Abstract
Ghd7 is an important rice gene that has a major effect on several agronomic traits, including yield. To reveal the origin of Ghd7 and sequence evolution of this locus, we performed a comparative sequence analysis of the Ghd7 orthologous regions from ten diploid Oryza species, Brachypodium distachyon, sorghum and maize. Sequence analysis demonstrated high gene collinearity across the genus Oryza and a disruption of collinearity among non-Oryza species. In particular, Ghd7 was not present in orthologous positions except in Oryza species. The Ghd7 regions were found to have low gene densities and high contents of repetitive elements, and that the sizes of orthologous regions varied tremendously. The large transposable element contents resulted in a high frequency of pseudogenization and gene movement events surrounding the Ghd7 loci. Annotation information and cytological experiments have indicated that Ghd7 is a heterochromatic gene. Ghd7 orthologs were identified in B. distachyon, sorghum and maize by phylogenetic analysis; however, the positions of orthologous genes differed dramatically as a consequence of gene movements in grasses. Rather, we identified sequence remnants of gene movement of Ghd7 mediated by illegitimate recombination in the B. distachyon genome.
Introduction
Comparative genomics is a powerful tool to study gene and genome evolution [1]. However, studies based on model genomes are largely insufficient to interpret the evolutionary history and mechanism of genomic changes. The genus Oryza provides a fantastic model to study gene and genome evolution with its well defined phylogenic relationships and rich genomic resources available [2]–[6]. Comparative genomics in Oryza have provided insights into genome evolution [7]–[9], genome size variation [10], [11] and dynamics of gene evolution, such as lineage specific gene deletions, repeat-mediated gene movements and de novo gene formation [4], [12]–[14].
Recently, whole genome sequences of several grass species have provided us insights into genome conservation and lineage-specific features [15]–[17]. Exceptions to gene collinearity have also been observed frequently [1], [18]–[20]. Comparisons between Oryza sativa (rice) and Brachypodium distachyon indicate ∼18% of genes are absent in collinear blocks; this value rises to ∼43% when comparing rice and Sorghum bicolor (sorghum) [16], [21]. Erosion of gene collinearity can be explained by gene movement events [18], [22]. However, there were few reported cases for movements of agronomically important genes [13], [18], [23], [24].
The genus Oryza, together with several completely sequenced grass species, is becoming a powerful system to elucidate the evolutionary origin of agronomically important genes. Ghd7, a CCT domain-containing gene located on the short arm of rice chromosome 7, controls the number of grains per panicle, plant height and heading date [25]. Enhanced expression of Ghd7 under long-day conditions plays a central role in the photoperiod pathway of flowering. The reduced function of Ghd7 is associated with adaptation of rice to regions with low temperatures and short growth seasons [25]. Ghd7 is thought to be an evolutionarily new gene, because it does not have homologs in Arabidopsis thaliana, and the protein sequence lacks a B-box domain, and the non-CCT portion differs from other CCT domain-containing proteins [25], [26]. Interestingly, comparative sequence analysis among rice, sorghum and maize indicated that Ghd7 was absent from orthologous regions in the Andropogoneae lineage [27]. In order to uncover the evolutionary history of the Ghd7 locus, we performed a comparative sequence analysis of the Ghd7 orthologous regions in ten diploid Oryza species and related regions from B. distachyon, sorghum and Zea mays (maize). The Ghd7 regions showed distinctive heterochromatic features compared to previously analyzed euchromatic regions (Adh1, Moc1, Hd1) in the genus Oryza [4], [13], [14]. The evolutionary history of Ghd7 and the mechanism of gene movements were interpreted and discussed.
Results
Identification and Sequencing of BAC Clones of the Ghd7 Orthologous Regions from the Genus Oryza
BAC clones covering the Ghd7 orthologous regions were isolated from Oryza rufipogon (AA), Oryza nivara (AA), Oryza glaberrima (AA), Oryza glumaepatula (AA), Oryza punctata (BB), Oryza officinalis (CC), Oryza australiensis (EE), and Oryza brachyantha (FF). Thirty-three BAC clones were sequenced using Illumina Genome Analyzer II and Roche/454 Genome Sequencer (Table S1). In total, we generated ∼5.83 Mb of DNA sequence, representing 3.86 Mb of the corresponding Ghd7 orthologous regions (Table S1). Having additional data points from the syntenic regions of non-Oryza species would be instrumental in reconstructing the evolution history of duplication, retention and syntenic gene order. Therefore, we also included the corresponding orthologous regions from O. sativa L. ssp. japonica (japonica), O. sativa L. ssp. indica (indica), B. distachyon, sorghum and maize for data analysis. A total of ∼7.6 Mb of genome sequence data were annotated (Table S2).
Gene Organization of the Ghd7 Regions
The Ghd7 region from japonica was used as a reference for all comparative analyses. Genes were reannotated as described in the experimental procedures. Twenty gene models and two pseudogenes were annotated in the 553 kb region (Tables S3, S4). The intron/exon structures of the annotated genes were corrected according to the full-length cDNA or EST sequences (Table S3). A total of 163 genes were annotated in other Oryza species, B. distachyon, sorghum and maize (Table S2). Genes from each species are denoted by the abbreviation of each species: J, japonica; I, indica; GLA, O. glaberrima; RUF, O. rufipogon; NIV, O. nivara; GLU, O. glumaepatula; P, O. punctata; O, O. officinalis; A, O. australiensis; B, O. brachyantha; BD, B. distachyon; SB, S. bicolor; ZM, Z. mays (Tables S5, S6, S7, S8).
The gene densities of the Ghd7-surrounding regions were much lower than the Moc1, Adh1 and Hd1 regions [4], [13], [14], ranging from 9 kb/gene in B. distachyon to 110 kb/gene in O. officinalis (Figure S2 and Table S2). Eleven pseudogenes were annotated (Figure S1). The pseudogenes were caused by the insertion of repetitive elements (J-19, GLA-19, GLU-19, SB-6 and ZM-15), premature terminations (J-2, I-2, I-17, GLA-8 and GLA-18) and mutation at a splice site and the initiation codon (GLU-17) (Figure S1). Most of the pseudogenes were detected as duplicated genes or lineage-specific genes. However, three conserved genes were also observed to be pseudogenized in O. glaberrima (GLA-8), sorghum (SB-6) and maize (ZM-15). In addition, six gene models were observed to have variations in intron/exon structures in Oryza species, and their structures were confirmed by RT-PCR experiments (P-9, P-11, A-11, B-9, B-11 and B-22); the gene structures of 11 predicted gene models in the non-Oryza species were found to differ from their orthologs in japonica (Figure S1).
High Gene Collinearity in the Diploid Oryza Species and a Loss of Conservation in B. distachyon, Sorghum and Maize
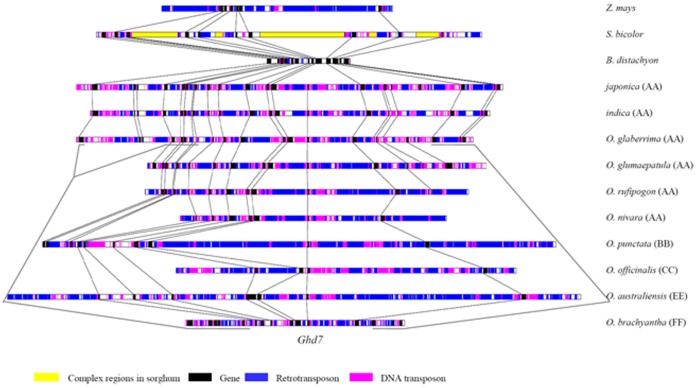
A comparison of the Ghd7 orthologous regions indicated high gene collinearity within the genus Oryza and a loss of sequence conservation in B. distachyon, sorghum and maize (Figure 1, Tables S6, S7, S8). Indica has the highest level of sequence identity with japonica, differing mainly by the insertion or deletion of several repetitive elements. Even within the AA genomes, O. glaberrima and O. glumaepatula contain lineage-specific genes or pseudogenes. The core gene Ghd7 was present in the diploid Oryza species only, but absent in the syntenic regions in B. distachyon, sorghum and maize. There were three pairs of tandem gene duplicates in japonica (Figure S1). Interestingly, these duplicated genes showed dramatic changes in distal species compared to rice, including copy number, the presence or absence of pseudogenes, gene structure and the direction of expression. In particular, one copy of a duplicated gene pair (J-18, I-18, GLA-18, GLU-18) contains a single exon and no introns, suggesting this duplicated gene copy may arise by reverse transcription of processed mRNA, with subsequent integration into the genome.
Figure 1. Synteny of the Ghd7 orthologous regions in Oryza species, B. distachyon, sorghum and maize. Lines connect orthologous genes.
The sizes of the Ghd7 orthologous regions varied tremendously among rice, B. distachyon, sorghum and maize. As shown in Figure 1 and Figure S1, B. distachyon, sorghum and maize only contained approximately half of the 22 genes or pseudogenes in japonica. The genome size of B. distachyon (∼272 Mb) [15] was smaller than that of rice (∼400 Mb) [28], and the corresponding orthologous region (107 kb) in B. distachyon was approximately 20% of the size of the rice syntenic region (553 kb). Annotation of the syntenic regions indicated that the reduced size was associated with the absence of ten genes, the high gene density (8.92 kb/gene) and low transposable element (TE) content (6.81%) in B. distachyon.
The genome size of maize is ∼2400 Mb [29], much larger than that of rice; however, the Ghd7 orthologous region of maize (230 kb) is ∼40% of rice (553 kb). The eight annotated genes/gene fragments present in maize were divided into three gene islands surrounded by blocks of repetitive sequences. Many of the intergenic retrotransposon blocks were nested insertions. The factors contributing to the unexpectedly short sequence in maize differed from B. distachyon: 13 genes were absent and a high content of repetitive elements (78.04%) were observed, especially LTR retrotransposons.
Large regions of non-conservation were observed between the sorghum and rice Ghd7 orthologous regions. The genome size of sorghum is ∼730 Mb [16]; however, the Ghd7 orthologous region is ∼1.91 Mb. The five complex regions divided the sorghum region into six parts (Figure 1). A series of tandem duplicated genes were identified in the five complex regions, and many of them are related to biotic and abiotic stress responses, such as F-box genes (Table S7).
Abundance and Variation of Transposable Elements Contributed to the Complexity of the Ghd7 Orthologous Regions
TEs were annotated as described in the experimental procedures (Tables 1 and S9, S10, S11). The Ghd7 orthologous regions showed a much higher levels of RNA TEs compared to the regions analyzed previously (Table S13), with only one exception of lower RNA TE content in B. distachyon (∼6%). Most of the RNA TEs are LTR retrotransposons (Table 1). The average insertion time of LTR retrotransposons from each species indicated that these LTRs spread after speciation (Table S10). In addition to the insertions, LTR retrotransposons were removed through illegitimate recombination (IR) and unequal homologous recombination (UR, converting LTR retrotransposons into solo-LTRs) in these orthologous regions [30]–[33]. However, we observed that the removal efficiency was not sufficient to counter the expansion caused by LTR retrotransposons in the Ghd7 orthologous regions in O. rufipogon, O. punctata, sorghum and maize (Table 2).
Table 1. Composition and sequence contribution of TEs within Ghd7 regions in Oryza species, B. distachyon, S. bicolor and Z. mays.
| JAA | IAA | GLAAA | GLUAA | RUFAA | NIVAA | PBB | OCC | AEE | BFF | BD | SB | ZM | |
| Class I | |||||||||||||
| -LTR | |||||||||||||
| –Ty1/Copia | 7.39 | 7.67 | 6.4 | 4.24 | 8.51 | 6.45 | 5.96 | 5.5 | 10.54 | 8.47 | 5.06 | 7.97 | 36.89 |
| –Ty3/Gypsy | 23.37 | 24.04 | 21.93 | 33.83 | 44.69 | 45.52 | 50.46 | 26.91 | 35.72 | 12.39 | 0.12 | 20.44 | 35.38 |
| -Intact LTR | 18.31 | 7.97 | 12.06 | 9.4 | 12.83 | 10.38 | 16.67 | 5.26 | 9.18 | 0 | 0 | 19.4 | 36.12 |
| -Non-LTR | |||||||||||||
| –LINE | 1.15 | 0.85 | 1.24 | 0.75 | 0.68 | 0.82 | 0.75 | 0.19 | 0.11 | 0.48 | 0.84 | 2.94 | 0.25 |
| –SINE | 0.54 | 0.54 | 0.62 | 0.3 | 0.25 | 0.27 | 0.06 | 0.2 | 0.03 | 0.13 | 0 | 0.035 | 0 |
| -Intact Non-LTR | 0.3 | 0.28 | 0.29 | 0 | 0.13 | 0.1 | 0 | 0.12 | 0 | 0 | 0 | 0.02 | 0 |
| Total (%) | 33.62 | 33.22 | 30.3 | 39.42 | 54.27 | 53.23 | 58.33 | 39.27 | 48.02 | 22.47 | 6.02 | 33.1 | 72.88 |
| Class II | |||||||||||||
| –MITE/Tourist | 1.62 | 1.51 | 1.51 | 1.7 | 1.24 | 1.14 | 0.69 | 1.05 | 0.13 | 1.98 | 0.65 | 0.802 | 1.18 |
| -MITE/Stowaway | 1.53 | 1.35 | 1.69 | 1.29 | 1.18 | 1.03 | 0 | 0.48 | 0.16 | 2.11 | 0 | 0.37 | 0 |
| –hAT | 0.94 | 0.91 | 1.02 | 0.4 | 0.29 | 0.06 | 0.08 | 0.11 | 0.75 | 0.51 | 0.14 | 0.67 | 0.58 |
| –En-Spm | 6.74 | 3.87 | 4.3 | 5.74 | 6.56 | 7.51 | 8.58 | 13.71 | 8.81 | 0.13 | 0 | 13.82 | 0.92 |
| –MuDR-IS905 | 2.5 | 2.31 | 2.2 | 2.38 | 1.36 | 1.44 | 1.64 | 5.99 | 0.59 | 1.12 | 0 | 0.112 | 2.08 |
| –Unclassified | 1.98 | 1.71 | 1.91 | 1.8 | 0.86 | 1.31 | 0.64 | 0.29 | 0.77 | 1.18 | 0 | 0.796 | 0.4 |
| -Intact DNA TE | 10.48 | 6.01 | 7.55 | 9.05 | 7.02 | 9.02 | 3.49 | 11.17 | 9.53 | 2.04 | 0 | 6.03 | 1.05 |
| Total (%) | 15.31 | 11.66 | 12.63 | 13.31 | 11.49 | 12.49 | 11.63 | 21.63 | 11.21 | 7.03 | 0.79 | 16.57 | 5.16 |
Table 2. The number of different categories of RNA TEs presents in Ghd7 orthologous regions.
| JAA | IAA | GLAAA | RUFAA | GLUAA | NIVAA | PBB | OCC | AEE | BFF | BD | SB | ZM | Mechanism | |
| Intact LTR | 9 | 7 | 7 | 12 | 5 | 8 | 14 | 2 | 9 | 0 | 1 | 44 | 19 | |
| Solo w/TSD | 16 | 13 | 14 | 10 | 10 | 8 | 12 | 7 | 10 | 2 | 0 | 5 | 2 | UR(Intra) |
| Solo w/o TSD | 3 | 4 | 3 | 3 | 3 | 3 | 1 | 5 | 1 | 0 | 0 | 0 | 3 | UR (Inter) |
| Intact LTR w/o TSD | 1 | 0 | 2 | 0 | 0 | 3 | 2 | 0 | 0 | 1 | 0 | 9 | 1 | UR(Intra) |
| Truncated Solo | 15 | 19 | 10 | 19 | 11 | 19 | 32 | 26 | 30 | 15 | 0 | 15 | 18 | UR(Inter)or UR(Intra)&IR |
| 3' LTR deleted | 1 | 5 | 3 | 6 | 7 | 4 | 11 | 6 | 10 | 1 | 0 | 1 | 11 | IR |
| 5' LTR deleted | 4 | 7 | 1 | 6 | 4 | 4 | 9 | 1 | 17 | 1 | 0 | 1 | 5 | IR |
| 5' and/or 3' partially deleted | 4 | 3 | 9 | 13 | 8 | 7 | 22 | 14 | 15 | 7 | 1 | 10 | 8 | IR |
| Recombination complex | 1 | 3 | 2 | 2 | 2 | 3 | 6 | 3 | 9 | 0 | 0 | 1 | 8 | UR (Inter) |
| Tendencya | C | C | C | E | C | C | E | C | C | C | E | E | E |
C means contraction and E means expansion.
Ghd7 is a Heterochromatic Gene in Rice
The Gypsy retrotransposon content of the Ghd7 orthologous regions was greater than Copia, especially in O. rufipogon, O. nivara and O. punctata (Table S12). However, this difference was not observed in B. distachyon or maize, suggesting the Ghd7 regions in different species may be located in distinct chromatin environments (Table S12). From our annotation information, the Ghd7 orthologous regions in most diploid Oryza species display features of heterochromatin compared to other well-characterized euchromatic regions (Figure S2, Tables S12 and S13). Previous cytological studies indicated that the Ghd7 locus in chromosome 7 of rice is likely to be sited within a high density, or at least a moderately condensed, region of chromatin [34], [35]. Furthermore, strict recombination suppression was detected in the Ghd7 region from QTL analysis in rice [25]. All of these results suggested that Ghd7 might be a heterochromatic gene in the genus Oryza [36]–[38].
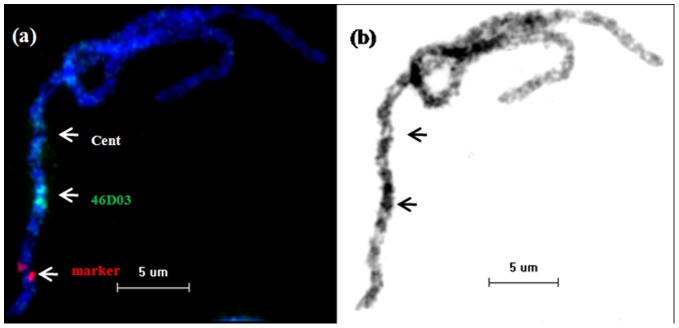
To confirm this possibility, we performed a fluorescence-in-situ-hybridization (FISH) experiment using the PAC (P1 artificial chromosome) clone 46D03, which contains the Ghd7 gene, as a probe. The result indicated that the PAC clone was located in a deeply stained region on the short arm of chromosome 7 of rice (Figure 2); indicating Ghd7 is a heterochromatic gene in rice.
Figure 2. Physical mapping of PAC 46D03 by pachytene chromosome FISH in japonica.
(a) The PAC 46D03 (green signal) is mapped to the heterochromatic region on the short arm of chromosome 7; “Cent” indicates the position of the centromere. The red signal is the marker for chromosome 7. (b) Inverted grayscale image of the same chromosome in (a). The black portion represents the heterochromatic region of chromosome 7 in rice.
The Evolutionary History of Ghd7 in the Grass Genomes
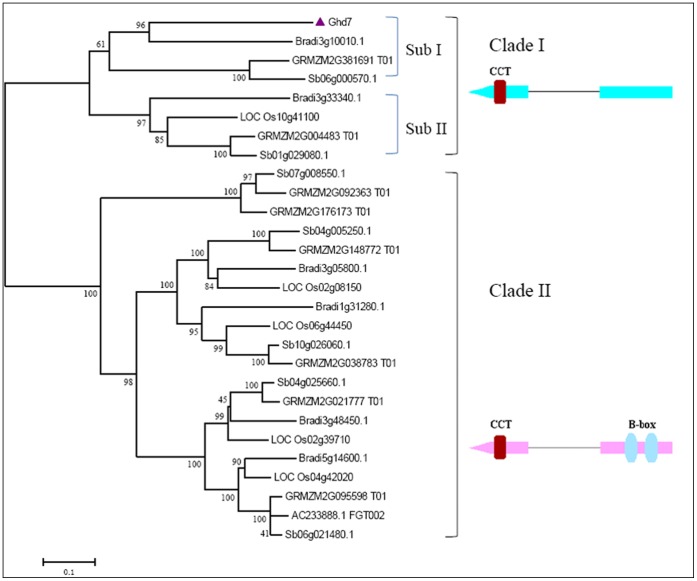
To trace the evolutionary history of Ghd7, the CCT domain-containing genes from rice, B. distachyon, sorghum and maize were identified by BLASTP based on homology to the Ghd7 CCT domain. A total of 28 homologous genes were identified (Table S14). As shown in Figure 3, the CCT domain-containing genes from different species can be classified into two clades by phylogenetic analysis. SMART and Pfam analyses indicated that genes in Clade I contain a CCT domain only, similar to Ghd7. In contrast, both a B-box and a CCT domain were detected for genes in Clade II, typified by the CCT family gene CO.
Figure 3. Unrooted phylogenetic tree for Ghd7 and its homologous genes from rice, B. distachyon, sorghum and maize.
The tree was built using the Neighbor-Joining method. The conserved motifs were shaded in different colors. The two clades can be distinguished by the motif distribution.
The Clade I can be further divided into two subgroups, each containing four genes from rice, B. distachyon, sorghum and maize. In subgroup II, LOC_Os10g41100, Bradi3g33340, Sb01g029080 and GRMZM2G004483_P01 are orthologous genes within a syntenic region. LOC_Os10g41100 and Ghd7 are duplicated genes in rice, and the gene duplication occurred prior to the divergence of rice, sorghum, maize and Brachypodium. In subgroup I, Ghd7 (LOC_Os07g15770), Bradi3g10010, Sb06g000570 and GRMZM2G381691_T01 were also defined as gene orthologs, as they met the following criteria: they formed a monophyletic clade; systematic information within this clade conformed to a generally accepted species trees; the bootstrap value was >55% [39]. These results indicated that Ghd7 has orthologs in non-Oryza species, but they are not located in syntenic regions (Table S15).
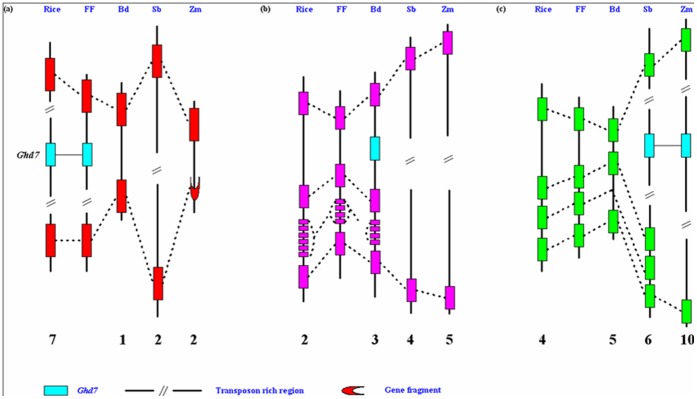
To investigate the origins of Ghd7 and its orthologs, we compared the corresponding orthologous regions from rice, B. distachyon, sorghum and maize. As a basal species of the genus Oryza, O. brachyantha (FF) had been subject to whole genome sequencing by our laboratory. To ensure a reasonable evolutionary gradient, the corresponding regions in O. brachyantha were included in the comparative analysis (Figure 4). Overall gene collinearity was observed in these genomic regions, but Ghd7 and its orthologs are located in three different syntenic regions in Oryza, B. distachyon and the Andropogoneae lineages (Figure 4), indicating Ghd7 was present prior to the divergence of rice, B. distachyon, sorghum and maize. Therefore, the Ghd7 orthologs must have moved to their current positions by undefined mechanisms in some species.
Figure 4. Comparative analysis of the corresponding orthologous regions from rice, O. brachyantha, B. distachyon, sorghum and maize.
(a) the Ghd7 orthologous regions; (b) the Bradi3g10010 orthologous regions; (c) the Sb06g000570 and GRMZM2G381691_T01 orthologous regions. FF: O. brachyantha; Bd: B. distachyon; Sb: S. bicolor; Zm: Z. mays. The number on the bottom depicts the chromosome number in each species.
The Mechanisms for Movements of Ghd7 and its Orthologs
A whole genome duplication event occurred ∼50–70 million years ago in grasses, prior to the divergence of major cereals [40], [41]. Many of the duplicated genes lost quickly after duplication by large-scale chromosomal rearrangements and deletions, leading to diploidization [42]–[45]. If the movement of Ghd7 or its orthologs was resulted from the differential fractionation of duplicated genes derived by the whole genome duplication, Ghd7 and its orthologs should be located on homoeologous chromosomes. Rice chromosome 3 is homoeologous to chromosome 7 where Ghd7 resides. However, the Ghd7 orthologs are located on chromosome 3 of B. distachyon, chromosome 6 of sorghum, and chromosome 10 of maize, which is homoeologous to chromosome 2 and chromosome 4 of rice, respectively (Figure 4), indicating Ghd7 and its orthologs are not located on homoeologous chromosomes. In fact, the duplicated region of the Ghd7 orthologous region could not be identified in rice at all, probably due to the heterochromatic features. Therefore, we conclude that the movements of Ghd7 and/or its orthologs were not resulted from the fractionation of duplicated genes following the whole genome duplication in grasses.
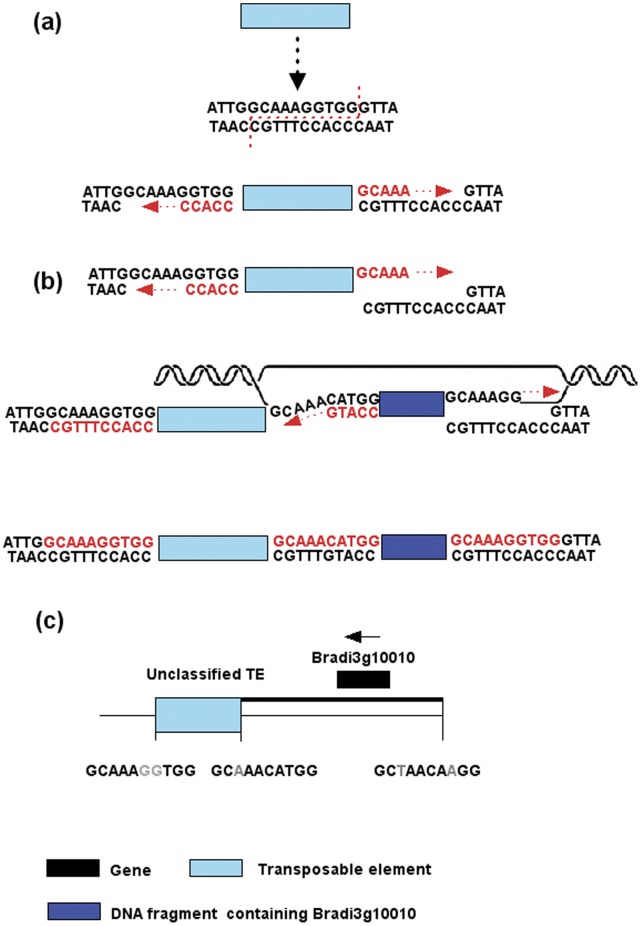
Based the model shown in Figure 5, we identified the evidence for the movement of Bradi3g10010, the Ghd7 ortholog in B. distachyon. We modeled that a DSB was caused by insertion of an unknown transposable element, and a DNA fragment containing Bradi3g10010 was used to fill the gap. A 10 bp target site duplication flanking the unknown transposable element and one side of Bradi3g10010 were detected (Figure 5c). Unfortunately, we could not detect clear evidence of Ghd7 movement in other species due to the high content of repetitive elements in the surrounding regions.
Figure 5. Molecular mechanism for the movement of Bradi3g10010.
(a) An unclassified intact transposable element cleaves the host DNA for insertion and the formation of target site duplication. (b) The fragment containing Bradi3g10010 was used to fill the double-stranded break. (c) The model for the movement of Bradi3g10010. This figure was modified according to Wicker et al. 2010.
Additional Gene Movements Mediated by DSB Repair in the Ghd7 Orthologous Regions
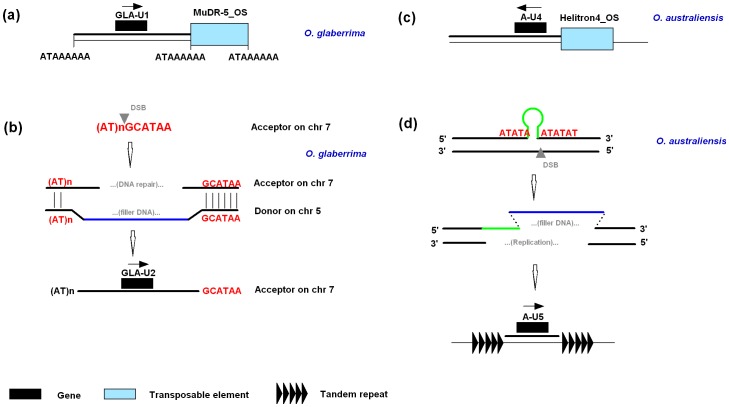
Four lineage-specific genes or gene fragments in the Ghd7 orthologous regions have signatures of DSB repair (Figure 6). GLA-U1 in O. glaberrima and A-U4 in O. australiensis were hypothesized to be moved to their current positions by repair of DSBs created by insertion of MuDR-5_OS and HELITRON4_OS, respectively (Figure 6a and 6c). Tandem repeat motifs are hotspots for recombination [46]. GLA-U2 located on chromosome 7 of O. glaberrima (the acceptor) and its homolog LOC_Os05g34200 located on chromosome 5 was flanked by an identical 6-bp sequence signature on one side (Figure 6b). In addition, simple sequence repeats surrounding A-U5 in O. australiensis suggested that the A-U5 movement was mediated by either unequal crossing-over or template slippage (Figure 6d). In particular, the donor regions of GLA-U1and GLA-U2 were also observed.
Figure 6. Examples of gene movements caused by DSB repair across the Ghd7 orthologous regions.
(a) The movement of GLA-U1 in O. glaberrima used a similar mechanism as Bradi3g10010 in B. distachyon (Figure 5). (b) The DSBs occur frequently in fragile sites, such as tandem repeats. A fragment containing GLA-U2 on chromosome 5 (donor) was used to fill the gap caused by DSB on chromosome 7 (acceptor) in O. glaberrima. The right borders for donor and acceptor are identical. (c) The fragment containing A-U4 might be captured by a Helitron element. (d) The A-U5 in O. australiensis is flanked by an array of tandem repeats on both sides. DSBs were possibly introduced during a template slippage of these tandem repeats, and the fragment containing A-U5 was used to fill the gap.
Gene Movements Mediated by Repeat Elements
The movements of four genes were found to be mediated by transposition of repeat elements (Figure S3). GLU-U7, A-U6 and B-U3 were embedded within different types of Pack-MULEs. GLU-U7, a GRF domain-containing gene, was evolved through capturing a gene fragment from elsewhere in the genome by TNR12/MuDR and inserted into the intron of pseudogene GLU-19. A-U6, a B-box-containing gene homologous to LOC_Os02g07930, moved to its current location by MERMITEA/MuDR. B-U3, which is located between B-8 and B-9 in O. brachyantha, was formed through transposition of OSTE1 carrying a DNA fragment from the second exon of LOC_Os10g34340. Finally, ZM7 moved to its current position by EnSpm-13_ZM, a CACTA family transposon in maize. Consistent with other studies, DNA TEs were found more frequently in capturing gene fragments through transposition [16], [17], [21].
Discussion
More and more species have been fully sequenced and provided insight into genome structural organization and evolution. Comparative analyses have shown the extensive conservation of gene order, but the loss or gain of genes or genomic segments can be easily detected in closely related species and are important for genome organization and evolution. For instance, comparison of grass genomes indicated gene expansion in the evolution of grass-specific-genes [15]. However, the gene loss in duplicated regions were implicated in returning a paleopolyploid to a diploid state after whole-genome duplication [16], [17]. Gene movement is a specific type of gene gain by gain of genes or genomic segments in the acceptor sites, but seems like “movement” due to the subsequent gene loss in the donor sites. The loss and gain of gene or genomic segment usually happened during the cell cycles when recombination occurs at double-strand break via homologous recombination (HR) or non-homologous end joining (NHEJ) pathways [46]-[49]. The mechanism was found to be involved in many human diseases [50], [51]. In addition, gain of genes or genomic segments could also be mediated by transposable element capture or retroposition. However, the frequency of these mechanisms are much lower than recombination [21].
Ghd7 is an agriculturally important gene that affects the number of grains per panicle, plant height and heading date. In our study, we found that Ghd7 was conserved throughout the genus Oryza, but absent in orthologous positions in non-Oryza species. However, Ghd7 was not deleted from the genome, but moved to different genomic locations in different subfamilies of grasses. In B. distachyon, features indicating Ghd7 movement was detected. DNA double-strand break (DSB) repair can take place at any position in the genome and is another mechanism for gene movement. Wicker et al. (2010) proposed a model that the DSB could be repaired with a foreign DNA fragment containing a gene after template slippage or an unequal crossing-over event through synthesis-dependent strand annealing. As a result of a DSB, which was induced by an unknown DNA TE, a DNA fragment containing Bradi3g10010 was used as a template for DSB repair by microhomology or non-homologous end joining [52]. Unfortunately, footprints for movements of Ghd7 in maize, sorghum, and foxtail millet have not been identified, possibly due to their being obscured by a high density of repetitive elements.
The whole genome analysis of rice and B. distachyon has indicated that “copy-and-paste” is the dominant duplication process for most non-collinear genes. The apparent movement of genes may result from subsequent deletions in the donor region [21]. The putative donor regions of Ghd7 and its orthologs were not observed in the species analyzed, suggesting that the original copy in the donor region was deleted.
The Ghd7 orthologs were identified in both short-day and long-day plants. The expressions of Ghd7 orthologs were detected by RT-PCR experiments in eight Oryza species grown under long-day conditions (Figure S4). Similar to Ghd7, the Ghd7 ortholog in maize (GRMZM2G381691_T01) was shown to regulate flowering time through determining photoperiod sensitivity [53]. We propose that the Ghd7 orthologs in sorghum (Sb06g000570) and Brachypodium (Bradi3g10010) have similar functions in controlling flowering time. Thus, the Ghd7 flowering pathway is not unique to rice; rather, it may regulate flowering properties in a wide range of grass species.
Heterochromatin is defined as densely coiled chromatin that generally replicates late during the S phase [38]. Low gene density, and large blocks of repetitive DNA, especially a high Gypsy content, are characteristics of heterochromatic regions [36]–[38]. Gypsy and Copia, which differ in the order of RT (reverse transcriptase) and INT (DDT integrase) in the POL, are two subfamilies of LTR retrotransposons [54]: Gypsy tends to insert into heterochromatin, while Copia inserts into euchromatin [55]. Most of the Ghd7 orthologous regions in Oryza species displayed features of characteristic heterochromatin, especially in O. rufipogon, O. nivara and O. punctata (Figure S2, Tables S9 and S11). In contrast, the low TE content and high gene density is suggestive of a euchromatic environment for the Ghd7 region of O. brachyantha. Heterochromatin is commonly characterized as silent chromatin; however, several hundreds of heterochromatic genes in Drosophila, plants and mammals are discovered recently and many of them have transcriptional activity [56]–[58]. In plants, agriculturally important heterochromatic genes have rarely been reported [59]–[61]. The expression of Ghd7 was detected in diploid Oryza species under long-day condition, suggestive of its conserved activity in a heterochromatic environment.
In summary, the dramatic position-shift of Ghd7 orthologs in the grass genomes and different allele distribution of Ghd7 in rice indicated plasticity of this agronomically important gene. The mechanism for Ghd7 movement in B. distachyon suggests that repetitive elements play an important role in gene and genome evolution in plants. Finally, as a heterochromatic gene, the regulation of Ghd7 might be an interesting model to understand the effect of chromatin environment on gene regulation in plants.
Materials and Methods
Materials, Growth Conditions and Gene Structure Verification
The seed dormancy of diploid Oryza species was broken by heat treatment (50–54°C for five days). Seeds were washed thoroughly and germinated on moist filter paper in petri dishes at 37°C. Seedlings were transferred to pots and placed in greenhouse seven days later following the methods described by Xue et al. (2008): neutral day-length conditions (12 h sunlight/day) for 30 days; then long-day conditions (15 h sunlight/day) for ten days. Finally, leaves were harvested and stored at −80°C. Total RNA was extracted with TRIzol reagent (TIANGEN, Cat#DP405-02). The cDNA was synthesized by reverse transcriptase (Promega, Cat.M170A). The PCR conditions were as follows: 5 min at 94°C; 35 cycles of 30 sec at 94°C, 30 sec at suitable primers temperature and 1 min at 72°C; 8 min at 72°C. Amplified products were cloned into a T vector (Promega, Cat#1360) and sequences were verified using an ABI 3730 automated capillary sequencer (Applied Biosystems).
Isolation of BAC Clones of Ghd7 Orthologous Regions from Wild Rice
Ten OMAP BAC libraries (O. nivara, O. rufipogon, O. glaberrima, O. glumaepatula, O. punctata, O. officinalis, O. australiensis, O. brachyantha, O. granulate and O. minuta) from nine diploid and one tetraploid species were used to isolate BAC clones of the Ghd7 orthologous regions. Primers for probes were designed for Ghd7 and its flanking genes. Each probe was hybridized to the wild rice BAC filters using protocols described in Arizona Genomics Institute Website (http://www2.genome.arizona.edu/research/protocols_bacmanual). Unfortunately, none of the positive clones were isolated in O. granulata (GG) and O. minuta (BBCC) because of insufficient coverage of the BAC clones in the target region. Validated BACs were purified using a QIAGEN Large-Construct Kit (Cat.No.12462) and sequenced using the next generation sequencing technology.
Sequencing the BAC Clones from the Ghd7 Orthologous Regions
For each species, paired-end sequencing libraries were constructed with an insertion size of approximately 500 bp and sequenced on Illumina Genome Analyzer II. Because of the high content of repetitive elements, the assembled sequences resulted in many scaffolds without ordering information. Therefore, we used the Roche/454 Genome Sequencer FLX Instrument as a complementary method to sequence these low quality BAC clones. Overlaps between neighboring BACs were determined using BLASTN, and the resultant pseudomolecules were constructed after careful inspection and verification of each overlap. The sequences of all BACs used in this analysis were deposited to the GeneBank datalibrary under the following accession numbers [GeneBank: JN873128-JN873135]. The sequences and related CDS (coding sequence) databases of japonica, indica, B. distachyon, sorghum and maize were downloaded from individual websites (http://rice.plantbiology.msu.edu; http://rice.genomics.org.cn/rice/index2.jsp; http://www.brachypodium.org; http://www.phytozome.net/sorghum; http://www.maizegdb.org).
Sequence Annotation of Protein-coding Genes and Transposable Elements
Sequences were annotated using the ab initio prediction programs FGENESH (http://www.softberry.com) for gene prediction [62]. In addition, candidate genes have to meet the following criteria: not transposon-related, and containing a known functional domain or having a homolog to known proteins or having a homolog at the syntenic position. All annotations were overlaid on individual BAC sequences and were visualized and edited using ACT v5 and Artemis [63]. The exon–intron structure of gene models were verified by aligning the genomic sequence with cDNAs or ESTs and experimental verification by sequencing RT-PCR products amplified using specific primers for each gene.
Transposable elements (TE) were identified by RepeatMasker (www.repeatmasker.org) and the signatures of each family of TE [54] using cross_match. The intact, truncated, recombination LTR elements and solo-LTRs were manually identified from the outputs of RepeatMasker and LTR_FINDER [64]. For sorghum and B. distachyon repeats analyses, the intact LTR retrotransposon sequences were isolated according to the results from LTR_FINDER, and then blasted against RepeatMasker TE libraries for subfamily classification.
Isolation of CCT Family Members from Rice, B. distachyon, Sorghum and Maize
The Ghd7 CCT domain protein sequence was used to search the rice, B. distachyon, sorghum and maize protein database using BLASTP with the following criterion: E-value ≤1e-5. The CDS of Ghd7 homolog candidates were aligned using MUSCLE [65] and imported into GeneDoc (http://www.nrbsc.org/gfx/gene-doc/index.html) for manual adjustment. The phylogenetic tree was built using MEGA4.0 [66]. The Neighbor-Joining method was used with the following parameters: pairwise deletion; bootstrap 1000 replicates and Kimura 2-parameter model. The domain of each gene was identified with SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1) and Pfam analysis. The dotplot analysis of Ghd7 homologous gene regions were carried out in Plant Genome Duplication Database [20] (http://chibba.agtec.uga.edu/duplication/index/dotplot).
Insertion Dating of LTR Retrotransposon
The insertion times of LTR retrotransposons were estimated by the divergence time (T) between two LTRs of single intact LTR retrotransposon [67], T = K/2r, where Ks refers to the distance between the two LTRs and r refers to the average substation rate. The two LTRs were aligned using MUSCLE [65]. The distance between each pair of LTRs (K) were calculated using the baseml program (runmode = 2; model = 4) described in PAML [68]. We used substitution rate (r) of 1.3×10-8 substitutions per site per year to estimate the divergence time of LTR as repeat elements were suggested to evolve much more rapid than coding regions [32], [69].
Analysis of the Molecular Mechanism of Gene Movement
Dot plot alignment was used to determine the borders of the repetitive elements in species without TE libraries. Target site duplications around genes and repetitive elements were identified using DOTTER [70]. Tandem repeat motifs around target genes were identified using Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.submit.options.html) and RepeatMasker.
Fluorescence In Situ Hybridization (FISH)
For japonica pachytene FISH, a PAC P0046D03 was used as probe (obtained from National Institute of Agrobiological Sciences in Japan; http://www.nias.affrc.go.jp/index_e.html). The chromosome 7 marker (BAC a0050F10) was obtained from Clemson University in USA (http://www.clemson.edu/). The FISH procedure applied to meiotic chromosomes was essentially the same as previously published protocols [71].
Supporting Information
Gene features of the Ghd7 regions in diploid Oryza species, B. distachyon , S. bicolor and Z. mays . Each gene is represented by a colored square. The yellow square with black borders indicates that the gene or gene fragment was captured by a transposable element. The numbers in purple rectangles represent the sequence length of the five complex regions in sorghum. The gene number is indicated above the black line and is summarized in Table S5. The “gap” indicates the non-overlapping regions. The abbreviation of each species is shown on the left.
(TIF)
A comparison of gene densities in the Ghd7 , Adh1 , Hd1 and Moc1 regions of Oryza species. O. glumaepatula is included for the comparative analysis of the Ghd7 region only.
(TIF)
Gene movements are mediated by repeat elements. The genes or gene fragments are in yellow. The gene number is in the yellow polygons. The name and types of transposable elements are shown. Target site duplications are shown by flanking the terminal inverted repeats.
(TIF)
RT-PCR results of Ghd7 orthologs in eight Oryza species.
(TIF)
BAC clones covering the Ghd7 regions in Oryza species.
(DOCX)
Genomic features of the Ghd7 orthologous regions.
(DOCX)
List of genes in the Ghd7 regions of O. sativa L. ssp. japonica .
(DOCX)
The gene models of O. sativa L. ssp. japonica derived from a comparative analysis.
(DOCX)
List of shared genes, unshared genes or gene fragments within the Ghd7 regions in Oryza species.
(DOCX)
List of genes in the corresponding orthologous region of B. distachyon .
(DOCX)
List of genes in the corresponding orthologous region of S. bicolor .
(DOCX)
List of genes in the corresponding orthologous region of Z. mays .
(DOCX)
Annotation of intact DNA transposable elements.
(DOCX)
List of intact retrotransposons, solo-LTRs and their conservation in Oryza species.
(DOCX)
List of intact DNA transposons and their conservation in Oryza species.
(DOCX)
Comparison of Gypsy and Copia content in the Ghd7 , Adh1 and Hd1 regions.
(DOCX)
Comparison of gene densities and TE contents in the Ghd7 , Adh1 , Hd1 and Moc1 regions.
(DOCX)
Accession number of CCT family genes from rice, B. distachyon , S. bicolor and Z. mays .
(DOCX)
Chromosomal location and sequence size of Ghd7 -related genes.
(DOCX)
Acknowledgments
We thank Drs. Yong-Bi Fu (Plant Gene Resources of Canada, Saskatoon Research Centre, Agriculture and Agri-Food Canada, Saskatoon, Canada) and Qian Qian (China National Rice Research Institute, Hangzhou, China) for their critical reading of the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant numbers 30770143, 30621001, 31171231) and the State Key Laboratory of Plant Genomics (grant number 2012B0301-02). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bennetzen JL (2007) Patterns in grass genome evolution. Curr Opin Plant Biol 10: 176–181. [DOI] [PubMed] [Google Scholar]
- 2. Ge S, Sang T, Lu BR, Hong DY (1999) Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci U S A 96: 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zou XH, Zhang FM, Zhang JG, Zang LL, Tang L, et al. (2008) Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biol 9: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu F, Ammiraju JS, Sanyal A, Zhang S, Song R, et al. (2009) Comparative sequence analysis of MONOCULM1-orthologous regions in 14 Oryza genomes. Proc Natl Acad Sci U S A 106: 2071–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ammiraju JS, Luo M, Goicoechea JL, Wang W, Kudrna D, et al. (2006) The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza . Genome Res 16: 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim H, Hurwitz B, Yu Y, Collura K, Gill N, et al. (2008) Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza . Genome Biol 9: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim H, San Miguel P, Nelson W, Collura K, Wissotski M, et al. (2007) Comparative physical mapping between Oryza sativa (AA genome type) and O. punctata (BB genome type). Genetics 176: 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma J, Wing RA, Bennetzen JL, Jackson SA (2007) Evolutionary history and positional shift of a rice centromere. Genetics 177: 1217–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang S, Gu Y, Singh J, Coleman-Derr D, Brar DS, et al. (2007) New insights into Oryza genome evolution: high gene colinearity and differential retrotransposon amplification. Plant Mol Biol 64: 589–600. [DOI] [PubMed] [Google Scholar]
- 10. Ammiraju JS, Zuccolo A, Yu Y, Song X, Piegu B, et al. (2007) Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza . Plant J 52: 342–351. [DOI] [PubMed] [Google Scholar]
- 11. Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, et al. (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16: 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ammiraju JS, Fan C, Yu Y, Song X, Cranston KA, et al. (2010) Spatio-temporal patterns of genome evolution in allotetraploid species of the genus Oryza . Plant J 63: 430–442. [DOI] [PubMed] [Google Scholar]
- 13. Ammiraju JS, Lu F, Sanyal A, Yu Y, Song X, et al. (2008) Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell 20: 3191–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sanyal A, Ammiraju JS, Lu F, Yu Y, Rambo T, et al. (2010) Orthologous comparisons of the Hd1 region across genera reveal Hd1 gene lability within diploid Oryza species and disruptions to microsynteny in Sorghum. Mol Biol Evol 27: 2487–2506. [DOI] [PubMed] [Google Scholar]
- 15. International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- 16. Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- 17. Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- 18. Wicker T, Mayer KF, Gundlach H, Martis M, Steuernagel B, et al. (2011) Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell 23: 1706–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lai J, Ma J, Swigonova Z, Ramakrishna W, Linton E, et al. (2004) Gene loss and movement in the maize genome. Genome Res 14: 1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang H, Bowers JE, Wang X, Ming R, Alam M, et al. (2008) Synteny and collinearity in plant genomes. Science 320: 486–488. [DOI] [PubMed] [Google Scholar]
- 21. Wicker T, Buchmann JP, Keller B (2010) Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res 20: 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freeling M, Lyons E, Pedersen B, Alam M, Ming R, et al. (2008) Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res 18: 1924–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang L, Bennetzen JL (2009) Distribution, diversity, evolution, and survival of Helitrons in the maize genome. Proc Natl Acad Sci U S A 106: 19922–19927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang W, Zheng H, Fan C, Li J, Shi J, et al. (2006) High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18: 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xue W, Xing Y, Weng X, Zhao Y, Tang W, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767. [DOI] [PubMed] [Google Scholar]
- 26. Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis . Plant Physiol 131: 1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M, Lu F, Jackson SA, Wing RA (2010) Dynamic Genome Evolution of Oryza, - A Genus-Wide Comparative Analysis. In DARWIN’S HERITAGE TODAY: Proceedings of the Darwin 200 Beijing International Conference: 76–83.
- 28. International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- 29. Wei F, Zhang J, Zhou S, He R, Schaeffer M, et al. (2009) The physical and genetic framework of the maize B73 genome. PLoS Genet 5: e1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bennetzen JL, Kellogg EA (1997) Do Plants Have a One-Way Ticket to Genomic Obesity? Plant Cell 9: 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Devos KM, Brown JK, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis . Genome Res 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P (2000) A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res 10: 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng Z, Buell CR, Wing RA, Gu M, Jiang J (2001) Toward a cytological characterization of the rice genome. Genome Res 11: 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohmido N, Fukui K, Kinoshita T (2010) Recent advances in rice genome and chromosome structure research by fluorescence in situ hybridization (FISH). Proc Jpn Acad Ser B Phys Biol Sci 86: 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y, Huang Y, Zhang L, Li Y, Lu T, et al. (2004) Structural features of the rice chromosome 4 centromere. Nucleic Acids Res 32: 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.John B (1988) The biology of heterochromatin. In: V R S, editor Heterochromatin: Molecular and Structural aspects Cambridge University Press P.1–128.
- 38. Brown SW (1966) Heterochromatin. Science 151: 417–425. [DOI] [PubMed] [Google Scholar]
- 39. Zhang J, Zhang YP, Rosenberg HF (2002) Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet 30: 411–415. [DOI] [PubMed] [Google Scholar]
- 40. Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A 101: 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tian CG, Xiong YQ, Liu TY, Sun SH, Chen LB, et al. (2005) Evidence for an ancient whole-genome duplication event in rice and other cereals. Yi Chuan Xue Bao 32: 519–527. [PubMed] [Google Scholar]
- 42. Xiong Y, Liu T, Tian C, Sun S, Li J, et al. (2005) Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol 59: 191–203. [DOI] [PubMed] [Google Scholar]
- 43. Wu Y, Zhu Z, Ma L, Chen M (2008) The preferential retention of starch synthesis genes reveals the impact of whole-genome duplication on grass evolution. Mol Biol Evol 25: 1003–1006. [DOI] [PubMed] [Google Scholar]
- 44. Wang X, Shi X, Hao B, Ge S, Luo J (2005) Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol 165: 937–946. [DOI] [PubMed] [Google Scholar]
- 45. Freeling M (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60: 433–453. [DOI] [PubMed] [Google Scholar]
- 46. Pfeiffer P, Goedecke W, Obe G (2000) Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 15: 289–302. [DOI] [PubMed] [Google Scholar]
- 47. Tan FJ, Hoang ML, Koshland D (2012) DNA Resection at Chromosome Breaks Promotes Genome Stability by Constraining Non-Allelic Homologous Recombination. PLoS Genet 8: e1002633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56: 1–14. [DOI] [PubMed] [Google Scholar]
- 49. Xu JH, Bennetzen JL, Messing J (2012) Dynamic gene copy number variation in collinear regions of grass genomes. Mol Biol Evol 29: 861–871. [DOI] [PubMed] [Google Scholar]
- 50. Zhang F, Gu W, Hurles ME, Lupski JR (2009) Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 10: 451–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gorbunova V, Levy AA (1999) How plants make ends meet: DNA double-strand break repair. Trends in Plant Science 4: 263–269. [DOI] [PubMed] [Google Scholar]
- 53. Ducrocq S, Giauffret C, Madur D, Combes V, Dumas F, et al. (2009) Fine mapping and haplotype structure analysis of a major flowering time quantitative trait locus on maize chromosome 10. Genetics 183: 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, et al. (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8: 973–982. [DOI] [PubMed] [Google Scholar]
- 55. Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, et al. (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, et al. (1999) Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468–2474. [DOI] [PubMed] [Google Scholar]
- 57. Weiler KS, Wakimoto BT (1995) Heterochromatin and gene expression in Drosophila. Annu Rev Genet 29: 577–605. [DOI] [PubMed] [Google Scholar]
- 58. Mudge JM, Jackson MS (2005) Evolutionary implications of pericentromeric gene expression in humans. Cytogenet Genome Res 108: 47–57. [DOI] [PubMed] [Google Scholar]
- 59. Guyot R, Cheng XD, Su Y, Cheng Z, Schlagenhauf E, et al. (2005) Complex organization and evolution of the tomato pericentric region at the FER gene locus. Plant Physiol 138: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, et al. (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36: 138–145. [DOI] [PubMed] [Google Scholar]
- 61. Fan C, Walling JG, Zhang J, Hirsch CD, Jiang J, et al. (2011) Conservation and purifying selection of transcribed genes located in a rice centromere. Plant Cell 23: 2821–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10: 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carver T, Berriman M, Tivey A, Patel C, Bohme U, et al. (2008) Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24: 2672–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35: W265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 67. SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20: 43–45. [DOI] [PubMed] [Google Scholar]
- 68. Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 69. Gaut BS, Morton BR, McCaig BC, Clegg MT (1996) Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL . Proc Natl Acad Sci U S A 93: 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sonnhammer EL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: 1–10. [DOI] [PubMed] [Google Scholar]
- 71. Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci U S A 92: 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene features of the Ghd7 regions in diploid Oryza species, B. distachyon , S. bicolor and Z. mays . Each gene is represented by a colored square. The yellow square with black borders indicates that the gene or gene fragment was captured by a transposable element. The numbers in purple rectangles represent the sequence length of the five complex regions in sorghum. The gene number is indicated above the black line and is summarized in Table S5. The “gap” indicates the non-overlapping regions. The abbreviation of each species is shown on the left.
(TIF)
A comparison of gene densities in the Ghd7 , Adh1 , Hd1 and Moc1 regions of Oryza species. O. glumaepatula is included for the comparative analysis of the Ghd7 region only.
(TIF)
Gene movements are mediated by repeat elements. The genes or gene fragments are in yellow. The gene number is in the yellow polygons. The name and types of transposable elements are shown. Target site duplications are shown by flanking the terminal inverted repeats.
(TIF)
RT-PCR results of Ghd7 orthologs in eight Oryza species.
(TIF)
BAC clones covering the Ghd7 regions in Oryza species.
(DOCX)
Genomic features of the Ghd7 orthologous regions.
(DOCX)
List of genes in the Ghd7 regions of O. sativa L. ssp. japonica .
(DOCX)
The gene models of O. sativa L. ssp. japonica derived from a comparative analysis.
(DOCX)
List of shared genes, unshared genes or gene fragments within the Ghd7 regions in Oryza species.
(DOCX)
List of genes in the corresponding orthologous region of B. distachyon .
(DOCX)
List of genes in the corresponding orthologous region of S. bicolor .
(DOCX)
List of genes in the corresponding orthologous region of Z. mays .
(DOCX)
Annotation of intact DNA transposable elements.
(DOCX)
List of intact retrotransposons, solo-LTRs and their conservation in Oryza species.
(DOCX)
List of intact DNA transposons and their conservation in Oryza species.
(DOCX)
Comparison of Gypsy and Copia content in the Ghd7 , Adh1 and Hd1 regions.
(DOCX)
Comparison of gene densities and TE contents in the Ghd7 , Adh1 , Hd1 and Moc1 regions.
(DOCX)
Accession number of CCT family genes from rice, B. distachyon , S. bicolor and Z. mays .
(DOCX)
Chromosomal location and sequence size of Ghd7 -related genes.
(DOCX)