Abstract
Legionella pneumophila, the agent of Legionnaires' disease pneumonia, is transmitted to humans following the inhalation of contaminated water droplets. In aquatic systems, L. pneumophila survives much of time within multi-organismal biofilms. Therefore, we examined the ability of L. pneumophila (clinical isolate 130b) to persist within biofilms formed by various types of aquatic bacteria, using a bioreactor with flow, steel surfaces, and low-nutrient conditions. L. pneumophila was able to intercalate into and persist within a biofilm formed by Klebsiella pneumoniae, Flavobacterium sp. or Pseudomonas fluorescens. The levels of L. pneumophila within these biofilms were as much as 4×104 CFU per cm2 of steel coupon and lasted for at least 12 days. These data document that K. pneumoniae, Flavobacterium sp., and P. fluorescens can promote the presence of L. pneumophila in dynamic biofilms. In contrast to these results, L. pneumophila 130b did not persist within a biofilm formed by Pseudomonas aeruginosa, confirming that some bacteria are permissive for Legionella colonization whereas others are antagonistic. In addition to colonizing certain mono-species biofilms, L. pneumophila 130b persisted within a two-species biofilm formed by K. pneumoniae and Flavobacterium sp. Interestingly, the legionellae were also able to colonize a two-species biofilm formed by K. pneumoniae and P. aeruginosa, demonstrating that a species that is permissive for L. pneumophila can override the inhibitory effect(s) of a non-permissive species.
Introduction
The aquatic bacterium Legionella pneumophila is the agent of Legionnaires' disease, a serious form of pneumonia that is occurring with increasing incidence [1]–[4]. L. pneumophila is ubiquitous in natural and man-made water systems, and infection can occur following the inhalation of L. pneumophila-containing droplets produced by a variety of devices [5]. The widespread distribution of L. pneumophila results from the bacterium's ability to flourish within multiple types of niches, including survival in the planktonic phase, infection of and replication within protozoan hosts, and persistence within multi-organismal biofilms that cover surfaces within water systems [6]–[9]. With the goal of developing strategies for minimizing disease transmission [10], investigators have been utilizing laboratory models to understand when and how L. pneumophila is able to exist within biofilms. The chemical and physical parameters that influence the behavior of the legionellae in biofilms include the properties of the surface, the flow rate and turbulence of the liquid over the surface, the ambient temperature, carbon and metal concentrations, and the presence of biocides [11]–[22]. The first biological parameter that influences the bacterium's impact in biofilms is the presence of protozoa that are permissive for intracellular growth of legionellae. Indeed, various types of amoebae, including Hartmannella vermiformis and Acanthamoebae castellanii, greatly promote the growth of L. pneumophila within biofilms [21], [23]–[25]. Some studies have further concluded that replication within the biofilm requires the presence of protozoan hosts [23]–[26]. However, others have argued that L. pneumophila can grow in the absence of amoebal hosts by utilizing the matrix and nutrients provided by other bacteria within the biofilm [27]–[30]. Thus, the second critical biological parameter that influences the presence of L. pneumophila within biofilms is the type of bacterial species that inhabit the biofilm. At the very least, these organisms provide the matrix to which L. pneumophila can attach and persist prior to encountering an amoebal host. Although many studies have used microbial consortia, both defined and undefined, obtained from water systems to establish and study Legionella-containing biofilms in the laboratory [14], [22], [24], [26], [31]–[34], little is known about the relationships between L. pneumophila and particular bacterial species within the context of a dynamic biofilm. In the course of documenting the importance of amoebae for biofilms, Murga et al found that L. pneumophila (strain RI243) barely persisted (i.e., ≤10 CFU/steel coupon that provided 2.5 cm2 of surface area) within a multi-species biofilm composed of Klebsiella pneumoniae, Pseudomonas aeruginosa, and Flavobacterium sp. [24]. Using the same “CDC bioreactor” and steel coupons as Murga et al did, a later study found that a different strain of L. pneumophila (i.e., clinical isolate 130b) persisted in the Klebsiella-Pseudomonas-Flavobacterium biofilm at a level of 100–1000 CFU/coupon for a period of 15 days [35]. These data suggested that one or more these heterologous bacteria are capable of providing a biofilm that is conducive to the long-term persistence of L. pneumophila. Utilizing the same steel coupons as the previous two studies and the CDC bioreactor, which assesses bacterial colonization on surfaces in the presence of significant flow and in the absence of planktonic replication, we now demonstrate that L. pneumophila is able to persist at high levels (e.g., 104–105 CFU/coupon) when in a biofilm that is formed by just K. pneumoniae, Flavobacterium sp., or Pseudomonas fluorescens but not P. aeruginosa.
Materials and Methods
Bacterial strains and growth media
L. pneumophila 130b (ATCC strain BAA-74), also known as AA100 or Wadsworth, served as our wild-type strain [36]. Mutants of 130b that were examined included the flaA mutant NU347 which lacks flagella, pilQ mutant NU278 which lacks type IV pili, and bbcB mutant NU388 which lacks surfactant [36]–[38]. In order to help distinguish 130b and its derivatives from other bacteria in the biofilms, the chloramphenicol-resistant vector pMMB2002 [37] was placed into the strains. Legionellae were routinely grown at 37°C in buffered yeast extract broth or on buffered charcoal yeast extract (BCYE) agar [36]. The heterologous bacteria that were used to create biofilms were Klebsiella pneumoniae strain DMDS 92-08-28a, Flavobacterium sp. strain CDC-65, Pseudomonas aeruginosa ATCC strain 7700, and Pseudomonas fluorescens ATCC strain 17569 [24], [39]. These organisms can be found in potable-water environments, along with a wide variety of other bacteria and in some instances along with L. pneumophila [24], [40]–[44]. The bacteria were maintained on R2A media, which consists of, per liter, 0.5 g each of yeast extract, (Becton Dickinson [BD], Franklin Park, NJ), bacto-peptone (BD), bacto-tryptone (BD), and glucose, 0.39 g K2PO3·3H2O, 0.3 g sodium pyruvate, and 0.05 g MgSO4· 7H2O, pH 7.2 [45]. R2A agar consisted of R2A media plus 0.5 g/l soluble starch and 15 g/l agar. Unless otherwise noted, chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Biofilm reactor
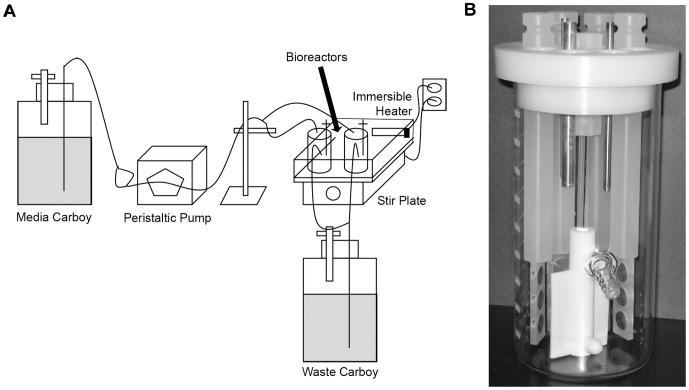
In order to assess the ability of L. pneumophila to exist in biofilms, we utilized the CDC biofilm reactor (BioSurface Technologies, Bozeman, MT) [15], [46]. The bioreactor consisted of a 1-liter glass beaker with 8 polypropylene rods suspended from a ported lid. Each rod held 3 circular, 1.25-cm diameter stainless steel 316L coupons (BioSurface Tech.) that were positioned perpendicularly to a rotating baffle. Thus, the exposed surface area for each two-sided coupon was 2.5 cm2. Stainless steel 316L is a low-carbon version of 316 steel, a chromium-nickel stainless steel containing molybdenum (AK Steel Co., West Chester, OH). Such material is commonly used for cooling towers and plumbing materials, and previous studies have shown that L. pneumophila can exist in biofilms formed on stainless steel [13], [15], [22], [24], [35]. A schematic of the bioreactor set-up appears in Fig. 1.
Figure 1. Schematic of the bioreactor system.
(A) Representation of system set-up. Medium was pumped from the media carboy to the reactors by using a peristaltic pump. The bioreactors themselves were in a water bath that was maintained at 30°C by the use of an immersible aquarium heater. The bioreactors were kept spinning at a constant rpm over the course of the experiment. Liquid exited the bioreactors by gravity into a waste carboy. (B) Close-up view of the CDC biofilm reactor. The bioreactor has openings for 8 polypropylene rods. Each rod (four shown here, for clarity) contains three spaces for stainless steel coupons which can be removed to assay for bacterial CFU. At the center of the bioreactor is a stirring baffle that maintains constant shear stress. There is a spout located about 1/3 the length of the vessel from the bottom to allow for the exit of media.
Biofilm experiments
On day-0, the bioreactor was inoculated with bacteria (i.e., 105 CFU of K. pneumoniae, 108 CFU of Flavobacterium sp., 108 of CFU P. aeruginosa, and/or 108 CFU of P. fluorescens) resuspended in 300 ml of R2A medium. The bioreactor was kept in static “batch” mode for 3 days at 30°C, 200 rpm [15]. On that third day, 1010 CFU of L. pneumophila were added to the bioreactor, and after 2 hours to allow for Legionella adherence to the biofilm, 1∶100 R2A (i.e., 1% R2A solution [vol/vol]) began to be pumped through the reactor at a rate of 1–2 ml/min. The bioreactors were run under continuous flow of 1∶100 R2A (30°C, 200 rpm) for up to an additional 12–14 days with samples being taken every 2–4 days. For sampling, one rod was removed and replaced with a sterile, blank rod. The coupons taken from the rod were washed twice in Butterfield buffer (42.5 mg/l KH2PO4) and then aseptically transferred to a 15-ml conical tube with 10 ml of Butterfield buffer for disaggregation. Each coupon was treated with 3 cycles of 30-sec sonication (Branson Sonifer 450D, 15% amplitude) followed by 1.5 min of vortexing [35]. Serial dilutions of the resulting suspension were then plated in triplicate on the appropriate media for enumerating bacterial CFU, resulting in a limit of detection equal to 10 CFU per coupon (i.e., 4 CFU/cm2). BCYE agar containing 100 U of polymyxin B and 6 mg/l chloramphenicol were used to assess L. pneumophila counts. To eliminate P. fluorescens from the sample, a heating step of 50°C for 30 min was required. Because L. pneumophila does not grow in R2A medium, the recovery of L. pneumophila CFU was a reflection of the organism's ability to attach to and persist in the biofilm. To enumerate the other species of bacteria, the sample was plated onto R2A agar. When K. pneumoniae and Flavobacterium sp. were used in the same reactor, 6 mg/l chloramphenicol was added to the R2A agar in order to assess Flavobacterium CFU.
Results
L. pneumophila colonizes and persists within monospecies biofilms of K. pneumoniae and Flavobacterium sp
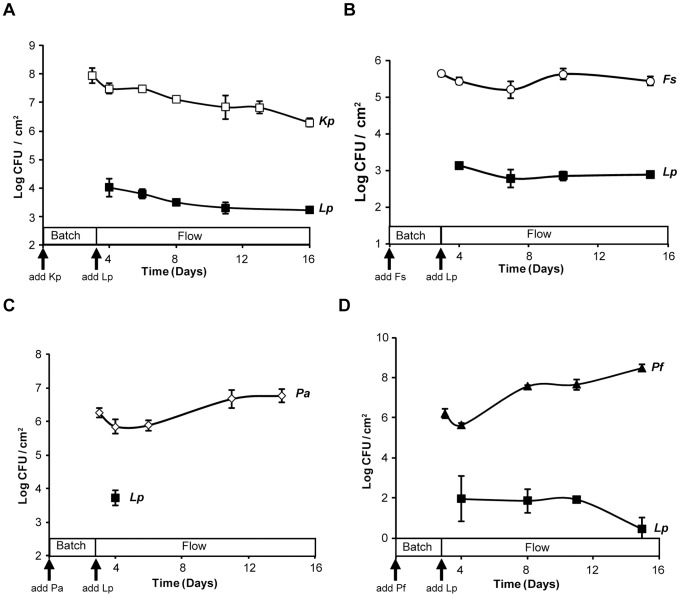
To begin to determine if L. pneumophila can adhere to and persist within biofilms that had been formed by heterologous bacteria, we inoculated our bioreactors with K. pneumoniae or Flavobacterium sp. and then later added strain L. pneumophila strain 130b. By three days post-inoculation and prior to the introduction of L. pneumophila, K. pneumoniae heavily attached to the steel coupons, achieving levels between 4×107 to 4×108 CFU per cm2 (Fig. 2A). On the third day, 1010 CFU of L. pneumophila were added to the reactor and the flow of liquid through the system was established. Over the next 12 days of the experiment, the numbers of K. pneumoniae on the coupons declined gradually but at no point did the level fall below 4×106 CFU per cm2 (Fig. 2A). Most importantly, L. pneumophila colonized the Klebsiella biofilm at ≥4×103 CFU per cm2 and then persisted at this level throughout the time course. In the next series of experiments, Flavobacterium sp. CDC-65 was found to be capable of establishing a biofilm on the steel coupons albeit not as robustly as the K. pneumoniae strain had done (Fig. 2B). L. pneumophila strain 130b effectively intercalated into the Flavobacterium biofilm and was maintained over the entire course of the experiment at approximately 4×103 CFU per cm2 (Fig. 2B). In two additional experiments, when 1010 CFU of L. pneumophila strain 130b were added to the bioreactor in the absence of any other bacteria, no CFU were recovered from the coupons (limit of detection = 10 CFU) indicating that L. pneumophila cannot establish its own biofilm in this model system. In summary, K. pneumoniae and Flavobacterium were both able to provide a biofilm environment that is conducive to colonization and high-level persistence by L. pneumophila.
Figure 2. Persistence of L. pneumophila in monospecies biofilms formed by K. pneumoniae, Flavobacterium sp., P. aeruginosa, or P. fluorescens.
A base biofilm of either K. pneumoniae strain DMDS 92-08-28a (□) (A), Flavobacterium sp. strain CDC-65 (Δ) (B), P. aeruginosa ATCC strain 7700 (◊) (C), or P. fluorescens ATCC strain 17569 (Δ) (D) pre-formed on stainless steel coupons was exposed to L. pneumophila strain 130b (▪) on day 3 and flow of 1∶100 R2A began at 1–2 ml/min. Each data point represents the averages and standard deviations of CFU obtained from the coupons within a single rod. The experiments shown are representative of four experiments for (A) and at least two for (B–D). In a repeat of the experiment using P. aeruginosa base biofilms, no legionellae were recovered at the initial sampling point.
L. pneumophila colonizes and persists within monospecies biofilms of P. fluorescens but not P. aeruginosa
We next sought to determine whether P. aeruginosa could provide a suitable base biofilm for L. pneumophila colonization and persistence. When the base biofilm consisted of only P. aeruginosa, initial attachment of L. pneumophila was often observed and at a level (approximately 3×103 CFU/cm2) that was similar to what had been seen with base biofilms consisting of Flavobacterium sp. or K. pneumoniae (Fig. 2C). However, no L. pneumophila CFU were recovered two days later or at subsequent time points. These data suggest that P. aeruginosa produces a factor(s) that prevents the maintenance of strain 130b. Furthermore, they indicate that the persistence of L. pneumophila that we had observed when using the K. pneumoniae and Flavobacterium sp. biofilms was not simply an artifact of the bioreactor system or our protocol. To determine if the result obtained with P. aeruginosa was typical for Pseudomonas species, we performed an experiment using P. fluorescens, another bacterium that is often found in water samples alongside L. pneumophila [40]. When the base biofilm consisted of only P. fluorescens, L. pneumophila was able to both intercalate and persist (Fig. 2D), albeit at a level that was approximately 10-fold less than we had observed with the Klebsiella or Flavobacterium species. Thus, L. pneumophila strain 130b was able to colonize and persist within dynamic biofilms formed by some but not all species of Pseudomonas.
L. pneumophila persists within various two-species biofilms
Given the ability of L. pneumophila to integrate into a monospecies biofilm of Klebsiella or Flavobacterium, we next sought to determine how strain 130b would fare when exposed to a biofilm consisting of both K. pneumoniae and Flavobacterium sp. On day-3 and prior to the addition of legionellae, the Klebsiella and Flavobacterium organisms demonstrated an ability to co-exist within the biofilm formed on steel coupons, with each achieving levels that were comparable to what had been observed in the monospecies experiments (Fig. 3A). More importantly, L. pneumophila integrated into the biofilm and persisted over the remaining 12-day course of the experiment. The presence of legionellae within the multi-species biofilm was maintained between 400 to 4000 CFU per cm2, which was comparable to that of the flavobacteria (Fig. 3A) and also similar to the degree to which the legionellae persisted when in combination with Klebsiella alone or Flavobacterium alone (Figs. 2A and 2B). Taken together, these data indicate that L. pneumophila is capable of persisting relatively well within biofilms that contain multiple heterologous species and that K. pneumoniae and Flavobacterium sp. alone or in combination are not inhibitory to L. pneumophila. When strain 130b was exposed to a multi-species biofilm consisting of K. pneumoniae and P. aeruginosa, the legionellae colonized and persisted at approximately 4000 CFU per cm2 for at least 8 days and then dropped in numbers about 10-fold by the end of the experiment (Fig. 3B). The decline in L. pneumophila appeared coincident with increasing numbers of P. aeruginosa in the biofilm. These data confirm that L. pneumophila can persist within different sorts of multi-species biofilms. Furthermore, they indicate that the dramatic inhibitory effect of P. aeruginosa on L. pneumophila that we had observed in earlier experiments was absent or greatly reduced when a third species (e.g., K. pneumoniae) is present in the mixed biofilm.
Figure 3. Persistence of L. pneumophila in two-species biofilms formed by K. pneumoniae and Flavobacterium sp. or K. pneumoniae and P. aeruginosa.
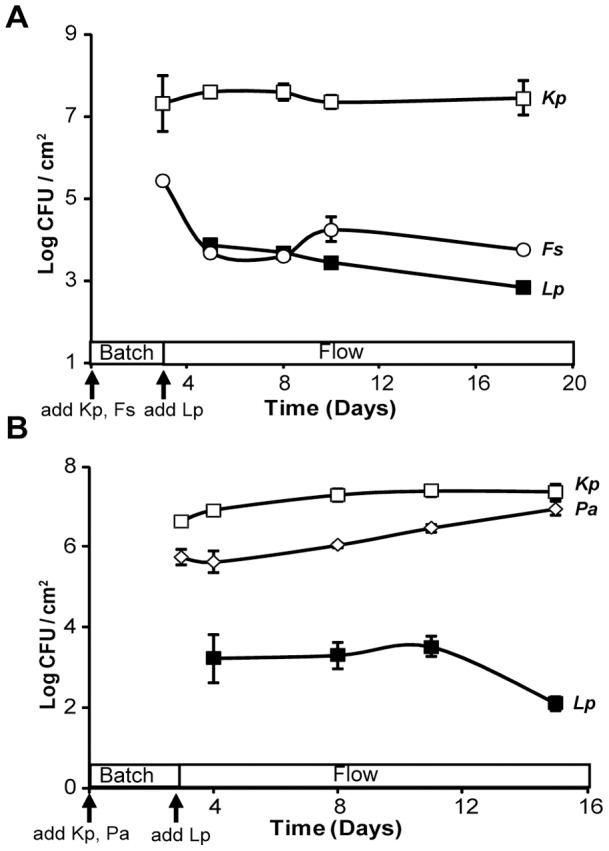
A base biofilm of either K. pneumoniae strain DMDS 92-08-28a (□) and Flavobacterium sp. strain CDC-65 (◊) (A) or K. pneumoniae strain DMDS 92-08-28a (□) and P. aeruginosa ATCC strain 7700 (◊) (B) pre-formed on stainless steel coupons was inoculated with L. pneumophila strain 130b (▪) on day 3 and flow of 1∶100 R2A began at 1–2 ml/min. Each data point represents the averages and standard deviations of colony counts from a single rod, and the experiments shown here are representative of two experiments.
L. pneumophila flagella, pili, and surfactant are not required for attachment to or persistence in a dynamic biofilm formed by K. pneumoniae
L. pneumophila is known to exhibit swimming via the action of a polar flagellum [47], twitching motility and adhesiveness that are associated with type IV pili [48]–[50], and sliding motility and anti-microbial activity that are dependent on a secreted biosurfactant [36], [38]. Because factors such as these are implicated in biofilm formation by other bacteria [7], [38], we separately tested flagella, pili, and surfactant mutants of strain 130b in a bioreactor that had been previously inoculated with K. pneumoniae. In each of the experiments using a mutant, the numbers of K. pneumoniae on the steel coupons were comparable to what we had observed when testing parental strain 130b (data not shown), indicating that Legionella flagella, pili, and surfactant do not significantly impact the behavior of the heterotroph. In the first experimental set-up which consisted of three reactors running in parallel, colonization and persistence by the flagella and pilus mutants mirrored that of the wild-type strain; i.e., all displayed approximately 4×103–4×104 CFU per cm2 over the entire time course (Fig. 4A). Although one or both of the mutants appeared to be less prominent compared to wild-type at several time points, these differences were not statistically significant. In the next round of experiments which consisted of two reactors running in parallel, the surfactant mutant behaved as wild-type did in terms of both colonization and persistence (Fig. 4B). In summary, these data indicate that flagella, type IV pili, and surfactant are not required for the ability of L. pneumophila to colonize and persist within a base biofilm consisting of K. pneumoniae.
Figure 4. Persistence of L. pneumophila mutants in a K. pneumoniae biofilm.
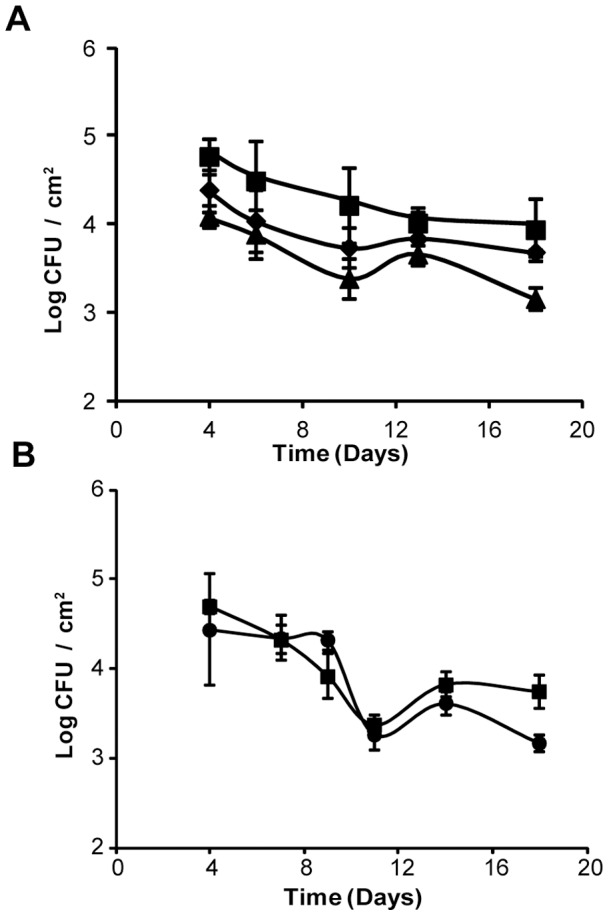
A base biofilm of K. pneumoniae strain DMDS 92-08-28a pre-formed on stainless steel coupons was inoculated in (A) with either L. pneumophila wild-type strain 130b (▪), flaA mutant NU347(pMMB2002) (♦), or pilQ mutant NU278(pMMB2002) (▴) and in (B) with either L. pneumophila wild-type strain 130b (▪) or bbcB mutant NU388 (•) on day 3 and flow of 1∶100 R2A began at 1–2 ml/min. Each data point represents the averages and standard deviations of colony counts from a single rod, and the experiments presented are representative of two repeats.
Discussion
Our data demonstrate, for the first time, that L. pneumophila is able to intercalate into and persist within a monospecies biofilm formed by K. pneumoniae, Flavobacterium sp. or P. fluorescens. K. pneumoniae (strain DMDS 92-08-28a) and Flavobacterium sp. (strain CDC-65) were comparable in terms of their ability to support L. pneumophila persistence, while P. fluorescens (ATCC strain 17569) was slightly less permissive. Importantly, our experiments used a dynamic bioreactor with flow conditions, steel surfaces, and low-nutrient media. Furthermore, the persistence that we observed reached as much as 4×104 CFU per cm2 and lasted at least 12 days. Currently, it is not possible to discern whether the bacterial load on the steel coupons reflects bacteria that persist but do not replicate or is the sum of growth plus loss due to the flow within the bioreactor; e.g., it is possible that, at some time point(s), the legionellae can replicate in 1∶100 R2A medium that has been “conditioned” by some of the biofilm communities.
Only one previous study examined the ability of L. pneumophila to engage a monospecies biofilm formed by K. pneumoniae, but the investigators found that the bacterium was not able to attach or persist [21]. The possible reasons for our differing results include differences in the Legionella strains used (130b vs. JR32), the Klebsiella strains (DMDS 92-08-28a vs. 21UHC), the substrata (i.e., steel vs. glass and plastic), the media, the flow rates and other physical aspects of the dynamic model system. Thus, it would appear that the ability of L. pneumophila to intercalate and persist within a K. pneumoniae biofilm is a variable trait. No previous study using a dynamic biofilm model examined the ability of L. pneumophila to attach to and persist within a monospecies biofilm formed by P. fluorescens, although one prior study found that P. fluorescens strain SSD (but not P. fluorescens ATCC 49838) inhibited the ability of L. pneumophila (strain “Lp-1”) to adhere to the wells of a polystyrene microtiter plate [42]. To our knowledge, no previous study using any sort of biofilm model examined the ability of L. pneumophila to attach to and persist in a monospecies biofilm made by Flavobacterium sp. Further review of the literature revealed that there are only a few other examples of a heterologous bacterium supporting the presence of L. pneumophila in biofilms. Using dynamic flow models, Mampel et al and Vervaeren et al found that biofilms formed by Empedobacter breve, Microbacterium sp., or Pseudomonas putida are able to provide a base that is conducive to L. pneumophila long-term persistence [21], [51]. Another study found that L. pneumophila intercalates into a dynamic biofilm formed by Sphingomonas sp. and can be isolated again 12 hours later; however, because no other time points were examined, it is difficult to conclude whether these data represent L. pneumophila persistence [52]. Because these past studies did not report CFU and used different protocols, it is not possible to rank K. pneumoniae, Flavobacterium sp., P. fluorescens, E. breve, Microbacterium sp., and P. putida (and Sphingomonas sp.) in terms of their capacity to support L. pneumophila persistence within dynamic biofilms. Finally, it is worthwhile to add that other past studies have reported that Acinetobacter lwoffii and Mycobacterium chelonae individually promote biofilm formation by L. pneumophila under static conditions; i.e., by assessing bacterial numbers bound to either the wells of a plastic microtiter plate or PVC coupons placed into the wells [42], [53].
In contrast to the results obtained with K. pneumoniae, Flavobacterium sp., and P. fluorescens, we observed that a monospecies biofilm formed by P. aeruginosa ATCC strain 7700 was not conducive to the persistence of L. pneumophila strain 130b. In some of our experiments, the legionellae initially associated with the biofilm but were lost within two days of further incubation, suggesting that L. pneumophila can attach to a matrix produced by P. aeruginosa but is unable to resist inhibitory substances and/or effectively compete for space or nutrients. Although the reason why P. aeruginosa impedes the persistence of L. pneumophila in our system remains to be determined, one possible speculation derives from the fact that purified homoserine lactones produced by P. aeruginosa inhibit another strain of L. pneumophila in a static (microtiter-plate) biofilm assay [54]. To our knowledge, only one previous study examined the interaction of L. pneumophila with a monospecies biofilm of P. aeruginosa under dynamic flow conditions; in that case, L. pneumophila strain JR32 never attached to the biofilm that had been formed by P. aeruginosa strain K [21]. In considering our result with those of this previous study, it would appear that P. aeruginosa biofilms can be unsuitable for L. pneumophila for a variety of reasons. P. aeruginosa is not alone in its refractory effect on L. pneumophila. Using a dynamic biofilm model, Mampel et al found that L. pneumophila JR32 was unable to attach to a biofilm formed by Corynebacterium glutamicum and unable to persist within a biofilm made by Acinetobacter baumannii [21]. Examining static biofilms in microtiter-plates, others found that L. pneumophila (i.e., strain Lp-1 and NCTC 12821) can be inhibited by Aeromonas hydrophila, Burkholderia cepacia, Acidovorax sp., and Sphingomonas sp. [42], [53].
In light of the ability of L. pneumophila to persist in a monospecies biofilms formed by either K. pneumoniae or Flavobacterium sp., it was not surprising that the organism also persisted well in a mixed biofilm consisting of both K. pneumoniae and Flavobacterium sp. Given this result and the fact that L. pneumophila persisted in a biofilm formed by P. fluorescens, we strongly suspect that L. pneumophila would intercalate into and persist within a two-species biofilm consisting of either K. pneumoniae and P. fluorescens or Flavobacterium sp. and P. fluorescens. An interesting and arguably more surprising result was the observation that L. pneumophila persisted relatively well within in a mixed biofilm formed by K. pneumoniae and P. aeruginosa, whereas L. pneumophila was completely unable to persist in a biofilm formed by P. aeruginosa alone. This experiment documented, for the first time, that the permissiveness of one species (e.g., K. pneumoniae) for L. pneumophila can be dominant over the non-permissiveness of another species (e.g., P. aeruginosa) for L. pneumophila. It will be interesting, in the future, to investigate how K. pneumoniae is able to erase the negative effect(s) of P. aeruginosa. Our data also help explain why L. pneumophila was able to persist to some degree in dynamic biofilms that consisted of K. pneumoniae, Flavobacterium sp., and P. aeruginosa [24], [35]; i.e., K. pneumoniae and Flavobacterium sp. likely provided factors that directly stimulated the persistence of L. pneumophila while at the same time dampening the inhibitory effect(s) of P. aeruginosa. In light of these results, it might be instructive to “deconstruct” the other past studies that had exposed L. pneumophila to biofilms consisting of a different combination of known bacteria. For example, several studies showed that L. pneumophila, though not replicating, could persist within a four-species biofilm that was composed of A. hydrophila, Escherichia coli, Flavobacterium breve, and P. aeruginosa [25], [55]. Another study found that a seven-species biofilm consisting of A. baumannii, C. glutamicum, E. breve, K. pneumoniae, Microbacterium sp., P. aeruginosa, and P. putida was not even conducive to persistence [21].
At this point, it is not clear how K. pneumoniae, Flavobacterium sp., and P. fluorescens are facilitating the integration and persistence of L. pneumophila within their biofilms. Since strains of these species/genera produce capsular, extracellular matrix material [56]–[58], it is likely that they are providing an appropriate substrate for the attachment of L. pneumophila. Additionally, they may be directly or indirectly providing nutrients that promote the survival and/or growth of the legionellae. Another important question is what factors encoded by L. pneumophila promote the organism's ability to attach and persist within the biofilms formed by these other bacteria under dynamic flow conditions. Using a set of specific L. pneumophila 130b mutants and a model biofilm derived from K. pneumoniae, we could not uncover a role for flagella, type IV pili, or surfactant. The lack of a role for flagella in attachment and persistence could be a reflection of the fact that our system provided a sufficient mechanism for bringing the legionellae into contact with the biofilm on the steel coupons. The lack of a required role for type IV pili suggests that L. pneumophila strain 130b has other surface molecules that mediate attachment to the K. pneumoniae biofilm. Previously, Lucas et al found that another type IV pilus mutant of L. pneumophila had a modestly reduced ability to colonize a mixed biofilm consisting of K. pneumoniae, Flavobacterium sp., and P. aeruginosa [35]. In trying to reconcile our results with this past study, it would appear that L. pneumophila attachment to the mixed biofilm is a combination of events, including non-type IV pilus-mediated attachment to that portion of the biofilm consisting of K. pneumoniae and/or K. pneumoniae matrix, and type IV pilus-mediated attachment to other portions that are not dominated by K. pneumoniae. Although our study and that of Lucas et al are the only ones to have investigated any L. pneumophila mutants in a mixed biofilm model with dynamic flow conditions, several groups have obtained results using just legionellae in the static, microtiter plate-based assay. Factors that have been identified as being important by that measure include the FliA sigma factor, a collagen-like adhesin, twin-arginine translocation, and nitric-oxide sensors [21], [59]–[62]. Factors that were not found to be important in the static biofilm model include flagella, type IV pili, Dot/Icm type IV secretion system, the Lvh type IV secretion system, the Lqs quorum-sensing system, and the regulatory factors, RpoS, LetA, and CsrA [21], [63]. In sum, very little is known about the factors encoded by L. pneumophila that mediate its attachment to and persistence within biofilms created by other bacteria. We posit that the mechanisms of L. pneumophila survival within natural biofilms are likely to be quite variable, depending upon the types of bacteria that constitute the biofilm base as well as the presence of different protozoan hosts and non-microbial environmental factors.
Acknowledgments
We thank both Barry Fields for providing us with K. pneumoniae, Flavobacterium sp., and P. aeruginosa strains and Alan Hauser for giving us the P. fluorescens strain. We also thank Claressa Lucas for advice on the use of the CDC reactor. We acknowledge Matt Barnes for technical assistance and the other members of the Cianciotto lab for helpful comments.
Funding Statement
This work was supported by National Institutes of Health grants AI043987 and AI034937 awarded to NPC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hicks LA, Garrison LE, Nelson GE, Hampton LM (2011) Legionellosis – United States, 2000–2009. MMWR 60: 1083–1086.21849965 [Google Scholar]
- 2.Edelstein PH, Cianciotto NP (2010) Legionella. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and Practice of Infectious Diseases, 7th ed. Philadelphia: Elsevier Churchill Livingstone. pp. 2969–2984.
- 3. Newton HJ, Ang DK, van Driel IR, Hartland EL (2010) Molecular pathogenesis of infections caused by Legionella pneumophila . Clin Microbiol Rev 23: 274–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li JS, OBrien ED, Guest C (2002) A review of national legionellosis surveillance in Australia, 1991 to 2000. Commun Dis Intell 26: 461–468. [PubMed] [Google Scholar]
- 5. Fields BS, Benson RF, Besser RE (2002) Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15: 506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lau HY, Ashbolt NJ (2009) The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J Appl Microbiol 107: 368–378. [DOI] [PubMed] [Google Scholar]
- 7. Declerck P (2010) Biofilms: the environmental playground of Legionella pneumophila . Environ Microbiol 12: 557–566. [DOI] [PubMed] [Google Scholar]
- 8. Taylor M, Ross K, Bentham R (2009) Legionella, protozoa, and biofilms: interactions within complex microbial systems. Microb Ecol 58: 538–547. [DOI] [PubMed] [Google Scholar]
- 9. Hilbi H, Hoffmann C, Harrison CF (2011) Legionella spp. outdoors: colonization, communication, and persistence. Environ Microbiol Reports 3: 286–296. [DOI] [PubMed] [Google Scholar]
- 10. Buse HY, Schoen ME, Ashbolt NJ (2012) Legionellae in engineered systems and use of quantitative microbial risk assessment to predict exposure. Water Res 46: 921–933. [DOI] [PubMed] [Google Scholar]
- 11. Wright JB, Ruseska I, Costerton JW (1991) Decreased biocide susceptibility of adherent Legionella pneumophila . J Appl Bacteriol 71: 531–538. [DOI] [PubMed] [Google Scholar]
- 12. Bezanson G, Burbridge S, Haldane D, Marrie T (1992) In situ colonization of polyvinyl chloride, brass, and copper by Legionella pneumophila . Can J Microbiol 38: 328–330. [DOI] [PubMed] [Google Scholar]
- 13. Turetgen I, Cotuk A (2007) Monitoring of biofilm-associated Legionella pneumophila on different substrata in model cooling tower system. Environ Monit Assess 125: 271–279. [DOI] [PubMed] [Google Scholar]
- 14. Rogers J, Dowsett AB, Dennis PJ, Lee JV, Keevil CW (1994) Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl Environ Microbiol 60: 1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donlan RM, Forster T, Murga R, Brown E, Lucas C, et al. (2005) Legionella pneumophila associated with the protozoan Hartmannella vermiformis in a model multi-species biofilm has reduced susceptibility to disinfectants. Biofouling 21: 1–7. [DOI] [PubMed] [Google Scholar]
- 16. Liu Z, Lin YE, Stout JE, Hwang CC, Vidic RD, et al. (2006) Effect of flow regimes on the presence of Legionella within the biofilm of a model plumbing system. J Appl Microbiol 101: 437–442. [DOI] [PubMed] [Google Scholar]
- 17. Lehtola MJ, Torvinen E, Kusnetsov J, Pitkanen T, Maunula L, et al. (2007) Survival of Mycobacterium avium, Legionella pneumophila, Escherichia coli, and caliciviruses in drinking water-associated biofilms grown under high-shear turbulent flow. Appl Environ Microbiol 73: 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hindre T, Bruggemann H, Buchrieser C, Hechard Y (2008) Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiol 154: 30–41. [DOI] [PubMed] [Google Scholar]
- 19. Piao Z, Sze CC, Barysheva O, Iida K, Yoshida S (2006) Temperature-regulated formation of mycelial mat-like biofilms by Legionella pneumophila . Appl Environ Microbiol 72: 1613–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pang CM, Liu WT (2006) Biological filtration limits carbon availability and affects downstream biofilm formation and community structure. Appl Environ Microbiol 72: 5702–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mampel J, Spirig T, Weber SS, Haagensen JA, Molin S, et al. (2006) Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl Environ Microbiol 72: 2885–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van der Kooij D, Veenendaal HR, Scheffer WJ (2005) Biofilm formation and multiplication of Legionella in a model warm water system with pipes of copper, stainless steel and cross-linked polyethylene. Water Res 39: 2789–2798. [DOI] [PubMed] [Google Scholar]
- 23. Kuiper MW, Wullings BA, Akkermans AD, Beumer RR, van der Kooij D (2004) Intracellular proliferation of Legionella pneumophila in Hartmannella vermiformis in aquatic biofilms grown on plasticized polyvinyl chloride. Appl Environ Microbiol 70: 6826–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murga R, Forster TS, Brown E, Pruckler JM, Fields BS, et al. (2001) Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiol 147: 3121–3126. [DOI] [PubMed] [Google Scholar]
- 25. Declerck P, Behets J, van Hoef V, Ollevier F (2007) Replication of Legionella pneumophila in floating biofilms. Curr Microbiol 55: 435–440. [DOI] [PubMed] [Google Scholar]
- 26. Declerck P, Behets J, Margineanu A, van Hoef V, De Keersmaecker B, et al. (2009) Replication of Legionella pneumophila in biofilms of water distribution pipes. Microbiol Res 164: 593–603. [DOI] [PubMed] [Google Scholar]
- 27. Rogers J, Keevil CW (1992) Immunogold and fluorescein immunolabelling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl Environ Microbiol 58: 2326–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keevil CW (2003) Rapid detection of biofilms and adherent pathogens using scanning confocal laser microscopy and episcopic differential interference contrast microscopy. Water Sci Technol 47: 105–116. [PubMed] [Google Scholar]
- 29. Wright JB, Ruseska I, Athar MA, Corbett S, Costerton JW (1989) Legionella pneumophila grows adherent to surfaces in vitro and in situ. Infect Control Hosp Epidemiol 10: 408–415. [DOI] [PubMed] [Google Scholar]
- 30. Temmerman R, Vervaeren H, Noseda B, Boon N, Verstraete W (2006) Necrotrophic growth of Legionella pneumophila . Appl Environ Microbiol 72: 4323–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moritz MM, Flemming HC, Wingender J (2010) Integration of Pseudomonas aeruginosa and Legionella pneumophila in drinking water biofilms grown on domestic plumbing materials. Int J Hyg Environ Health 213: 190–197. [DOI] [PubMed] [Google Scholar]
- 32. Messi P, Anacarso I, Bargellini A, Bondi M, Marchesi I, et al. (2011) Ecological behaviour of three serogroups of Legionella pneumophila within a model plumbing system. Biofouling 27: 165–172. [DOI] [PubMed] [Google Scholar]
- 33. Storey MV, Langmark J, Ashbolt NJ, Stenstrom TA (2004) The fate of legionellae within distribution pipe biofilms: measurement of their persistence, inactivation and detachment. Water Sci Technol 49: 269–275. [PubMed] [Google Scholar]
- 34.Farhat M, Moletta-Denat M, Frere J, Onillon S, Trouilhe MC, et al. (2012) Legionella, Eukarya and biofilm: disinfection effects in hot water system. Appl Environ Microbiol: DOI:10.1128/AEM.00831-00812. [DOI] [PMC free article] [PubMed]
- 35. Lucas CE, Brown E, Fields BS (2006) Type IV pili and type II secretion play a limited role in Legionella pneumophila biofilm colonization and retention. Microbiol 152: 3569–3573. [DOI] [PubMed] [Google Scholar]
- 36. Stewart CR, Rossier O, Cianciotto NP (2009) Surface translocation by Legionella pneumophila: A form of sliding motility that is dependent upon type II protein secretion. J Bacteriol 191: 1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rossier O, Starkenburg S, Cianciotto NP (2004) Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect Immun 72: 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stewart CR, Burnside DM, Cianciotto NP (2011) The surfactant of Legionella pneumophila is secreted in a TolC-dependent manner and is antagonistic toward other Legionella species. J Bacteriol 193: 5971–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stanier RY, Palleroni NJ, Doudoroff M (1966) The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43: 159–271. [DOI] [PubMed] [Google Scholar]
- 40. Merault N, Rusniok C, Jarraud S, Gomez-Valero L, Cazalet C, et al. (2011) Specific real-time PCR for simultaneous detection and identification of Legionella pneumophila serogroup 1 in water and clinical samples. Appl Environ Microbiol 77: 1708–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stojek NM, Dutkiewicz J (2011) Co-existence of Legionella and other Gram-negative bacteria in potable water from various rural and urban sources. Ann Agric Environ Med 18: 330–334. [PubMed] [Google Scholar]
- 42. Guerrieri E, Bondi M, Sabia C, de Niederhausern S, Borella P, et al. (2008) Effect of bacterial interference on biofilm development by Legionella pneumophila . Curr Microbiol 57: 532–536. [DOI] [PubMed] [Google Scholar]
- 43.Committee on Public Water Supply Distribution Systems: Assessing and Reducing Risks NRC (2006) Public Health Risk from Distribution System Contamination. Drinking Water Distribution Systems: Assessing and Reducing Risks. Washington, D.C.: The National Academies Press. pp. 87–141.
- 44. Wingender J, Flemming HC (2011) Biofilms in drinking water and their role as reservoir for pathogens. Int J Hyg Environ Health 214: 417–423. [DOI] [PubMed] [Google Scholar]
- 45. Reasoner DJ, Geldreich EE (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goeres DM, Loetterle LR, Hamilton MA, Murga R, Kirby DW, et al. (2005) Statistical assessment of a laboratory method for growing biofilms. Microbiol 151: 757–762. [DOI] [PubMed] [Google Scholar]
- 47. Heuner K, Steinert M (2003) The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int J Med Microbiol 293: 133–143. [DOI] [PubMed] [Google Scholar]
- 48. Stone BJ, Abu Kwaik Y (1998) Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun 66: 1768–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Coil DA, Anne J (2009) Twitching motility in Legionella pneumophila . FEMS Microbiol Lett 293: 271–277. [DOI] [PubMed] [Google Scholar]
- 50. Coil DA, Anne J (2010) The role of fimV and the importance of its tandem repeat copy number in twitching motility, pigment production, and morphology in Legionella pneumophila . Arch Microbiol 192: 625–631. [DOI] [PubMed] [Google Scholar]
- 51. Vervaeren H, Temmerman R, Devos L, Boon N, Verstraete W (2006) Introduction of a boost of Legionella pneumophila into a stagnant-water model by heat treatment. FEMS Microbiol Ecol 58: 583–592. [DOI] [PubMed] [Google Scholar]
- 52. Manz W, Amann R, Szewzyk R, Szewzyk U, Stenstrom TA, et al. (1995) In situ identification of Legionellaceae using 16S rRNA-targeted oligonucleotide probes and confocal laser scanning microscopy. Microbiol 141: 29–39. [DOI] [PubMed] [Google Scholar]
- 53. Giao MS, Azevedo NF, Wilks SA, Vieira MJ, Keevil CW (2011) Interaction of Legionella pneumophila and Helicobacter pylori with bacterial species isolated from drinking water biofilms. BMC Microbiol 11: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kimura S, Tateda K, Ishii Y, Horikawa M, Miyairi S, et al. (2009) Pseudomonas aeruginosa Las quorum sensing autoinducer suppresses growth and biofilm production in Legionella species. Microbiol 155: 1934–1939. [DOI] [PubMed] [Google Scholar]
- 55.Declerck P, Behets J, Margineanu A, van Hoef V, De Keersmaecker B, et al.. (2007) Replication of Legionella pneumophila in biofilms of water distribution pipes. Microbiol Res. [DOI] [PubMed]
- 56. Wu MC, Lin TL, Hsieh PF, Yang HC, Wang JT (2011) Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS One 6: e23500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Basson A, Flemming LA, Chenia HY (2008) Evaluation of adherence, hydrophobicity, aggregation, and biofilm development of Flavobacterium johnsoniae-like isolates. Microb Ecol 55: 1–14. [DOI] [PubMed] [Google Scholar]
- 58. Kives J, Orgaz B, Sanjose C (2006) Polysaccharide differences between planktonic and biofilm-associated EPS from Pseudomonas fluorescens B52. Colloids Surf B Biointerfaces 52: 123–127. [DOI] [PubMed] [Google Scholar]
- 59. Duncan C, Prashar A, So J, Tang P, Low DE, et al. (2011) Lcl of Legionella pneumophila is an immunogenic GAG binding adhesin that promotes interactions with lung epithelial cells and plays a crucial role in biofilm formation. Infect Immun 79: 2168–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carlson HK, Vance RE, Marletta MA (2010) H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila . Mol Microbiol 77: 930–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. De Buck E, Maes L, Meyen E, Van Mellaert L, Geukens N, et al. (2005) Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. Biochem Biophys Res Commun 331: 1413–1420. [DOI] [PubMed] [Google Scholar]
- 62. Molofsky AB, Shetron-Rama LM, Swanson MS (2005) Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect Immun 73: 5720–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tiaden A, Spirig T, Sahr T, Walti MA, Boucke K, et al. (2010) The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila . Environ Microbiol 12: 1243–1259. [DOI] [PubMed] [Google Scholar]