Abstract
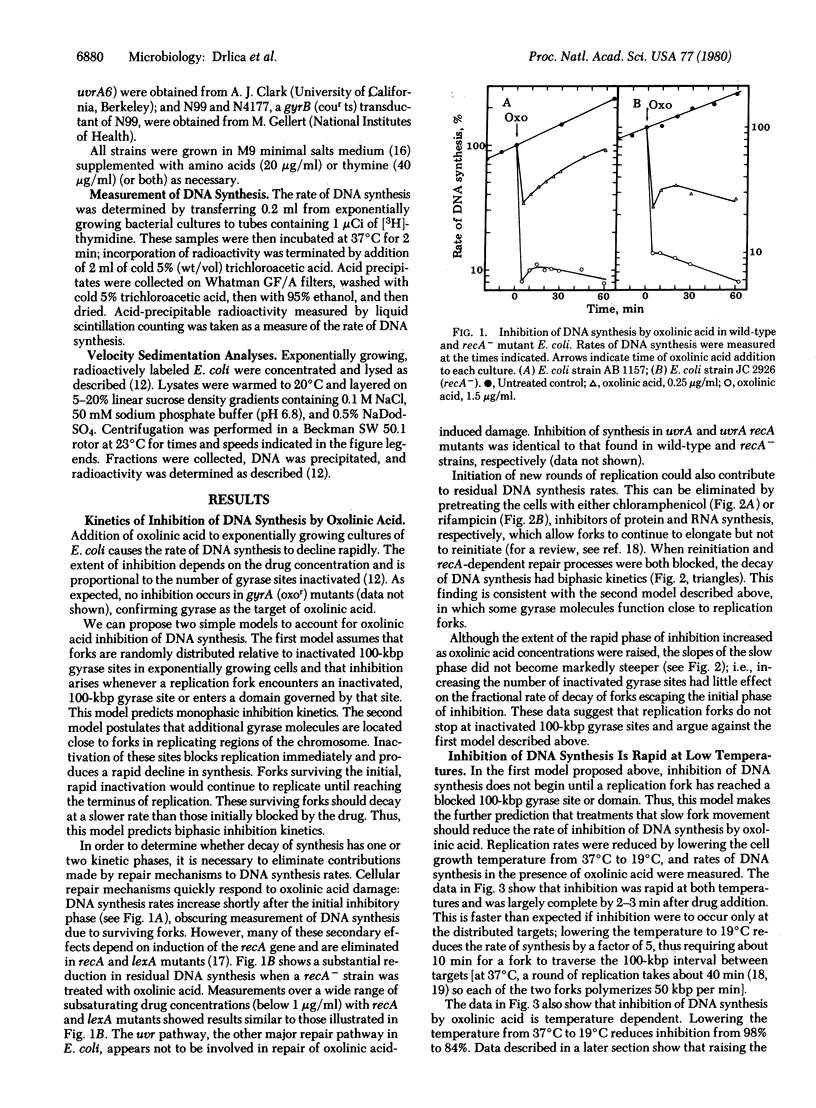
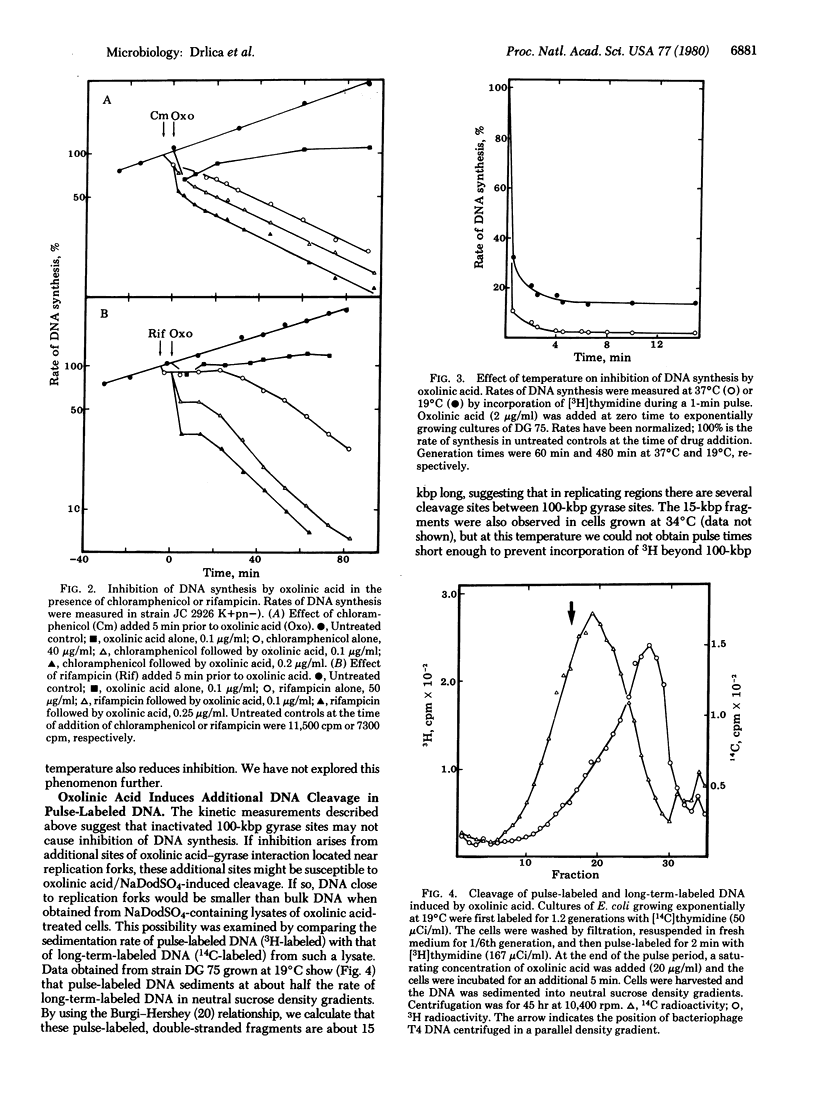
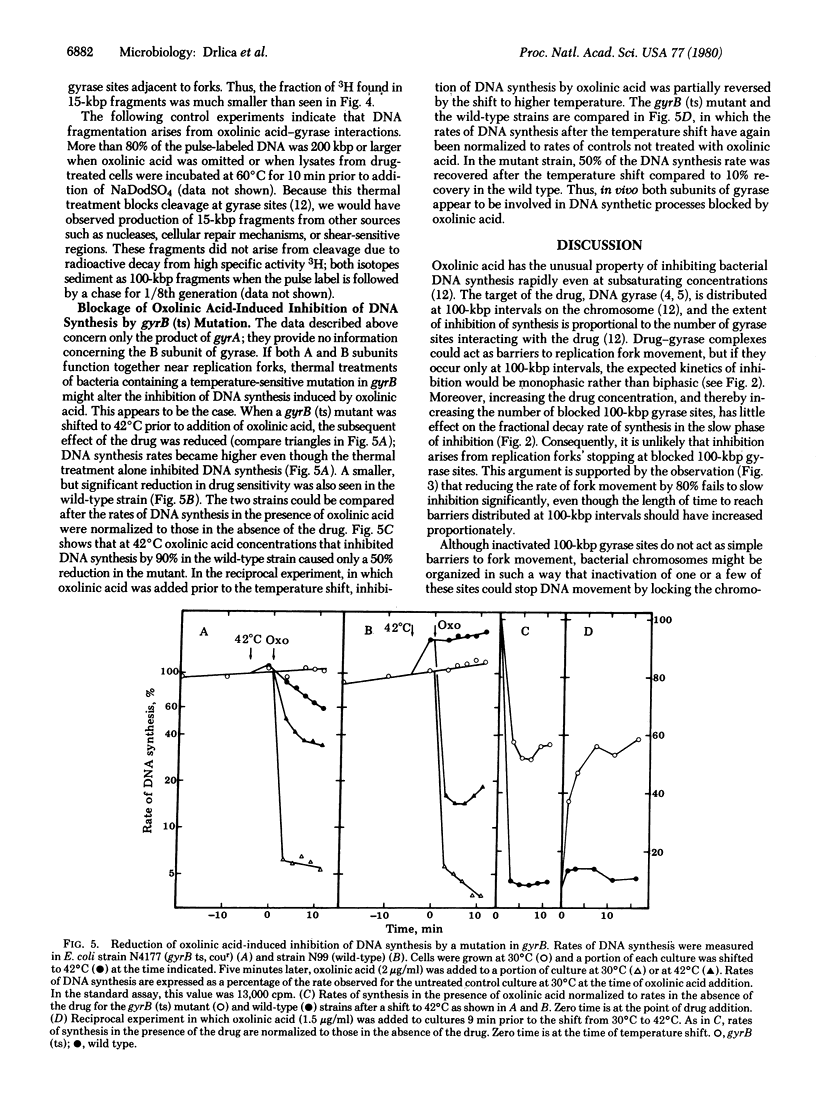
In previous studies we have shown that oxolinic acid, a specific inhibitor of the A subunit of DNA gyrase, induces DNA cleavage at 100,000-base-pair intervals on the Escherichia coli chromosome. At subsaturating drug concentrations, cleavage is induced at a fraction of these sites and DNA synthesis is partially inhibited. This partial inhibition is surprisingly rapid even when few sites have been inactivated. We now report kinetic measurements suggesting that inactivation of 100,000-base-pair gyrase sites by oxolinic acid does not inhibit DNA synthesis by simply producing barriers to replication fork movement. Slowing the rate of fork movement, thus increasing the time for a fork to reach a barrier, fails to proportionately slow inhibition of DNA synthesis. Moreover, the initial, rapid phase of inhibition is followed by a slower decline that is not accelerated by increasing the frequency of barriers by raising drug concentrations. These data, when added to the observation that additional oxolinic acid-induced cleavage occurs in replicating regions of the chromosome, suggest that gyrase may function at replication forks as well as at 100,000-base-pair intervals on the chromosome.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. O., Peebles C. L., Cozzarelli N. R. A topoisomerase from Escherichia coli related to DNA gyrase. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6110–6114. doi: 10.1073/pnas.76.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Snyder M. Superhelical Escherichia coli DNA: relaxation by coumermycin. J Mol Biol. 1978 Apr 5;120(2):145–154. doi: 10.1016/0022-2836(78)90061-x. [DOI] [PubMed] [Google Scholar]
- Gellert M., Fisher L. M., O'Dea M. H. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6289–6293. doi: 10.1073/pnas.76.12.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Flanders P. Genes that control DNA repair and genetic recombination in Escherichia coli. Adv Biol Med Phys. 1968;12:299–317. doi: 10.1016/b978-1-4831-9928-3.50016-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. Integrative recombination of bacteriophage lambda: requirement for supertwisted DNA in vivo and characterization of int. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1099–1109. doi: 10.1101/sqb.1979.043.01.122. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Cozzarelli N. R. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol. 1979 Nov;140(2):424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell. 1979 May;17(1):175–184. doi: 10.1016/0092-8674(79)90305-2. [DOI] [PubMed] [Google Scholar]
- Morrison A., Higgins N. P., Cozzarelli N. R. Interaction between DNA gyrase and its cleavage site on DNA. J Biol Chem. 1980 Mar 10;255(5):2211–2219. [PubMed] [Google Scholar]
- Orr E., Fairweather N. F., Holland I. B., Pritchard R. H. Isolation and characterisation of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol Gen Genet. 1979;177(1):103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- Peebles C. L., Higgins N. P., Kreuzer K. N., Morrison A., Brown P. O., Sugino A., Cozzarelli N. R. Structure and activities of Escherichia coli DNA gyrase. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):41–52. doi: 10.1101/sqb.1979.043.01.008. [DOI] [PubMed] [Google Scholar]
- Snyder M., Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979 Jun 25;131(2):287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf B., Newman A., Glaser D. A. On the origin and direction of replication of the Escherichia coli K12 chromosome. J Mol Biol. 1968 Mar 28;32(3):611–629. doi: 10.1016/0022-2836(68)90346-x. [DOI] [PubMed] [Google Scholar]



