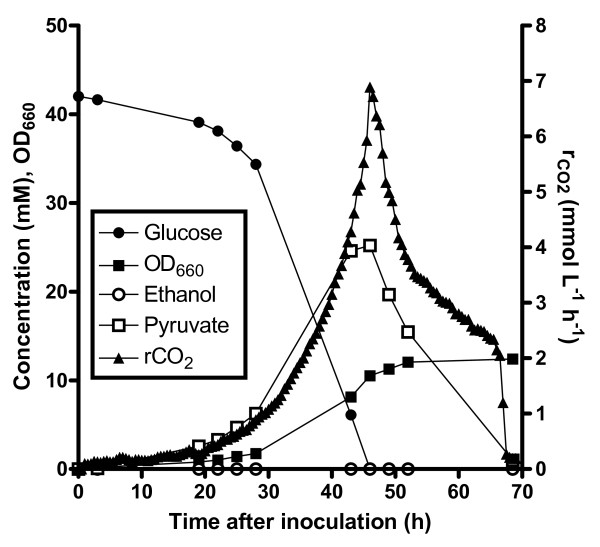
Although the characterization on plates provided a qualitative demonstration that the MTH1-ΔT allele confers glucose tolerance to Pdc- strains, quantitative analysis of growth and product formation required cultivation under controlled conditions. Therefore, strains IMI078 (Pdc-MTH1-ΔT) and IMI083 (evolved Pdc-) were grown in aerobic, pH-controlled bioreactors on synthetic medium supplemented with 7.5 g l-1 glucose and 0.3% (v/v) ethanol (Figure 3). Under these conditions, the specific growth rates of strain IMI078 (Pdc-MTH1-ΔT; 0.24 ± 0.00 h-1) and strain IMI083 (evolved Pdc-; 0.23 ± 0.00 h-1) were virtually the same. The observation that reverse engineering of MTH1-ΔT resulted in near-identical specific growth rates in glucose-ethanol grown batch cultures as observed with the evolved Pdc- strain, is consistent with the observation that the glucose tolerance was monogenic.
Figure 3 .
Growth profile of IMI078 (A; Pdc-MTH1-ΔT) and IMI083 (B; evolved Pdc-) on high concentrations of glucose. Cultivation in an aerated pH-controlled (pH 5) bioreactor with 7.5 g l-1 glucose and 0.3% (v/v) ethanol. The results are from one representative experiment. Duplicate experiments deviated <5% in titers, CO2 production and OD660.
During the first growth phase, in which ethanol and glucose were simultaneously consumed, the yield of pyruvate on substrate was higher in the evolved strain (0.30 ± 0.04 gpyr gglc+etoh-1) than in the reverse engineered strain IMI078 (Pdc-MTH1-ΔT; 0.07 ± 0.03 gpyr gglc+etoh-1) (p-value = 0.02; student’s t-test; n = 2). Apparently, there is/are additional mutation(s) that affect the extracellular accumulation of pyruvate. Both Pdc- strains showed a decrease of the rate of CO2 production when the ethanol added to the medium was depleted, which was caused by a decrease in the specific glucose consumption rate. Whilst the remaining glucose was consumed, the biomass concentration increased and as a consequence the volumetric CO2 production rate also increased again. After all the glucose was consumed, the volumetric CO2 production rate rapidly decreased while pyruvate, the main metabolite produced during the glucose consumption phase, was consumed (Figure 3).
In both strains, the optical density of the cultures increased by ca. 50 % after ethanol had been depleted. This result was expected for the evolved strain IMI083, which was specifically selected for its ability to grow on glucose in the absence of externally added C2-sources [25]. The biomass formation of strain IMI078 (Pdc-MTH1-ΔT) in this growth phase could either indicate a redistribution of lipids and lysine over newly synthesized cells or indicate that, in addition to increasing the glucose tolerance of Pdc-S. cerevisiae, presence of the MTH1-ΔT allele had an additional impact on the C2-compound auxotrophy of Pdc- strains. This observation is consistent with growth on plates with glucose as the sole carbon source.
Introduction of the MTH1-ΔT allele partially alleviates the C2-compound auxotrophy of Pdc-S. cerevisiae
In the experiments described above, ethanol was included in the growth media to meet the requirement of Pdc- strains that has been documented before and which has been attributed to a key role of pyruvate decarboxylase in the synthesis of cytosolic acetyl-coenzyme A [23-25]. Plate growth experiments indicated that both the Pdc-MTH1-ΔT strain and a Pdc- strain overexpressing the wild-type MTH1 gene grew on 20 g l-1 glucose without addition of ethanol as external C2 source (Figure 2). Although pure agarose was used in the plate experiments, a contamination with C2 compounds could not be entirely excluded and a further analysis was performed in aerobic, pH-controlled bioreactors on 7.5 g l-1 glucose without ethanol. This experiment confirmed that introduction of the MTH1-ΔT mutation was sufficient to enable growth in batch cultures on glucose as sole carbon source (Figure 4). In four replicate experiments a reproducible specific growth rate of 0.097 ± 0.007 h-1 was observed. This specific growth rate is lower than that of the evolved Pdc- strain under these conditions (0.20 h-1) [25], suggesting that this strain may harbor additional mutations which could also contribute to the growth on glucose in the absence of added C2-compounds.
Figure 4 .
Growth profile of IMI078 (Pdc-MTH1-ΔT) on glucose as the sole carbon source. Cultivation in an aerated pH-controlled (pH 5) bioreactor growing on 7.5 g l-1 glucose without external C2-sources. The depicted results are from one experiment of a set of four replicates, which all had identical specific growth rates (0.097 ± 0.007 h-1). The lag phase (rCO2 > 1 mmol l-1 h-1) of the replicates varied between 20 and 80 h after inoculation.
Discussion
In this study the molecular basis of the ability of a S. cerevisiae strain evolved from a Pdc- strain to grow on glucose was investigated. An internal deletion in the MTH1 gene was identified in an evolved Pdc- strain able to grow at high glucose concentrations. This internal deletion was introduced into a non-evolved Pdc- strain by chromosomal integration of the identified mutation. The observation that both the evolved strain and the newly created strain have the same ability to grow on glucose supplemented with ethanol, supports the conclusion that the internal deletion within MTH1 is solely responsible for the glucose positive growth in the evolved strain. The previously evolved Pdc- strain grew without a C2 source with a specific growth rate of 0.20 h-1[25]. In this study we show that introduction of the MTH1-ΔT allele in the ancestral strain results in a consistent specific growth rate of 0.097 ± 0.007 h-1 in aerobic batch cultures on glucose as the sole carbon source. The mechanism(s) underlying the different levels of C2 independence between the evolved and MTH1-ΔT Pdc- strains remains unknown. Unfortunately, the original transcriptome analysis on the evolved C2-source independent Pdc- mutant did not provide additional clues to the mechanism [25].
MTH1 encodes a protein that plays a critical role in the transcriptional regulation of glucose transporters in S. cerevisiae (for a review see [36]). Independent screens to isolate suppressors of glucose toxicity in other glycolytic mutants, such as tpi1[48,49,52], pyc1 pyc2[45], tps1[47], pgk1[46] or pgm1 mutants [46,47] have uncovered different mutant alleles of MTH1, indicating its important role in glucose metabolism. A common trait of these suppressor mutants is a low glucose influx [32,45-49] that results in decreased carbon catabolite repression [32,46-49] and consequently in an increased respiration [46]. The MTH1 alleles in these other studies carried mutations in either codon 85 or in both codon 85 and 102 [47]. These mutations are covered by the 225bp deletion spanning codons 57-131 found in the MTH1-ΔT allele. Two important features of Mth1 are altered in the protein resulting from the internal deletion: the alteration of a region with putative PEST sequences and the elimination of a phosphorylation site which spans from codon 118-137 [38]. PEST sequences are usually present in proteins with a short intracellular half-life [50,53]. Phosphorylation of Mth1 is also related with its degradation since only after Mth1 has been phosphorylated by Yck1 it is ubiquitinated by the SCFgrr complex and subsequently degraded by the proteasome [38]. Mth1 interacts with Rgt1 to repress transcription of glucose transporters and only its degradation allows their transcription. Decreased degradation of Mth1 is known to result in decreased mRNA levels for the HXT1 gene [38]. The internal deletion of MTH1-ΔT shall therefore interfere with Mth1 degradation and cause a lowered expression of hexose transporters. Indeed it was found that the evolved strain presented a lowered expression of several glucose transporters [25].
The observed increase of glucose tolerance in Pdc-S. cerevisiae upon expression of a mutated allele of MTH1 does not, in itself, explain the mechanism by which Pdc- strains become glucose sensitive. Pdc- strains are pyruvate hyperproducers and, in contrast to ethanol, pyruvate is unlikely to be exported by passive diffusion through the yeast plasma membrane. If it is assumed that the MTH1 mutation mainly acts by restricting glucose transport and, therefore, glycolytic flux, it might prevent intracellular accumulation of pyruvate to toxic levels. A possible alternative explanation is related to redox metabolism. The major function of alcoholic fermentation in wild-type S. cerevisiae is the fast reoxidation of NADH formed in glycolysis. Unrestricted glycolytic activity in a Pdc- strain, in combination with a limited capacity of the mitochondrial respiratory chain for reoxidation of cytosolic NADH, might lead to a reduction of the cytosolic NADH/NAD+ pool and thereby inhibit key reactions in biosynthesis.
Conclusions
Pyruvate decarboxylase negative (Pdc-) S. cerevisiae strains are attractive metabolic engineering platforms for pyruvate-derived products, but their application was hindered by the inability to grow on high glucose concentrations and a C2 auxotrophy. In this study, overexpression of MTH1 or introduction of the newly discovered allele MTH1-ΔT into a Pdc-S. cerevisiae strain enabled growth on glucose as the sole carbon source at industrially relevant growth rates. Furthermore, introduction of this modification partially relieves the C2 auxotrophy of Pdc- yeasts. Therefore, S. cerevisiae with a disruption of the pyruvate decarboxylase genes together with expression of a more stable MTH1 allele further increases the flexibility of S. cerevisiae as a platform micro-organism for the production of bio-based chemicals and fuels.
Methods
Strains and maintenance
Strains constructed in the present study (see Table 1) were derived from S. cerevisiae RWB837 [25], which contains targeted deletions of the three pyruvate decarboxylase genes PDC1PDC5 and PDC6 as well as a defective URA3 allele. RWB837 was constructed in the CEN.PK background [54,55]. Strains were maintained on YP medium (demineralized water; 10 g l-1 yeast extract [BD Difco, Franklin Lakes, NJ, USA]; 20 g l-1 peptone [BD Difco]) with 2% (v/v) glycerol and 3% (v/v) ethanol. Culture stocks were prepared from shake flask cultures, which were incubated at 30°C and stirred at 200 rpm, by the addition of 20% (v/v) glycerol and were stored at -80°C.
Table 1.
Strains used in this study
| Strain | Description and Genotype | Source |
|---|---|---|
| CEN.PK113-7D |
MATaLEU2 URA3 MAL2-8C |
P. Kötter, Germany |
| RWB837 |
MATapdc1Δ(-6,-2)::loxP pdc5Δ(-6,-2)::loxP pdc6Δ(-6,-2)::loxP ura3-52 |
[25] |
| TAM |
MATapdc1Δ (-6,-2)::loxP pdc5Δ (-6,-2)::loxP pdc6Δ (-6,-2)::loxP ura3-52, selected for C2 independence in glucose-limited chemostat cultures and glucose-tolerant growth in batch culture |
[25] |
| MY2243 |
MATα ura3-52 his3Δ1 trp1-289 pdc1Δ(-6,-2)::loxP pdc5Δ(-6,-2)::loxP |
Microbia Inc, U.S.A. |
| MY2280 |
MATa/MATα pdc1Δ/pdc1Δ pdc5Δ/pdc5Δ PDC6/pdc6Δ ura3-52/ura3-52 MTH1/MTH1-ΔT |
This study |
| IMI073 |
MATapdc1Δ(-6,-2)::loxP pdc5Δ(-6,-2)::loxP pdc6Δ(-6,-2)::loxP ura3-52 MTH1-ΔT::pUD143 (URA3)::MTH1 |
This study |
| IMI076 |
IMI073 ura3-52 MTH1-ΔT |
This study |
| IMI078 |
IMI076 URA3 MTH1-ΔT |
This study |
| IMI082 |
RWB837 URA3 |
This study |
| IMI083 |
TAM URA3 |
This study |
| IMZ103 |
RWB837 pvv214 |
This study |
| IMZ104 | RWB837 pEXp214-MTH1.2 | This study |
Segregation analysis and sequencing
The evolved S. cerevisiae Pdc- strain TAM [25] was crossed with strain MY2243 by incubating a mix of equal numbers of cells together on a YPD plate at 30°C for several hours, and selecting for robust growth on YNB plus 2% glycerol (v/v), 200 μM uracil and 2 g l-1 casamino acids. These conditions, under which TAM grows poorly and the other parent not at all, were used to select the diploid MY2280. Sporulation was performed by incubating a culture of MY2243 on an agar plate with 20 g l-1 potassium acetate and 50 μM uracil for 3-4 days at 23°C. Spores were segregated on YP agar plates supplemented with 2% ethanol (v/v) using a standard micromanipulator, incubated at 30°C and subsequently tested for growth on YP 2% agar (w/v) medium with 20 g l-1 glucose.
Oligonucleotides used in this study are shown in Table 2. The MTH1 gene was amplified by PCR from genomic DNA of strains TAM and RWB837 using oligonucleotides MTH1fw and MTHrv. The PCR products were sequenced by Baseclear BV (Baseclear, Leiden, The Netherlands). The mutated MTH1 allele of TAM was named MTH1-ΔT. Putative PEST regions within the Open Reading Frame of MTH1 were identified by using the online tool ePESTfind (http://emboss.bioinformatics.nl/cgi-bin/emboss/epestfind).
Table 2.
Oligonucleotides used in this study
| name | confirmation of strain | sequence (5'- > 3') |
|---|---|---|
|
primers for cloning |
|
|
| MTH1BamHI |
GCGATCGCGGATCCTTGAGGAGGTAGGGAACATC |
|
| MTH1HindIII |
CTGACGCCAAGCTTAAACGGCGACTGGTGGTAAG |
|
| URA3fw |
|
GCTGCTACTCATCCTAGTCC |
| URA3rv |
|
CTTTAGCGGCTTAACTGTGC |
| MTH1fw |
|
CACCATGTTTGTTTCACCACCACCAGCAACTTCG |
| MTH1rv |
|
TCAGGATACTGAATCCGGCTGCCAATCCA |
|
primers for diagnostic PCR | ||
| MTH1fw |
IMI073, IMI076 |
CACCATGTTTGTTTCACCACCACCAGCAACTTCG |
| MTH1rv |
IMI073, IMI076 |
TCAGGATACTGAATCCGGCTGCCAATCCA |
| m13fw |
IMI073, IMI076, IMZ103, IMZ104 |
GTAAAACGACGGCCAG |
| m132rv |
IMI073, IMI076, IMZ103, IMZ104 |
GGAAACAGCTATGACCATG |
| URA3-A |
IMI078, IMI082, IMI083 |
TGCATGAGTCTCAGCTCTAC |
| URA3-B | IMI078, IMI082, IMI083 | CCAAGGGTAGAGATCCTAAG |
Plasmid and strain construction
Chromosomal introduction of the wild type MTH1 allele in strain RWB837 was done by integrating plasmid pUD143 into the MTH1 locus after linearization of the plasmid pUD143 by EcoRI. Construction of the plasmid pUD143 was done by amplifying the MTH1-ΔT allele from the evolved Pdc- strain (TAM) PCR using primers MTH1bamHI and MTH1hindIII (Table 2) and cloning the fragment into plasmid pRS406 [56] by ligating HindIII and BamHI treated plasmids and inserts using T4 DNA ligase (Roche, Basel, Switzerland) to produce plasmid pUD143 (Table 3). HindIII and BamHI restriction sites were added to MTH1-ΔT allele by amplifying the gene from TAM genomic DNA with the primers MTH1BamHI and MTH1HindIII (Table 2) using Phusion Hot Start High Fidelity Polymerase (Finnzymes). After recombination of the plasmid pUD143 into the chromosome, which generated two copies of MTH1, chromosomal replacement of the wild type MTH1 allele by the MTH1-ΔT allele was done using the pop-in/pop-out method [57]. A uracil auxotrophic transformant was selected on synthetic medium with trace elements and vitamins as described before [58] containing 3% (v/v) ethanol, 2% (v/v) glycerol, uracil and 5-FOA, which was named IMI076. Correct replacement of MTH1 was confirmed by PCR using primers MTH1fw, MTH1rv, m13fw and m132rv. To obtain prototrophic strains, IMI076, RWB837 and TAM were transformed with URA3 obtained by PCR from pRS406 [56], using Phusion Hot Start High Fidelity Polymerase (Finnzymes, Espoo, Finland). The resulting strains were named respectively IMI078, IMI082 and IMI083.
Table 3.
Plasmids used in this study
| Plasmid | Characteristic | Reference |
|---|---|---|
| pENTR/D-TOPO |
Gateway entry clone |
Invitrogen, USA |
| pvv214 |
2μ ori, URA3, PPGK1-ccdB-TCYC1 |
[59] |
| pENTR-D-TOPO-MTH1 |
Gateway entry clone, MTH1 |
This work |
| pEXp214-MTH1.2 |
2μ ori, URA3, PPGK1- MTH1-ΔT-TCYC1 |
This work |
| pRS406 |
Integration plasmid, URA3 |
[56] |
| pUD143 | Integration plasmid, URA3, PMTH1- MTH1-ΔT-TMTH1 | This work |
Construction of plasmid pEXp214-MTH1.2 was achieved by amplification of MTH1 from genomic DNA of CEN.PK113-7D with primers MTH1fw and MTH1rv using Phusion Hot Start High Fidelity Polymerase (Finnzymes) and subsequent cloning in pVV214 using Gateway Technology [59]. RWB837 was transformed with either pEXp214-MTH1.2 or the empty vector pVV214 resulting in strain IMZ104 and IMZ103 respectively.
Transformation and amplification of plasmids was done in E. coli One Shot TOP10 competent cells (Invitrogen, Grand Island, NY, USA) according to manufacturer's instructions. Plasmids were isolated from E. coli with the Sigma GenElute Plasmid Miniprep Kit (Sigma, St. Louis, USA).
Transformation of plasmids or linear DNA fragments in S. cerevisiae was done according to the lithium-based transformation protocol described by Gietz and Woods [60]. Transformants were selected on synthetic agar medium supplemented with uracil drop-out medium (Y1501, Sigma, St. Louis USA) containing 3% (v/v) ethanol and 2% (v/v) glycerol. Single colony isolates were confirmed to have the correct insert by PCR (Table 2) on colony material suspended in 0.02M NaOH and boiled for ten minutes. Plasmids were isolated from the yeast strains with Zymoprep yeast miniprep kit II (Zymo Research, Orange, CA, USA) and sequenced (Baseclear, Leiden, The Netherlands) for confirmation.
Cultivation procedures
Cultivations were performed at 30°C in synthetic medium with glucose, trace elements and vitamins as described before [58]. Ethanol (3 ml l-1) was added when relevant. Cultivation on solid media was performed on medium containing 20 g l-1 glucose and 20 g l-1 of agarose (Sigma). To minimize the chance of C2-contamination in the medium, agarose instead of agar was chosen for the solid growth media assays. Cultivation in bioreactors was performed in medium containing 7.5 g l-1 glucose supplemented with Antifoam Emulsion C (Sigma), which has been autoclaved separately (120°C) as a 20% (w/v) solution and added to a final concentration of 0.2 g l−1.
Batch cultivations were performed in 2-liter bioreactors (Applikon, Schiedam, Netherlands) at a working volume of 1 liter. The pH was controlled by automatic addition of 2M KOH or 2M H2SO4 at a value of 5.5. Bioreactors were sparged with 500 ml min-1 air and stirred at 800 rpm. For growth rate measurements, the initial optical density at 660 nm (OD660) after inoculation was 0.1. Maximum specific growth rates were determined from duplicate cultures (errors are given as mean deviations) and were based on the OD660. Preculture shake flasks with synthetic medium containing 3% (v/v) ethanol were inoculated with 1 ml aliquots of frozen stock cultures. Cells from exponentially growing shake-flask precultures were washed twice with demineralized water and used to inoculate batch cultures.
Determination of culture dry weight and optical density
Culture samples were filtered over preweighed nitrocellulose filters (pore size: 0.45 μm; Gelman Laboratory, Ann Arbor, MI). Culture dry weight was determined by weighing the filters after two washes with demineralized water and dried in a microwave oven (Bosch, Stuttgart, Germany) for 20 min at 350W. Duplicate determinations varied by less than 1%. Measurement of optical density was done at a wavelength of 660 nm in a Libra S11 spectrophotometer (Biochrom, Cambridge, UK).
Gas and metabolite analysis
Exhaust gas was cooled in a condenser (2°C) and dried with a Permapure type MD-110-48P-4 dryer (Permapure, Toms River, NJ). Oxygen and carbon dioxide concentrations were determined with an NGA 2000 analyzer (Rosemount Analytical, Orrville, OH, USA). In calculations of rates a correction was made for sample volumes.
Glucose, ethanol and pyruvate concentrations were determined in culture supernatants with a high-performance liquid chromatography (HPLC) on a Waters Alliance 2690 HPLC (Waters, Milford, MA) containing a Bio-Rad HPX 87H column (Bio-Rad, Hercules, CA). The HPLC was operated at 60°C with 5 mM H2SO4 as mobile phase at a flow rate of 0.6 ml min−1. Detection was by means of a Waters 2410 refractive-index detector and a Waters 2487 UV detector.
Competing interests
The authors declare no competing financial interests.
Authors’ contributions
BO designed and carried out the sequencing of MTH1, the strain constructions, the cultivation experiments, analyzed the results and drafted the manuscript. CLF, XZ, JT and CG performed the crossing studies, identified MTH1 as a candidate gene and revised the manuscript. JMD, JTP and AvM supervised the design, revised the manuscript and coordinated the study. All authors read and approved the final manuscript.
Contributor Information
Bart Oud, Email: B.Oud@tudelft.nl.
Carmen-Lisset Flores, Email: clflores@iib.uam.es.
Carlos Gancedo, Email: cgancedo@iib.uam.es.
Xiuying Zhang, Email: xzhang15@yahoo.com.
Joshua Trueheart, Email: joshua.trueheart@dsm.com.
Jean-Marc Daran, Email: J.G.Daran@tudelft.nl.
Jack T Pronk, Email: J.T.Pronk@tudelft.nl.
Antonius JA van Maris, Email: A.J.A.vanMaris@tudelft.nl.
Acknowledgements
The PhD project of BO was carried out within the research programme of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative / Netherlands Organization for Scientific Research. The work of CLF and CG was helped by grant BFU2010-19628-CO2-O2 from the Spanish Ministry of Science and Innovation (MICINN).
References
- Abbott DA, Zelle RM, Pronk JT, van Maris AJA. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res. 2009;9:1123–1136. doi: 10.1111/j.1567-1364.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- van Maris AJA, Abbott DA, Bellissimi E, van den Brink J, Kuyper M, Luttik MAH. et al. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Van Leeuwenhoek. 2006;90:391–418. doi: 10.1007/s10482-006-9085-7. [DOI] [PubMed] [Google Scholar]
- Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol R. 2008;72:379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KK, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci. 2012;69:2671–2690. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong B, Siewers V, Nielsen J. Systems biology of yeast: enabling technology for development of cell factories for production of advanced biofuels. Curr Opin Biotech. 2012;23:624–630. doi: 10.1016/j.copbio.2011.11.021. [DOI] [PubMed] [Google Scholar]
- van Maris AJA, Winkler A, Kuyper M, de Laat W, van Dijken J, Pronk JT. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv Biochem Eng Biot. 2007;108:179–204. doi: 10.1007/10_2007_057. [DOI] [PubMed] [Google Scholar]
- Weber C, Farwick A, Benisch F, Brat D, Dietz H, Subtil T. et al. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl Microbiol Biot. 2010;87:1303–1315. doi: 10.1007/s00253-010-2707-z. [DOI] [PubMed] [Google Scholar]
- Arsenault PR, Wobbe KK, Weathers PJ. Recent advances in artemisinin production through heterologous expression. Curr Med Chem. 2008;15:2886–2896. doi: 10.2174/092986708786242813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BE, Hart-Wells E, Matsuda SPT. Metabolic engineering to produce sesquiterpenes in yeast. Org Lett. 2003;5:1629–1632. doi: 10.1021/ol034231x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Halls C, Zhang J, Matsuno M, Zhang Y, Yu O. Stepwise increase of resveratrol biosynthesis in yeast Saccharomyces cerevisiae by metabolic engineering. Metab Eng. 2011;13:455–463. doi: 10.1016/j.ymben.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R. et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA. 2012;109:111–118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelle RM, de Hulster E, Kloezen W, Pronk JT, van Maris AJA. Key process conditions for production of C(4) dicarboxylic acids in bioreactor batch cultures of an engineered Saccharomyces cerevisiae strain. Appl Environ Microb. 2010;76:744–750. doi: 10.1128/AEM.02396-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelle RM, Harrison JC, Pronk JT, van Maris AJA. Anaplerotic role for cytosolic malic enzyme in engineered Saccharomyces cerevisiae strains. Appl Environ Microb. 2011;77:732–738. doi: 10.1128/AEM.02132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok S, Nijkamp JF, Oud B, Roque FC, de Ridder D, Daran JM. et al. Laboratory evolution of new lactate transporter genes in a jen1Δ mutant of Saccharomyces cerevisiae and their identification as ADY2 alleles by whole-genome resequencing and transcriptome analysis. FEMS Yeast Res. 2012;12:359–374. doi: 10.1111/j.1567-1364.2011.00787.x. [DOI] [PubMed] [Google Scholar]
- van Maris AJA, Winkler AA, Porro D, van Dijken JP, Pronk JT. Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy-dependent lactate export. Appl Environ Microb. 2004;70:2898–2905. doi: 10.1128/AEM.70.5.2898-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maris AJA, Konings WN, van Dijken JP, Pronk JT. Microbial export of lactic and 3-hydroxypropanoic acid: implications for industrial fermentation processes. Metab Eng. 2004;6:245–255. doi: 10.1016/j.ymben.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Pronk JT, Steensma HY, Van Dijken JP. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(SICI)1097-0061(199612)12:16<1607::AID-YEA70>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Gaucher E, Burgan MF, De Kee DW, Li T, Aris JP. et al. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Gen. 2005;37:630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Rozpêdowska E, Hellborg L, Ishchuk OP, Orhan F, Galafassi S, Merico A. et al. Parallel evolution of the make-accumulate-consume strategy in Saccharomyces and Dekkera yeasts. Nat Commun. 2011;2:302. doi: 10.1038/ncomms1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom J, De Mattos MJ, Grivell L. Redirection of the respiro-fermentative flux distribution in Saccharomyces cerevisiae by overexpression of the transcription factor Hap4p. Appl Environ Microb. 2000;66:1970–1973. doi: 10.1128/AEM.66.5.1970-1973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderich J, Raamsdonk LML, Kuiper A, Kruckeberg AL, Berden J, Teixeira de Mattos MJJ. et al. Effects of a hexokinase II deletion on the dynamics of glycolysis in continuous cultures of Saccharomyces cerevisiae. FEMS Yeast Res. 2002;2:165–172. doi: 10.1111/j.1567-1364.2002.tb00081.x. [DOI] [PubMed] [Google Scholar]
- Flikweert MT, Van Der Zanden L, Janssen WM, Steensma HY, Van Dijken JP, Pronk JT. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast. 1996;12:247–257. doi: 10.1002/(SICI)1097-0061(19960315)12:3<247::AID-YEA911>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Flikweert MT, de Swaaf M, Van Dijken JP, Pronk JT. Growth requirements of pyruvate-decarboxylase-negative Saccharomyces cerevisiae. FEMS Microbiol Lett. 1999;174:73–79. doi: 10.1111/j.1574-6968.1999.tb13551.x. [DOI] [PubMed] [Google Scholar]
- van Maris AJA, Geertman JM, Vermeulen A, Groothuizen MK, Winkler AA, Piper MDW. et al. Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl Environ Microb. 2004;70:159–166. doi: 10.1128/AEM.70.1.159-166.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CJ, Rasmussen JJ, Rønnow, Olsson L, Nielsen J. Investigation of the impact of MIG1 and MIG2 on the physiology of Saccharomyces cerevisiae. J Biotechnol. 1999;68:197–212. doi: 10.1016/S0168-1656(98)00205-3. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol. 1991;173:7963–7969. doi: 10.1128/jb.173.24.7963-7969.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JE, Sburlati A, Hatzimanikatis V, Lee K, Renner WA, Tsai PS. Inverse metabolic engineering: a strategy for directed genetic engineering of useful phenotypes. Biotechnol Bioeng. 1996;52:109–121. doi: 10.1002/(SICI)1097-0290(19961005)52:1<109::AID-BIT11>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Oud B, van Maris AJA, Daran JM, Pronk JT. Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast. FEMS Yeast Res. 2012;12:183–196. doi: 10.1111/j.1567-1364.2011.00776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Polish J, Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH. DNA-binding properties of the yeast Rgt1 repressor. Biochimie. 2009;91:300–303. doi: 10.1016/j.biochi.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniak A, Xue Z, Macool D, Kim JH, Johnston M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:221–231. doi: 10.1128/EC.3.1.221-231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttykrishnan S, Sabina J, Langton LL, Johnston M, Brent MR. A quantitative model of glucose signaling in yeast reveals an incoherent feed forward loop leading to a specific, transient pulse of transcription. Proc Natl Acad Sci USA. 2010;107:16743–16748. doi: 10.1073/pnas.0912483107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. The early steps of glucose signalling in yeast. FEMS Microbiol Rev. 2008;32:673–704. doi: 10.1111/j.1574-6976.2008.00117.x. [DOI] [PubMed] [Google Scholar]
- Flick KM, Spielewoy N, Kalashnikova TI, Guaderrama M, Zhu Q, Chang HC. et al. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol Biol Cell. 2003;14:3230–3241. doi: 10.1091/mbc.E03-03-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci USA. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabina J, Johnston M. Asymmetric signal transduction through paralogs that comprise a genetic switch for sugar sensing in Saccharomyces cerevisiae. J Biol Chem. 2009;284:29635–29643. doi: 10.1074/jbc.M109.032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan J, Mosley AL, Ozcan S. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr Genet. 2003;44:19–25. doi: 10.1007/s00294-003-0423-2. [DOI] [PubMed] [Google Scholar]
- Polish J, Kim JH, Johnston M. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics. 2005;169:583–594. doi: 10.1534/genetics.104.034512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás-Cobos L, Sanz P. Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem J. 2002;368:657–663. doi: 10.1042/BJ20020984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouandot D, Roy A, Kim JH. Functional dissection of the glucose signaling pathways that regulate the yeast glucose transporter gene (HXT) repressor Rgt1. J Cell Biochem. 2011;112:3268–3275. doi: 10.1002/jcb.23253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez M, Gamo FJ, Gancedo C. A mutation affecting carbon catabolite repression suppresses growth defects in pyruvate carboxylase mutants from Saccharomyces cerevisiae. FEBS Lett. 1995;377:197–200. doi: 10.1016/0014-5793(95)01337-7. [DOI] [PubMed] [Google Scholar]
- Gamo FJ, Lafuente MJ, Gancedo C. The mutation DGT1-1 decreases glucose transport and alleviates carbon catabolite repression in Saccharomyces cerevisiae. J Bacteriol. 1994;176:7423–7429. doi: 10.1128/jb.176.24.7423-7429.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente MJ, Gancedo C, Jauniaux JC, Gancedo JM. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol Microbiol. 2000;35:161–172. doi: 10.1046/j.1365-2958.2000.01688.x. [DOI] [PubMed] [Google Scholar]
- Schulte F, Ciriacy M. HTR1/MTH1 encodes a repressor for HXT genes. Yeast. 1995;11:S239. [Google Scholar]
- Schulte F, Wieczorke R, Hollenberg CP, Boles E. The HTR1 gene is a dominant negative mutant allele of MTH1 and blocks Snf3- and Rgt2-dependent glucose signaling in yeast. J Bacteriol. 2000;182:540–542. doi: 10.1128/JB.182.2.540-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Daran-Lapujade P, Daran JM, van Maris AJA, de Winde JH, Pronk JT. Chemostat-based micro-array analysis in baker's yeast. Adv Microb Physiol. 2009;54:257–311. doi: 10.1016/S0065-2911(08)00004-0. [DOI] [PubMed] [Google Scholar]
- Ozcan S, Freidel K, Leuker A, Ciriacy M. Glucose uptake and catabolite repression in dominant HTR1 mutants of Saccharomyces cerevisiae. J Bacteriol. 1993;175:5520–5528. doi: 10.1128/jb.175.17.5520-5528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C. et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Tech. 2000;26:706–714. doi: 10.1016/S0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- Entian KD, Kötter P. Yeast Genetic Strain and Plasmid Collections. Methods Microbiol. 2007;36:629–666. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA, Van Dijken JP. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- Van Mullem V, Wery M, De Bolle X, Vandenhaute J. Construction of a set of Saccharomyces cerevisiae vectors designed for recombinational cloning. Yeast. 2003;20:739–746. doi: 10.1002/yea.999. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]