Abstract
Background
Information on polymorphic DNA in organelle genomes is essential for evolutionary and ecological studies. However, it is challenging to perform high-throughput investigations of chloroplast and mitochondrial DNA polymorphisms. In recent years, EcoTILLING stands out as one of the most universal, low-cost, and high-throughput reverse genetic methods, and the identification of natural genetic variants can provide much information about gene function, association mapping and linkage disequilibrium analysis and species evolution. Until now, no report exists on whether this method is applicable to organelle genomes and to what extent it can be used.
Methodology/Principal Findings
To address this problem, we adapted the CEL I-based heteroduplex cleavage strategy used in Targeting Induced Local Lesions in Genomes (TILLING) for the discovery of nucleotide polymorphisms in organelle genomes. To assess the applicability and accuracy of this technology, designated ORG-EcoTILLING, at different taxonomic levels, we sampled two sets of taxa representing accessions from the Brassicaceae with three chloroplast genes (accD, matK and rbcL) and one mitochondrial gene (atp6). The method successfully detected nine, six and one mutation sites in the accD, matK and rbcL genes, respectively, in 96 Brassica accessions. These mutations were confirmed by DNA sequencing, with 100% accuracy at both inter- and intraspecific levels. We also detected 44 putative mutations in accD in 91 accessions from 45 species and 29 genera of seven tribes. Compared with DNA sequencing results, the false negative rate was 36%. However, 17 SNPs detected in atp6 were completely identical to the sequencing results.
Conclusions/Significance
These results suggest that ORG-EcoTILLING is a powerful and cost-effective alternative method for high-throughput genome-wide assessment of inter- and intraspecific chloroplast and mitochondrial DNA polymorphisms. It will play an important role in evolutionary and ecological biology studies, in identification of related genes associated with agronomic importance such as high yield and improved cytoplasmic quality, and for identifying mitochondrial point mutations responsible for diseases in humans and other animals.
Introduction
The fields of ecology and phylogeny are currently experiencing a renaissance spurred by the rapid development of molecular detection techniques. By detecting genetic variation in both nuclear and organelle genomes, molecular markers have made a profound and significant contribution to studies of evolution, domestication, speciation, evolution of genomes, genetic diversity, population structure, levels of gene flow, patterns of historical biogeography and analyses of parentage assignments, genetic variability and inbreeding [1]–[6]. Mitochondria and chloroplasts are important organelles in eukaryotic organisms, and both genomes contain two vital sets of genes [7]. Plant mitochondrial (mt) DNAs are extremely variable in size (200–2400 kb) [8], [9], whereas animal mtDNAs are essentially invariant in gene order among all vertebrates [10]. Chloroplasts (cps) contain their own small genomes, which averages 120 to 200 kilobase pairs (kb) among almost all chloroplast-containing organisms [11]. The plant chloroplast genome shares many features with animal mtDNA and the two have been referred to as ‘natural counterparts’ [12]. In animals, mtDNA is characterized by a small size, high copy number, relatively conserved gene order, ready availability of primers and rapid substitution rates [13], whereas in plants, the chloroplast genome is associated with a conserved gene order, widespread availability of primers and a general lack of heteroplasmy and recombination [14]. As a result, the nonrecombinant, uniparentally inherited and effectively haploid nature of chloroplast and mitochondrial genomes makes them useful tools for studies on plant and animal evolution, respectively [14]. Chloroplast genomes are predominantly maternally inherited (mainly transmitted through the embryos of seeds) and so can reveal maternal lineages [15]–[18], enabling divergent patterns of variation to be detected in these genomes compared with those revealed by nuclear markers. For example, investigation of chloroplast genomes can be used to document the maternal parent of hybrid plants [19], define organelle haplotypes [20] or detect introgression [21], [22].
In order to detect informative polymorphisms for phylogenetic studies, a large number of molecular technologies have been developed for assessing chloroplast and mitochondrial genomes. Conventional methods include DNA restriction mapping [23], [24], the use of hybridization-based restriction fragment length polymorphisms (RFLP) [25], PCR-RFLP [26]–[28] and nucleotide sequence analysis [29], [30]. However, each of these methods have some disadvantages, including low resolution, labor intensity and the requirement for a large amount of isolated DNA. For instance, although Chloroplast microsatellites (cpSSR) have proven to be useful markers for gaining insights into the genetic relationships of closely related species and populations [31], [14], they are limited to study of closely related taxa where there are no or few nucleotide substitutions, especially in the coding regions of the chloroplast genome [32].
Because single-base substitutions and small insertions and deletions (INDELs) are the most common forms of genetic variation in organelle genomes, nucleotide sequence analysis of specific chloroplast and mitochondrial genes has been widely used in phylogenetic and ecological studies. Several key genes, such as chloroplast rbcL, matK and accD, mitochondrial atp6 and a mitochondrial fragment COX1 play important roles in clarifying phylogenetic relationships amongst plants [33]–[37]. However, rbcL, matK and accD only represent about 2.9% of the genome. Next-generation sequencing technologies, such as Roche/454, Illumina/Solexa and Life/APG are able to generate large volumes of sequence data at relatively low cost [38], enabling complete coverage of organelle genomes. However, at present this is only feasible for relatively low level sampling of individuals [39], [40]. This has led to a debate over the relative worth of taxon sampling (investigating many lineages) compared with site sampling (investigating many sites from many genes from a few crucial lineages) in building phylogenetic trees from sequence data [41], [42]. Thus there is a need for new methods that have the features of high resolution, high throughput, and ease of use, accuracy and cost effectiveness.
TILLING (Targeting Induced Local Lesions in Genomes) and EcoTILLING were developed as a means for high-throughput discovery of DNA polymorphisms in nuclear genomes in EMS-mutagenized [43]–[47] and natural populations [48]. The initial development of these methods employed the mismatch-specific endonuclease CEL I to discover point mutations in genes, in conjunction with PCR-based screening [49], [50]. Subsequently there have been modifications of the approach using other detection methods, including the use of new generation sequencing coupled with multidimensional pooling for the identification of rare alleles in populations of rice and wheat [51].
Given the ability of CEL1 and a PCR-based strategy to detect single nucleotide polymorphisms (SNPs), we decided to explore whether this approach could be applied to organelle genomes. We therefore developed a method, designated “ORG- EcoTILLING”, for high-resolution, high-throughput detection of multiple polymorphisms in chloroplast and mitochondrial genes.
In order to assess the applicability and accuracy of ORG-EcoTILLING we sampled two sets of plant taxa representing different levels of variation within the Brassicaceae. The Brassicaceae include approximately 340 genera and 3350 species [52], of which the model plant Arabidopsis thaliana and domesticated species such as oilseed rape, Chinese cabbage, broccoli, turnip, radish, mustard, and other Brassica crops, are perhaps the most familiar [53]. Plants within the Brassicaceae have therefore been regarded as ideal materials for the study of evolutionary relationships [54], [55]. This study aimed to (1) develop a strategy for ORG-EcoTILLING to discriminate genetic SNPs and INDELs in chloroplast and mitochondrial genes of numerous Brassicaceae plant accessions; (2) assess the accuracy of ORG-EcoTILLING for the identification of cytoplasmic DNA Polymorphism; and (3) provide an effective, low-cost, high-throughput technology for identification of genes associated with agronomic importance such as high yield and improved cytoplasmic efficiency, analyzing phylogenetic relationships in plants, and identifying point mutations of mitochondrial genes responsible for diseases in humans and other animals.
Materials and Methods
Plant material
To evaluate the applicability and accuracy of ORG-EcoTILLING for detecting DNA variation in chloroplast genes at different taxonomic levels, two sets of plant specimens were sampled. The first set, composed of 91 representative accessions from 45 species and 29 genera of seven tribes (Trib. Brassiceae Hayek, Trib. Sisymbrieae DC, Trib. Matthioleae O.E. Schulz, Trib. Alysseae Gren. Et Godr, Trib. Arabideae DC, Trib. Hesperideae Prantl, and Trib. Lepidieae DC) (Table 1) was used to assess the applicability and accuracy at intertribal, intergeneric and interspecific levels. The second set sampled 90 accessions of cultivars of Brassica napus, and three accessions each of B. rapa and B. oleracea (Table 2), which were used to evaluate the applicability and accuracy of this method at intraspecific and interspecific levels.
Table 1. 91 accessions from Cruciferae used for ORG- EcoTILLING.
| Sample code | Materials name | Origin | Trib | Genus | Species |
| A1 | Changyouxiaoheiyoucai | China | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A2 | Xishui | China | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A3 | Guangfuqing | China | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A4 | Baichengbaiyoucai | China | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A5 | Ling chuan youcai | China | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A6 | Qu xu | China | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A7 | Jiningtianjinlv | China | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A8 | Piaoerbai | China | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A9 | Niuyezhongshucaixin | China | Trib. Brassiceae Hayek | Brassica | B. parachinensis L.H.Bailey |
| A10 | Wutacai | China | Trib. Brassiceae Hayek | Brassica | B. narinosa |
| A11 | Taicai | China | Trib. Brassiceae Hayek | Brassica | B.rapa |
| A12 | Lelingwuqing | China | Trib. Brassiceae Hayek | Brassica | B.rapa |
| A13 | Majrova “Petrowski” | Sweden | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A14 | Majrova “Purple Top Milan” | Sweden | Trib. Brassiceae Hayek | Brassica | B. rapa |
| A15 | Tradkal“Jersey Walking Stick” | Sweden | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A16 | Kalrot Lanttu “Wilhelmsburger” | Sweden | Trib. Brassiceae Hayek | Brassica | B. napus |
| A17 | Majrova Nauris “Goldhall” | Sweden | Trib. Brassiceae Hayek | Brassica | B. napus |
| A18 | Midasi | Canada | Trib. Brassiceae Hayek | Brassica | B. napus |
| A19 | H47 | Russia Union | Trib. Brassiceae Hayek | Brassica | B. napus |
| A20 | Qikuzhen | Japan | Trib. Brassiceae Hayek | Brassica | B. napus. |
| A21 | Zhongshuan-4 | China | Trib. Brassiceae Hayek | Brassica | B. napus. |
| A22 | Zhongshuan-4NSA | China | Trib. Brassiceae Hayek | Brassica | B. napus |
| A23 | Baoziganlan | China | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A24 | Ziganlan | China | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A25 | Shimianlianhuabai | China | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A26 | Baipilan | China | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A27 | Tuanyexiaohuacai | China | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A28 | Lilvqinghuacai | China | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A29 | Zhonghuajielan | China | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A30 | Yeshengganlan | China | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A31 | Yuyiganlan | China | Trib. Brassiceae Hayek | Brassica | B. oleracea |
| A32 | Nanfangren | Russia Union | Trib. Brassiceae Hayek | Brassica | B. juncea |
| A33 | Bangcai02 | China | Trib. Brassiceae Hayek | Brassica | B. juncea |
| A34 | Banyedatoucai | China | Trib. Brassiceae Hayek | Brassica | B. juncea |
| A35 | Donghaigaojiaofengweicai | China | Trib. Brassiceae Hayek | Brassica | B. juncea |
| A36 | Pusa Bold | India | Trib. Brassiceae Hayek | Brassica | B. juncea |
| A37 | CMS (Mri) | India | Trib. Brassiceae Hayek | Brassica | B. juncea |
| A38 | Oiebra | Sweden | Trib. Brassiceae Hayek | Brassica | B. nigra |
| A39 | Heijie | Germany | Trib. Brassiceae Hayek | Sinapis L. | B. nigra |
| A40 | Xinjiangyeshengyoucai A | China | Trib. Brassiceae Hayek | Sinapis L. | S. arvensis |
| A41 | Xinjiangyeshengyoucai B | China | Trib. Brassiceae Hayek | Sinapis L. | S. arvensis |
| A42 | Baijie | China | Trib. Brassiceae Hayek | Sinapis L. | S. alba |
| A43 | Maojiaocaizi | China | Trib. Brassiceae Hayek | Brassica | S. alba |
| A44 | Ethiopia jie | Ethiopia | Trib. Brassiceae Hayek | Brassica | B. carinata |
| A45 | Huangziaijie | Ethiopia | Trib. Brassiceae Hayek | Brassica | B. carinata |
| A46 | 77-1304 | USA | Trib. Brassiceae Hayek | Brassica | B. carinata |
| A47 | 77-1305 | USA | Trib. Brassiceae Hayek | Brassica | B. carinata |
| A48 | 88-212463 | USA | Trib. Brassiceae Hayek | Brassica | B. carinata |
| A49 | 88-203221 | USA | Trib. Brassiceae Hayek | Brassica | B. carinata |
| A50 | Yuewangluobozi | China | Trib. Brassiceae Hayek | Raphanus L. | R. sativus |
| A51 | Lanhuazi | China | Trib. Brassiceae Hayek | Raphanus L. | R. sativus |
| A52 | Luobozhi (Ogu CMS) | Japan | Trib. Brassiceae Hayek | Raphanus L. | R. sativus |
| A53 | Eryuelan | China | Trib. Brassiceae Hayek | Orychophragmus Bunge | C. Violaceua |
| A54 | Celezhahong | China | Trib. Brassiceae Hayek | Eruca Mill | E. sativa |
| A55 | Yunjie | China | Trib. Brassiceae Hayek | Eruca Mill | E. sativa |
| A56 | Hancai | China | Trib. Arabideae DC | Rorippa | R. india |
| A57 | INIA 0863-66 | Spain | Trib. Brassiceae Hayek | Moricandia DC | M. arvensis |
| A58 | Banlangen | China | Trib. Lepidieae DC | Isatis | I. indigotica |
| A59 | Ziluolan | China | Trib. Matthioleae O.E.Schulz | Matthiola | M. incana |
| A60 | Tri | Sweden | Trib. Arabideae DC | Arabidopsis | A. thaliana |
| A61 | KAS | Sweden | Trib. Arabideae DC | Arabidopsis | A. thaliana |
| A62 | Bsch | Sweden | Trib. Arabideae DC | Arabidopsis | A. thaliana |
| A63 | mr | Sweden | Trib. Arabideae DC | Arabidopsis | A. thaliana |
| A64 | Col | Sweden | Trib. Arabideae DC | Arabidopsis | A. thaliana |
| A65 | Zihuabangguojie | China | Trib. Hesperideae Prantl | Sterigmostemum M.Bieb | S. matthioloides |
| A66 | Yiguolan | China | Trib. Matthioleae O.E. Schulz | Diptychocarpus Trautv | D. strictus |
| A67 | Choujie | China | Trib. Lepidieae DC | Coronopus J.G.Zinn | C. didymus |
| A68 | Dasuanjie | China | Trib. Sisymbrieae DC. | Sisymbrium L. | S. altissimum |
| A69 | Duoxingdasuanjie | China | Trib. Sisymbrieae DC. | Sisymbrium L. | S. polymorphum |
| A70 | Xianghuajie | China | Trib. Hesperideae Prantl | Hesperis L. | H. trichosepala |
| A71 | Ququhua | China | Trib. Lepidieae DC | Iberis L. | I. amara |
| A72 | Guizhuxiang | China | Trib. Hesperideae Prantl | Cheiranthus L. | Ch. cheiri |
| A73 | Hancai | China | Trib.Arabideae DC | Rorippa Scop. | R. india |
| A74 | Keshigaoyuanjie | China | Trib.Arabideae DC | Christolea Camb. | Ch. kashgarica |
| A75 | Doubancai | China | Trib. Arabideae DC | Nasturium R.Br | N. officinale |
| A76 | Lizijie | China | Trib. Matthioleae O. E. Schulz | Chorispora DC | Ch. tenella |
| A77 | Xianyiruijie | China | Trib. Arabideae DC | Dimorphostemon Kitag | D. glandulosus |
| A78 | Ganxinnianzhujie | China | Trib. Sisymbrieae DC. | Torularia (Coss)O.E.Schulz | T. korolkowii |
| A79 | Qiganjie | China | Trib. Arabideae DC | Turritis L. | T. glabra |
| A80 | Sejie | China | Trib. Hesperideae Prantl | Malcolmia R.Br | M. africana |
| A81 | Suimijie | China | Trib. Arabideae DC | Cardamine L. | C. impatiens |
| A82 | Songlan | China | Trib. Lepidieae DC | Iberis L. | S. altissimum |
| A83 | Silengjie | China | Trib. Hesperideae Prantl | Goldbachia DC | G. laevigata |
| A84 | Xiaoyesuimijie | China | Trib. Arabideae DC | Cardamine L. | C. microzyga |
| A85 | Tuanshanjie | China | Trib. Alysseae Gren.et Godr | Berteroa DC | B. incana |
| A86 | Tiaoyetingjie | China | Trib. Alysseae Gren.et Godr | Alyssum L. | A. linifolium |
| A87 | Xianguojie | China | Trib. Brassiceae Hayek | Conringia Adans | C. planisiliqua |
| A88 | Xiangxueqiu | China | Trib. Alysseae Gren.et Godr | Lobularia Desv. | L. maritima |
| A89 | Yanjie | China | Trib. Sisymbrieae DC. | Thellungiella | T. salsuginea |
| A90 | Xiaoguoyamajie | China | Trib. Sisymbrieae DC. | Camelina Crantz | C. microcarpa |
| A91 | Ziluolan | China | Trib. Matthioleae O.E.Schulz | Matthiola R.Br | M. incana |
Table 2. 90 accessions of cultivars of B. napus, and each of 3 accessions of B. rapa and B. oleracea used for ORG-EcoTILLING.
| Sample code | Materials name | Ploidy level | Genome | Origin |
| B1 | Midas | 4× | AACC (n = 19) | Canada |
| B2 | Oro | 4× | AACC (n = 19) | Canada |
| B3 | Major | 4× | AACC (n = 19) | France |
| B4 | Primor | 4× | AACC (n = 19) | France |
| B5 | Yaojin Rape | 4× | AACC (n = 19) | Italy |
| B6 | Marnoo | 4× | AACC (n = 19) | Australia |
| B7 | Ujfertadi | 4× | AACC (n = 19) | Hungary |
| B8 | SavariA | 4× | AACC (n = 19) | Hungary |
| B9 | Expander | 4× | AACC (n = 19) | Germany |
| B10 | Ledos | 4× | AACC (n = 19) | Germany |
| B11 | H47 | 4× | AACC (n = 19) | Russia |
| B12 | H51 | 4× | AACC (n = 19) | Russia |
| B13 | Janpol | 4× | AACC (n = 19) | Poland |
| B14 | Start | 4× | AACC (n = 19) | Poland |
| B15 | Mikado | 4× | AACC (n = 19) | England |
| B16 | P20 | 4× | AACC (n = 19) | England |
| B17 | Lingot | 4× | AACC (n = 19) | England |
| B18 | Wipot | 4× | AACC (n = 19) | Norway |
| B19 | Regent | 4× | AACC (n = 19) | Canada |
| B20 | Tower | 4× | AACC (n = 19) | Canada |
| B21 | Shiralee | 4× | AACC (n = 19) | Australia |
| B22 | Viking | 4× | AACC (n = 19) | Denmark |
| B23 | Cobra | 4× | AACC (n = 19) | Germany |
| B24 | Parter | 4× | AACC (n = 19) | Germany |
| B25 | Falcon | 4× | AACC (n = 19) | Germany |
| B26 | Nevin | 4× | AACC (n = 19) | France |
| B27 | Samouran | 4× | AACC (n = 19) | France |
| B28 | Roman-1 | 4× | AACC (n = 19) | Netherlands |
| B29 | Tornado | 4× | AACC (n = 19) | Sweden |
| B30 | Legend | 4× | AACC (n = 19) | Sweden |
| B31 | Grant | 4× | AACC (n = 19) | Sweden |
| B32 | Celebra | 4× | AACC (n = 19) | Canada |
| B33 | Triton | 4× | AACC (n = 19) | Canada |
| B34 | Profit | 4× | AACC (n = 19) | Canada |
| B35 | Startigh | 4× | AACC (n = 19) | Sweden |
| B36 | Bounty | 4× | AACC (n = 19) | Sweden |
| B37 | Garrison | 4× | AACC (n = 19) | Sweden |
| B38 | Gcsunder | 4× | AACC (n = 19) | Germany |
| B39 | Disamant | 4× | AACC (n = 19) | Germany |
| B40 | Mar | 4× | AACC (n = 19) | Poland |
| B41 | Star | 4× | AACC (n = 19) | Denmark |
| B42 | Shengli Qinggen | 4× | AACC (n = 19) | Shanghai, China |
| B43 | Jiuer rape | 4× | AACC (n = 19) | Zhejiang, China |
| B44 | Hanfeng-1 | 4× | AACC (n = 19) | Shanxi, China |
| B45 | Huayou-13 | 4× | AACC (n = 19) | Wuhan, China |
| B46 | Aijia zao | 4× | AACC (n = 19) | Sichuan, China |
| B47 | Southeast-302 | 4× | AACC (n = 19) | Sichuan, China |
| B48 | Yunyou-49 | 4× | AACC (n = 19) | Yunnan, China |
| B49 | Qingyou-6 | 4× | AACC (n = 19) | Qinghai, China |
| B50 | Nonglin-18 | 4× | AACC (n = 19) | Japan |
| B51 | F01*J6 1-1 | 4× | AACC (n = 19) | Hubei, China |
| B52 | Ganyou-5 | 4× | AACC (n = 19) | Wuhan, China |
| B53 | Zhongyou-821 | 4× | AACC (n = 19) | Wuhan, China |
| B54 | Xiangyou-5 | 4× | AACC (n = 19) | Hunan, China |
| B55 | Dong-Hae23 | 4× | AACC (n = 19) | Japan |
| B56 | Ganpol | 4× | AACC (n = 19) | Zhejiang, China |
| B57 | Norin16 | 4× | AACC (n = 19) | Japan |
| B58 | Zheyouyou-2 | 4× | AACC (n = 19) | Zhejiang, China |
| B59 | Yuyou-2 | 4× | AACC (n = 19) | Henan, China |
| B60 | Zhongyoudijie-1 | 4× | AACC (n = 19) | Wuhan, China |
| B61 | Qingyou-12 | 4× | AACC (n = 19) | Qinghai, China |
| B62 | Qikuzhen | 4× | AACC (n = 19) | Japan |
| B63 | Zhongshuang-4 | 4× | AACC (n = 19) | Wuhan, China |
| B64 | ISN-705 | 4× | AACC (n = 19) | India |
| B65 | H0302 | 4× | AACC (n = 19) | Hubei, China |
| B66 | 2000-5 | 4× | AACC (n = 19) | Hubei, China |
| B67 | H9944 | 4× | AACC (n = 19) | Hubei, China |
| B68 | 05 Za-V2 | 4× | AACC (n = 19) | Chongqing, China |
| B69 | 01 Za-654 | 4× | AACC (n = 19) | Sichuan, China |
| B70 | HY8 | 4× | AACC (n = 19) | Jiangsu, China |
| B71 | Youyan-10 | 4× | AACC (n = 19) | Guizhou, China |
| B72 | Qianyou-20 | 4× | AACC (n = 19) | Guizhou, China |
| B73 | 6766 | 4× | AACC (n = 19) | Hubei, China |
| B74 | H0202 | 4× | AACC (n = 19) | Hubei, China |
| B75 | Zheyou-5002 | 4× | AACC (n = 19) | Zhejiang, China |
| B76 | Zhongyouza-2 | 4× | AACC (n = 19) | Hubei, China |
| B77 | Za-839 | 4× | AACC (n = 19) | Hunan, China |
| B78 | Hongyou-3 | 4× | AACC (n = 19) | Jiangsu, China |
| B79 | Zashuang-5 | 4× | AACC (n = 19) | Henan, China |
| B80 | 7633 | 4× | AACC (n = 19) | Shanxi, China |
| B81 | Qinyou-7 | 4× | AACC (n = 19) | Shanxi, China |
| B82 | Rape-23 | 4× | AACC (n = 19) | Shanghai, China |
| B83 | Ganyou-4 | 4× | AACC (n = 19) | Hubei, China |
| B84 | Huayou-3 | 4× | AACC (n = 19) | Hubei, China |
| B85 | Huayou-8 | 4× | AACC (n = 19) | Hubei, China |
| B86 | ChuannongChangjiao | 4× | AACC (n = 19) | Sichuan, China |
| B87 | Chuanyou-7 | 4× | AACC (n = 19) | Sichuan, China |
| B88 | Luzhou-5 | 4× | AACC (n = 19) | Sichuan, China |
| B89 | Nanyang-41 | 4× | AACC (n = 19) | Henan, China |
| B90 | F11*J2 1-1 | 4× | AACC (n = 19) | Hubei, China |
| B91 | Fenyang rape | 2× | AA(n = 10) | Shanxi, China |
| B92 | Xishui rape | 2× | AA(n = 10) | Hubei, China |
| B93 | Wenjiang Qixingjian | 2× | AA(n = 10) | Sichuan, China |
| B94 | 2006 Holland-2 | 2× | CC (n = 9) | Netherlands |
| B95 | Chinese kale yellow/brown seeds | 2× | CC (n = 9) | Guangdong, China |
| B96 | C2-7 | 2× | CC (n = 9) | Spain |
ORG-EcoTILLING procedure
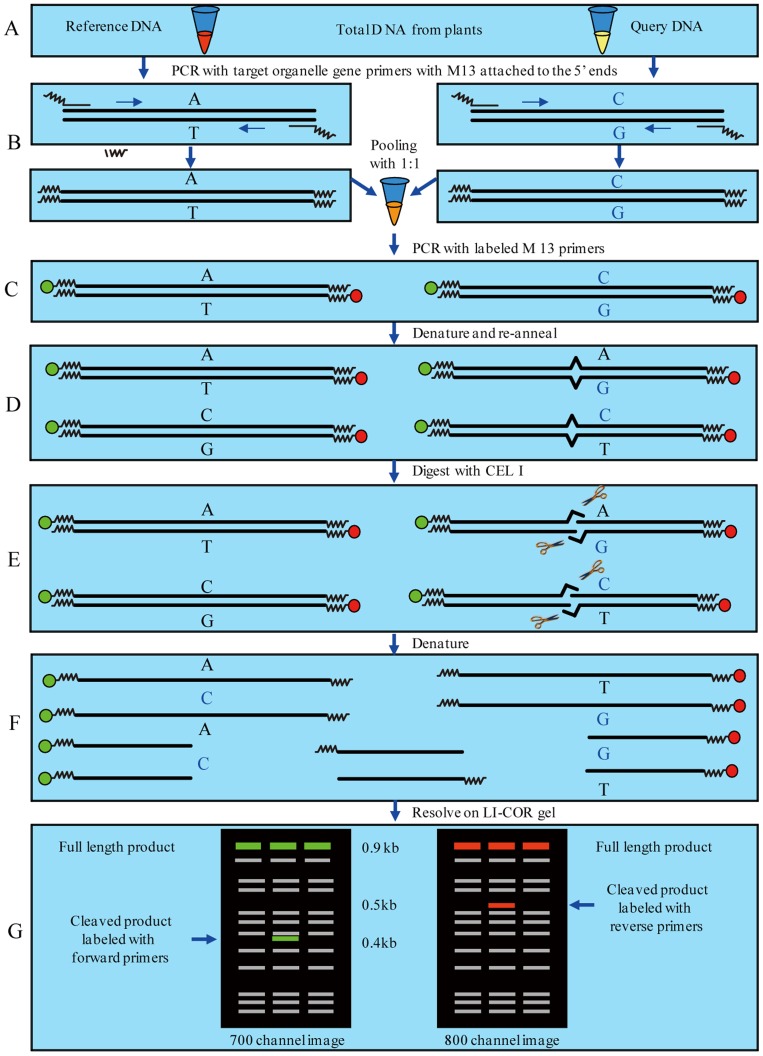
The ORG-EcoTILLING procedure includes six basic steps (Figure 1), modified from Haughn and Gilchrist (2006) [56]. Firstly, total DNA is extracted from above 181 accessions from the natural population and is normalized. Query DNA and reference DNA are by amplified gene-specific primers with M13F and M13R adaptors, respectively. Then the PCR products of query DNA and reference DNA are mixed in a 1∶1 ratio. The mixture is amplified using a forward primer with 700 nm dye label and a reverse primer with an 800 nm dye label attached to the 5′ ends. Thirdly, the PCR products are heated and cooled to form heteroduplexes between query DNA and reference DNA in the pool. Finally, any mismatches (SNPs or small INDELs) are detected by CEL I (a mismatch endonuclease) which cleaves the heteroduplex into two products, which are detected in the 700 and 800 dye channel of a LI-COR DNA analyzer. The additive size of the two cleaved fragments should equal the total length of the entire product. Because the nuclease cleaves either of the two strands arbitrarily, cleavage products can be detected in both the IRD700 and IRD800 channels of the gel image.
Figure 1. Overview of the ORG-EcoTILLING procedure for detecting DNA polymerphisms in Chloroplast and Mitochondrial genes, modified from Haughn and Gilchrist (2006).
A, Total DNA is extracted from plants. B, primers in which a sequence from the universal primer M13F (CACGACGTTGTAAAACGAC) and M13R (GGA TA A CAAT TTCACACAGG) are added to the 5′ end of a target organelle gene primers as adaptors, and query DNA and the reference DNA are amplified, respectively. Then the PCR products of query DNA and reference DNA with the ratio of one to one are pooled. C, the mixture is amplified using the universal primers tagged fluorescently. The forward and reverse primers are labeled with different fluorophors (IRD700 and IRD800) that each label a different end of the PCR fragments. D, the amplified products are denatured by heating and then allowed to cool slowly so that they randomly re-anneal. Heteroduplex molecules form when single DNA strands of mutant and wild-type PCR products anneal together. E, these heteroduplexes become targets for a single-strand-specific nuclease as found in celery juice extract. The nuclease cleaves these heteroduplex fragments at one of the two strands, 3′ to the site of the mismatch in the DNA. F, the PCR products that retain one of the labeled primers can be detected on polyacrylamide denaturing LI-COR gels. G, because the nuclease cleaves either of the two strands randomly, cleavage products can be detected in both the IRD700 and IRD800 channels of the gel image. The position of the mutation within the PCR amplicon can be calculated from the size of the fragments carrying the IRD700-labeled forward primer and the IRD800-labeled reverse primer. The additive size of the cleaved fragments should equal the total length of the entire product.
DNA extraction
Total DNA was extracted from leaves from seedlings according to the methods described by Murray and Thompson [57]. DNA from each accession of 50 pooled plants was normalized to a final concentration of 30 ng·µL−1. Following extraction, DNA samples were arrayed in a 96-well format.
Primer design
Using Primer Premier (PREMIER Biosoft International, http://www.premierbiosoft.com/index.html), two types of primers were designed for PCR amplification: gene-specific primers directly labeled at the 5′ end with IRD 700 and IRD 800 (such as the primer for atp6) or primers in which a sequence from the universal primer M13F (CACGACGTTGTAAAACGAC) was added to the 5′ end of a gene-specific primer as an adaptor, and M13R (GGATAACAAT TTCACACAGG) was also added to gene-specific primers (such as the primers for the accD gene, matK gene, and rbcL gene). The universal primers M13F and M13R were labeled at the 5′ end with IRD 700 and IRD 800 separately (MWG Biotech, Inc., Ebersberg, Germany). Altogether, 7 pairs of primers were designed against for 3 genes (accD, matK and rbcL) based on DNA sense strand, except a pair of primer against for atp6 according to DNA antisense strand. All of the primers are listed in Table 3 and the positions of these primers are shown in Figure S1.
Table 3. Primers used in the PCR amplification#.
| Taxon | Genes | GenBank No. | Gene length | Primer name | Primer type | Primer sequence | PCR products length |
| B. napus B. rapa B.oleracea | accD | GQ861354 | 1470 bp | accD-1 | M13-A1 Forward | M13F-tgactattcatctattgttatt | 974 bp |
| M13-A1 Reverse | M13R-gctttttataaggttcct | ||||||
| accD-2 | M13-A2 Forward | M13F-tgaccagttcagttcagagagaatt | 789 bp | ||||
| M13-A2 Reverse | M13R-tttttcagggttttcgtttaat | ||||||
| matK | GQ861354 | 1575 bp | matK-1 | M13-M1 Forward | M13F-tgaggtatttagtgctttcg | 959 bp | |
| M13-M1 Reverse | M13R-cttgagcaaccctaagagcg | ||||||
| matK-2 | M13-M2 Forward | M13F-gcaagcagtcttctcatttacg | 948 bp | ||||
| M13-M2 Reverse | M13R-ttttgttatctccgcattcc | ||||||
| rbcL | GQ861354 | 1440 bp | rbcL-1 | M13-R1 Forward | M13F-caacatatattactgtcaagag | 974 bp | |
| M13-R1 Reverse | M13R-ttcacattctcatcatctttgg | ||||||
| rbcL-2 | M13-R2 Forward | M13F-cagtttatgaatgtctacgtgg | 1081 bp | ||||
| M13-R2 Reverse | M13R-cttctttttttttcagattttg | ||||||
| Cruciferae | accD | AP000423 | 1476 bp | CA | M13-CAForward | M13F-tgaaagttacatttataatt | 827 bp |
| M13-CAReverse | M13R-cttttttcaatgtttgttca | ||||||
| atp6 | NM_126768.1 | 786 bp | UPA | IRD700 Forward | 700*-ggagatttatagcatcattcaag | 733 bp | |
| IRD800 Reverse | 800*-attgtcccattgattcctat |
Universal primers in this study included M13-IRD700 Forward and M13-IRD800 Reverse, and the corresponding primer sequences were 700*-cacgacgttgtaaaacgac and 800*-ggataacaatttcacacagg, respectively.
PCR amplification and mutation detection
PCR amplification and mutation detection followed the method of Barkley and Wang [58] with some modifications. For the accD, matK and rbcL genes, we used a two-step approach to amplify DNA because the primer pairs carried the universal primers M13F and M13R. The first PCR amplification was performed in a 15 µL reaction volume using 60 ng DNA template, 1.5 µL 10× Ex-buffer, 0.2 mM dNTPs, 0.75 U Ex-Taq polymerase and 0.25 µL 10 µM each primer [Ex-Taq polymerase and Ex-buffer both purchased from TAKARA Biotechnology (Dalian) CO., LTD.]. Cycling conditions were as follows: 94°C for 5 min, 35 cycles of 94°C for 1 min, primer-specific annealing temperature for 1 min (the annealing temperatures for the accD, matK and rbcL genes were 50°C, 55°C and 57°C, respectively) and 72°C for 1 min, followed by a final extension for 10 min at 72°C. After the first PCR amplification, 1 µL of the PCR products was diluted to a 50-µl volume, and then 2 µL of the diluted PCR products was extracted, mixed with 2 µL of the control PCR sample to serve as a template for the second PCR amplification. The reference samples of accD, matK and rbcL in Brassica species were B26, B26 and B85, respectively. However, the reference sample of accD in Brassiceae was A64. The second PCR was performed in 25-µL reaction volume using 2.5 µL 10× Ex-buffer, 0.32 mM dNTPs, 1.25 U Ex-Taq polymerase, and 1.0 µL 1 µM M13F and M13R universal primers labeled at the 5′ end with IRD 700 and IRD 800. PCR amplifications were conducted using the same reaction program as for the first PCR, except that 55°C was used for the annealing temperature. However, for the atp6 gene, PCR amplification was performed in the same way as the first PCR amplification of the two-step approach, except that 0.25 µL 1 µM each primer directly labeled with IRD 700 and IRD 800 was used. Sample A24 was served as the reference DNA in the atp6 gene of Brassicaceae.
After PCR amplification, denaturation, slow reannealing conditions were applied to the PCR fragments to form heteroduplexes. The products were denatured and annealed in a thermal cycler, as follows: 99°C for 10 min, 70 cycles of 70°C for 20 sec, with a decrement of 0.5°C per cycle, and finally, 8°C for 5 min. A total of 30 µL of the processed PCR fragments was incubated in 2 µL of 10×CEL I buffer (10×CEL I buffer include 5 mL 1 M MgSO4, 100 µL 10% Triton X-100, 5 mL 1 M Hepes (pH 7.4), 5 µL 20 mg/ml bovine serum albumen, 2.5 ml 2 M KCl, 37.5 ml ddH2O), 0.2 µL purified CEL I extracted and purified according to the method described by Oleykowski et al (1998) [59] and 10 µL of heteroduplexes at 45°C for 30 min. CEL I cuts with partial efficiency, allowing the detection of multiple mismatches in a DNA duplex [48]. The CEL I digestion reaction was stopped with 5 µL of 0.225 mM EDTA. Sample purification was performed using Sephadex G50 (medium coarse) columns, and 2 µL of blue stop solution (MWG Biotech, Inc., Ebersberg, Germany) was added to each sample. Then, all of the samples were concentrated to a final volume of 2–3 µL for 60 min at 85°C. A total of 0.8 µL was loaded onto 6.5% polyacrylamide gels on a LI-COR 4300 DNA analyzer. To confirm the polymorphisms identified by ORG-EcoTILLING, each gel image of ORG-EcoTILLING was generated from two replicate rans. Because the fragment patterns for the two replicates were always concordant, we selected the better quality gel image for analysis. Furthermore, the accessions exhibiting polymorphism on the gels were randomly selected for a second round of amplification with the corresponding primer pairs using proofreading Ex-Taq polymerase. The PCR products were sequenced with an ABI 3730 and chromatograms checked to identify PCR or sequencing errors.
Mutation frequency and detection rate
A total of 141,120 bp, 151,200 bp and 138,240 bp were screened for the accD, matK and rbcL genes separately, which was calculated by the multiplication of the number of samples and the total length of the gene sequence. The mutation frequency was determined by dividing the total base pairs screened by the total mutants detected. The detection rate was calculated by the following formula: Detection rate = A1/A2 (A1 was the number of mutations detected by ORG- EcoTILLING, and A2 was the number of mutations checked by sequencing).
Phylogenetic tree construction
To construct the phylogenetic tree for chloroplast genes, accD, matK and rbcL sequences of 90 accessions of B. napus and each of 3 accessions of B. rapa and B. oleracea were generated (GenBank accession numbers JF807893 to JF807913 inclusive), CLUSTAL W, version 1.81 [60] was used for sequence alignment and Phylip 3.68 with a neighbour-joining (N-J) algorithm (http://evolution.genetics.washington.edu/phylip.html) [61] for phylogenetic and molecular evolutionary analyses. The evolutionary distance (D) was computed using Kimura's two-parameter method [62], and the trees tested with 1000 bootstrap replicates. The corresponding bootstrap values (>750) for each data partition were displayed in each node on the tree.
Results
Applicability of ORG-EcoTILLING in the discovery of chloroplast DNA polymorphism for inter- and intra-specific analysis of Brassica
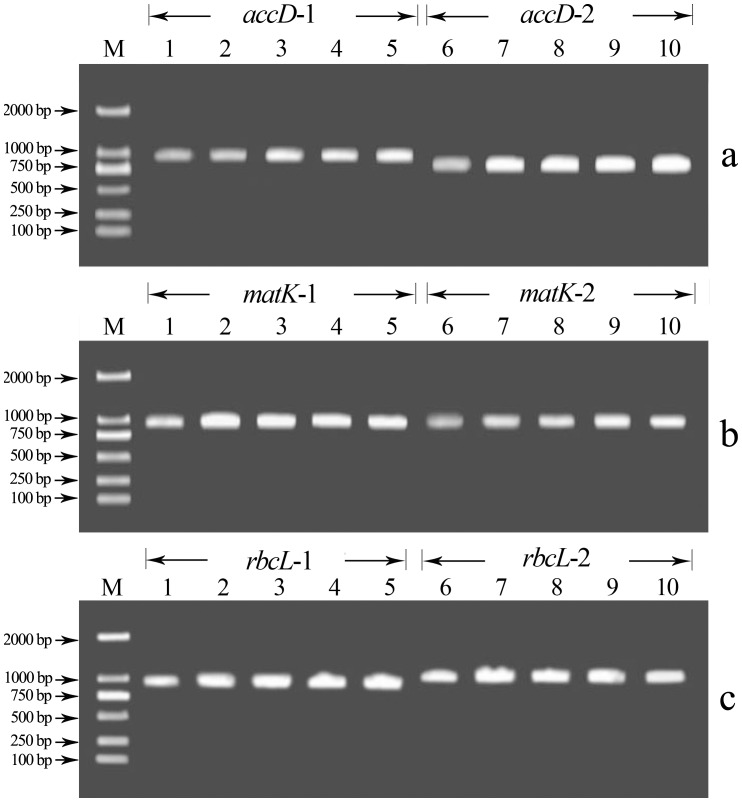
A total of 90 B. napus samples and three each of B. rapa and B. oleracea were analyzed. The accD, matK and rbcL genes were amplified using two pairs of primers each. Amplified DNA fragments were of expected length for the three genes, as detected by agarose gel electrophoresis (Figure 2), and the DNA yield was suitable for ORG-EcoTILLING analysis. The analysis reliably located the position of mutated base pairs in the accD, matK and rbcL genes.
Figure 2. Detection of amplified fragments of selectcd chloroplast genes by agarose gel electrophoresis.
a. accD gene, the size of PCR products in accD-1 and accD-2 amplified by two pairs of primers were 974 and 789 bp, respectively. b. matK gene, the size of PCR products in matK-1 and matK-2 amplified by two pairs of primers were 959 and 948 bp, respectively. c. rbcL gene, the size of PCR products in rbcL-1 and rbcL-2 amplified by two pairs of primers were 974 and 1081 bp, respectively. The sizes of the DNA fragments produced for the three genes corresponded with what we expected, and the abundance of DNA was fit for ORG-EcoTILLING analysis. The samples from 1 to 5 were B12, B20, B26, B85 and B94 in accD-1 gene fragment, and the same in other gene fragments. The molecular size of DNA is shown at the left with arrowheads. M represented the DNA ladder DL 2000 (Promega. Inc).
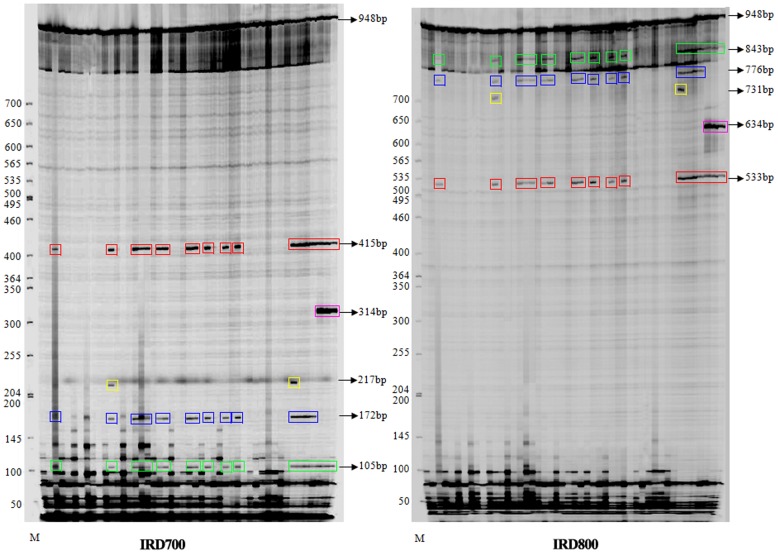
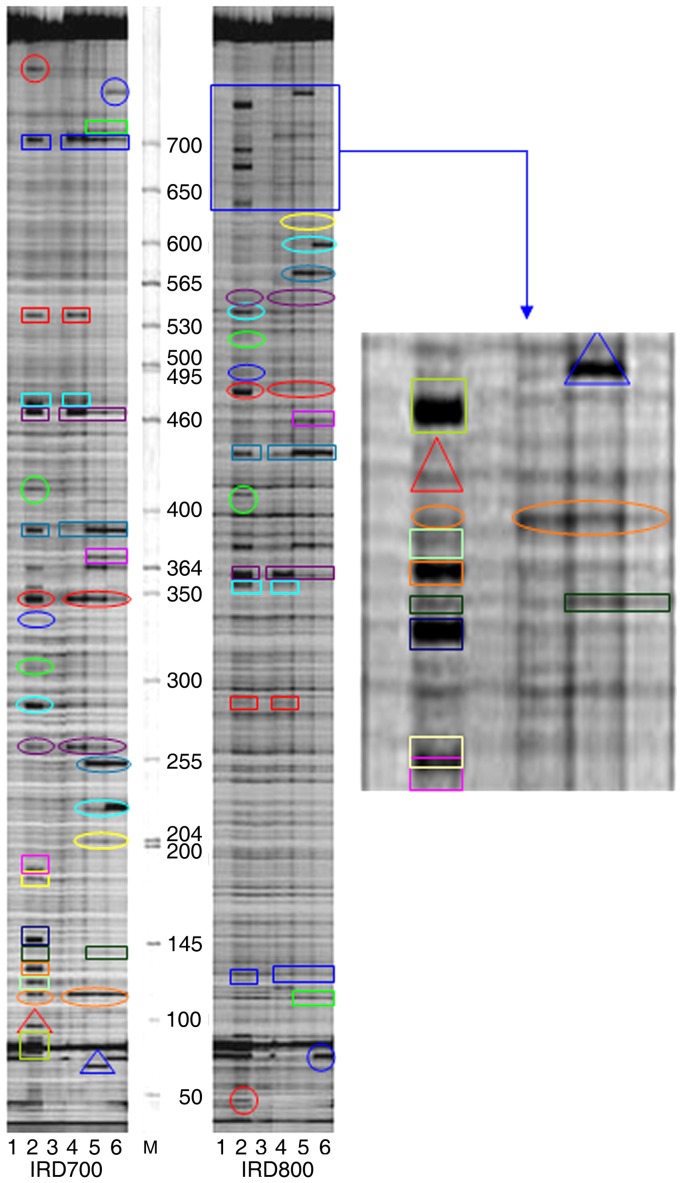
As an example, the IRD 700 and IRD 800 channels for the matK gene fragment (Primer: M13-M2 Forward and M13-M2 reverse, Table 3) are shown in Figure 3. This demonstrates that mismatches were detected by CEL I and cleaved into two separate products, which were distinctly detected in both the IRD700 and IRD800 channels of the gel image. The combined lengths of the IRD700-labeled and IRD800-labeled cleaved fragments corresponded to the length of the PCR fragments (948 bp) in the matK gene. Similarly, clear gel images were obtained for the accD and rbcL genes, and the IRD700-labeled and the combined length of the IRD800-labeled cleaved fragments corresponded to the PCR fragments of the two genes (Table S1).
Figure 3. Overview of mutation detections in matK gene of matK-2 fragment by ORG- EcoTILLING.
Samples were resolved by denaturing polyacrylamide gel electrophoresis and visualized with the LI-COR DNA analyzer. The sample in each lane from left to right was B49–B96 samples. The IRD 700 and IRD 800 channels were shown respectively. CEL I-cleaved heteroduplexes appeared as intense bands (circled in different color rectangles of IRD 700 channel and corresponding same color ones of IRD 800 channel). The size of the specific cleavage products in the IRD700 labeled plus the size of the IRD800 labeled (indicated by arrowheads in the same color geometries, respectively) was equal to that of uncleaved fragment (948 bp). The fragment sizes also indicated the position of the mutation present in the locus matK gene of the representative samples. The reference sample was B26.
ORG-EcoTILLING detected nine, six and a single distinct mutated base pair positions in the accD, matK and rbcL genes, respectively, corresponding to 25, 25 and 19 mutated out of the 96 samples (Table S1). Two primer pairs were designed to amplify each of the three genes; their target regions were denominated accD-1, accD-2, matK-1, matK-2, rbcL-1 and rbcL-2, respectively (Table 3). For the accD gene, most samples had one or two mutation points within accD-1, whereas there were five mutation points in accD-2, such as in samples B94, B95 and B96, B. oleracea, the ancestral parents of B. napus. In contrast, three or four mutation points were present in matK-2, but only one mutation point was found in matK-1. For the rbcL gene, a single mutation was observed in rbcL-1, and none in rbcL-2.
Interestingly, the three varieties of B. rapa possessed the same mutation positions as B. napus in the accD, matK and rbcL genes, respectively, whereas different mutation positions in the accD and matK genes were discovered in the three varieties of B. oleracea. For example, samples B94, B95 and B96 all had specific mutation positions in the accD-1 gene fragment, such as positions 409 and 871 in the IRD700 channel and five mutation positions in the accD-2 gene fragment, which were not observed in the B. napus and B. rapa varieties. Similarly, although the mutation positions in the matK-2 gene fragment were approximately the same in all mutation samples, only the three samples of B. oleracea presented one mutation position each in the matK-1 gene fragment. For the rbcL gene, the three samples of B. oleracea showed no mutation. Moreover, the samples mutated in the accD gene corresponded with those mutated in the matK gene, except sample B68. Similarly, the samples mutated in the rbcL gene were, without exception, distributed around the samples mutated of the accD and matK genes. Thus, our results effectively indicated that ORG-EcoTILLING could be effectively applied in chloroplast genes and that ORG-EcoTILLING could detect chloroplast DNA polymorphisms among the three species of Brassica, and very importantly among varieties of B. napus.
Application of ORG-EcoLLING for detecting SNPs within the accD gene across the Brassicaceae family
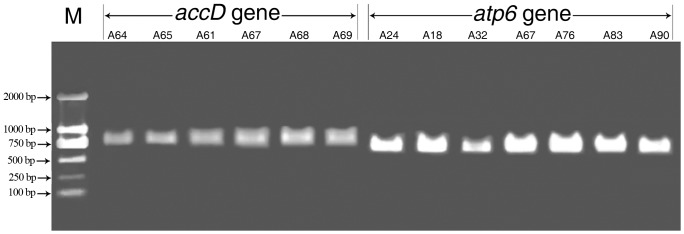
The key chloroplast gene accD was used to study the application of ORG-EcoTILLING in members of the Brassicaceae family (91 samples of different taxa). We designed the primers for the accD gene from a conservative region according to BLAST searches of different Brassicaceae taxa. The size of the accD fragment amplified by PCR was 827 bp in all samples.
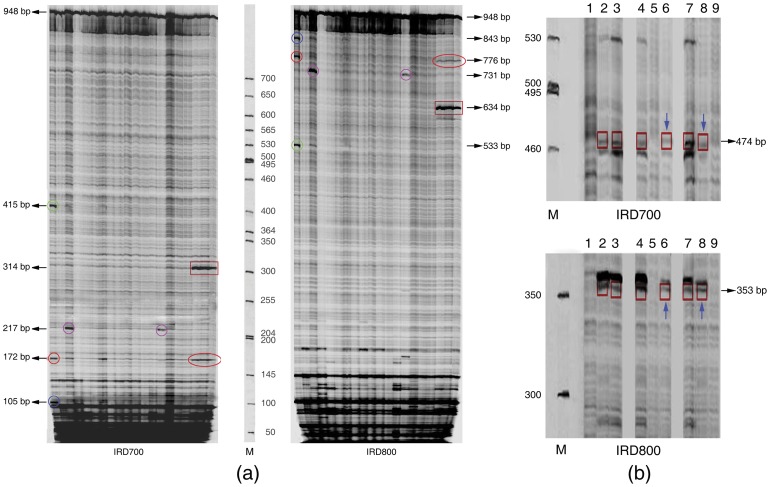
Sufficient DNA was generated for subsequent analysis (Figure 4). Overall, 44 mutation points in the accD gene were detected in 44 plant samples (Table S2). Analysis of a subset of CEL 1 digests in the representative samples (Figure 5) revealed sample A61 and the reference sample A64 both lacked cleaved positions, whilst samples A65, A67, A68 and A69 were characterized by 29, 9, 15 and 16 mutation sites, respectively. Furthermore, it was clear that the sizes of the 2 fragments produced for all mutation points indicated the accurate position of the mismatch, and thus the site of the mutation or nucleotide change. Detailed ORG-EcoTILLING analysis for these four mutation samples is shown in Table 4. Distinct SNPs were observed in the accD gene of the Brassicaceae family. For instance, these four mutation samples all shared with mutation site 6, 20, 25, 29, 32 and 37. However, sample A65 had 20 specific polymorphic sites which were not shared with other samples (Table 4). The results demonstrated that ORG-EcoTILLING could not only be applied for detecting DNA polymorphisms in chloroplast genes in different tribes and genera, but also among and between species.
Figure 4. Examples of DNA amplified regions detection of plastidic genes separated by agarose gel electrophoresis.
The picture shows the results from PCR analyses of plastidic gene accD (primer pair: M13-CAF and M13-CAR), and atp6 (primer pair: M13-CP F and M13-CP R), respectively. The samples of accD gene and atp6 came from 91 various species in Cruciferae. The sizes of the DNA fragments in the two genes detected by agarose gel electrophoresis were consistent with the result of primers design, and the abundance of DNA was fit for ORG-EcoTILLING analysis. The molecular size of DNA is shown at the left with arrowheads. M represented the DNA ladder DL 2000 (Promega. Inc).
Figure 5. Overview of mutation detections in Brassicaceae samples by ORG- EcoTILLING Electrophoresis on a 6.5% KBplus Gel of CEL I-digested products of APCR fragment of accD gene, 827 bp in ALI-COR 4300 DNA Analyzer were displayed here.
The sample in each lane from one to six was A64, A65, A61, A67, A68 and A69, respectively. CEL I-cleaved heteroduplexes appeared as intense bands (circled in different color geometric figures of IRD 700 channel and corresponding same color ones of IRD 800 channel). The fragment sizes indicated the position of the mutation present in the locus accD gene of the collected samples. The reference sample in this gene was A64.
Table 4. Analysis of ORG-EcoTILLING information in 700 and 800 channel from Figure 5.
| Mutation site No. | Geometric figure | IDY 700 | IDY 800 | Molecular Size (bp) | Sample code. |
| 1 | blue triangle | 73 | 754 | 827 | A68 |
| 2 | lime square | 86 | 741 | 827 | A65 |
| 3 | lime square | 87 | 740 | 827 | A65 |
| 4 | lime square | 88 | 739 | 827 | A65 |
| 5 | red triangle | 100 | 727 | 827 | A65 |
| 6 | orange ellipse | 119 | 708 | 827 | A65, A67,A68, A69 |
| 7 | light green rectangle | 127 | 700 | 827 | A65 |
| 8 | orange rectangle | 134 | 693 | 827 | A65 |
| 9 | orange rectangle | 135 | 692 | 827 | A65 |
| 10 | green rectangle | 144 | 683 | 827 | A65, A68, A69 |
| 11 | blue rectangle | 151 | 676 | 827 | A65 |
| 12 | blue rectangle | 152 | 675 | 827 | A65 |
| 13 | yellow rectangle | 187 | 640 | 827 | A65 |
| 14 | purple rectangle | 191 | 636 | 827 | A65 |
| 15 | yellow ellipse | 207 | 620 | 827 | A68, A69 |
| 16 | turquoise circular | 225 | 602 | 827 | A68, A69 |
| 17 | turquoise circular | 227 | 600 | 827 | A69 |
| 18 | cyan ellipse | 250 | 577 | 827 | A68, A69 |
| 19 | cyan ellipse | 251 | 576 | 827 | A68, A69 |
| 20 | dark purple ellipse | 260 | 567 | 827 | A65, A67, A68, A69 |
| 21 | turquoise ellipse | 283 | 544 | 827 | A65 |
| 22 | emerald green ellipse | 305 | 522 | 827 | A65 |
| 23 | blue ellipse | 339 | 488 | 827 | A65 |
| 24 | red ellipse | 345 | 482 | 827 | A65 |
| 25 | red ellipse | 346 | 481 | 827 | A65, A67, A68, A69 |
| 26 | purple rectangle | 372 | 455 | 827 | A68, A69 |
| 27 | cyan rectangle | 388 | 439 | 827 | A65 |
| 28 | cyan rectangle | 388 | 439 | 827 | A68, A69 |
| 29 | cyan rectangle | 389 | 438 | 827 | A65, A67, A68, A69 |
| 30 | green circular | 416 | 411 | 827 | A65 |
| 31 | dark purple rectangle | 468 | 359 | 827 | A65, A67 |
| 32 | dark purple rectangle | 469 | 358 | 827 | A65, A67, A68, A69 |
| 33 | dark purple rectangle | 470 | 357 | 827 | A65 |
| 34 | turquoise rectangle | 474 | 353 | 827 | A65 |
| 35 | turquoise rectangle | 474 | 353 | 827 | A67 |
| 36 | red rectangle | 543 | 284 | 827 | A65, A67 |
| 37 | blue rectangle | 704 | 123 | 827 | A65, A67, A68, A69 |
| 38 | emerald green rectangle | 713 | 114 | 827 | A68, A69 |
| 39 | blue circular | 755 | 72 | 827 | A69 |
| 40 | red circular | 781 | 46 | 827 | A65 |
The sizes of the two fragments of IRD 700 and 800 indicated the accurate position of the mismatch. Descriptions of different color geometric figures were corresponding with that of Figure 5.
Application of ORG-EcoTILLING for the detecting of SNPs within the atp6 gene across the Brassicaceae family
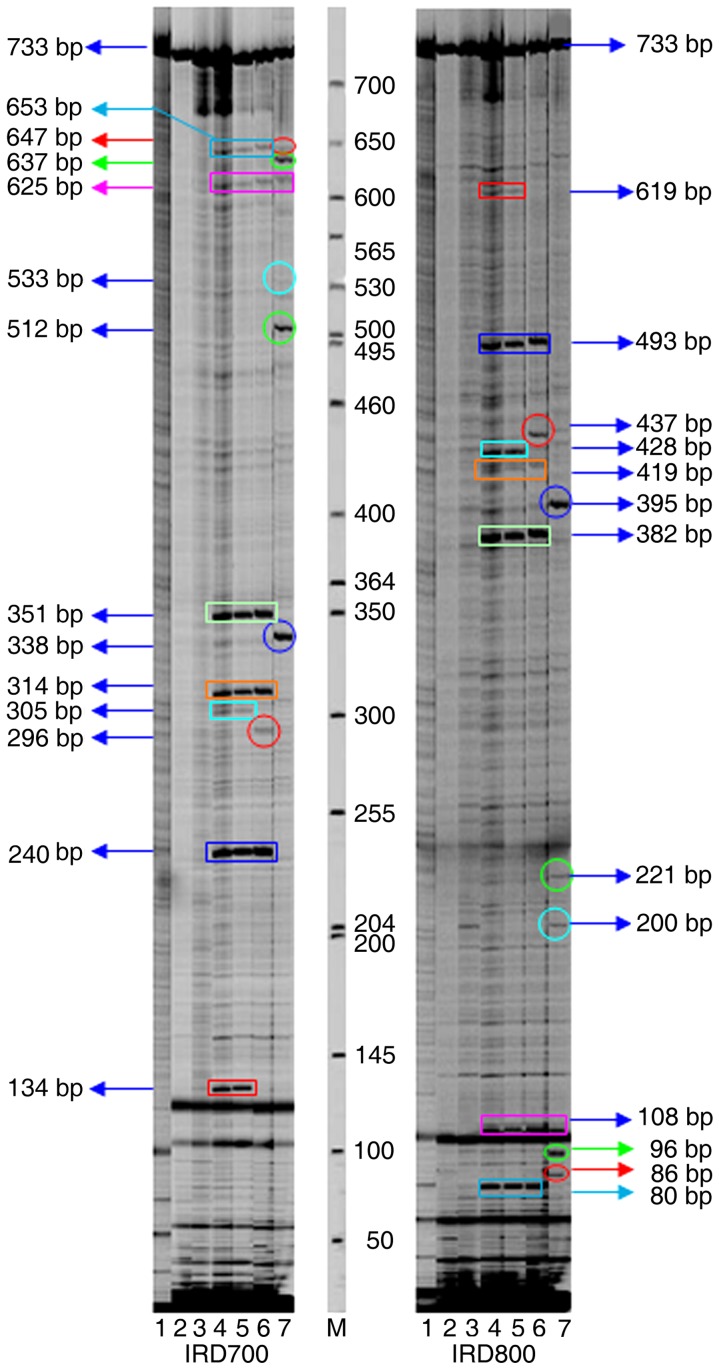
A key mitochondrial gene, atp6, was chosen to study the application of ORG-EcoTILLING in 91 different taxa of Brassicaceae. Initially, the primer for the atp6 gene was designed from a conserved region based on BLAST searches of different Brassicaceae taxa, with an expected PCR product length of 733 bp, which was confirmed in all 91 accessions by agarose gel electrophoresis (Figure 4). Analysis of all products of CEL 1 digests indicated that there were 17 mutation points distributed in 18 plant samples (Table 5). The distribution of the mutation points in the atp6 gene indicated that this atp6 gene was conserved amongst genera of Trib. Brassiceae, since only three mutation points were observed in sample A87, but no point mutations in other Trib. Brassiceae accession. In contrast, abundant mutations of the atp6 gene were detected in Trib. Lepidieae and Trib. Sisymbrieae, the former shared 11 mutation points among four plant accessions and the latter having eight SNPs in two plant samples. In addition, seven mutation points were detected in both of Trib. Hesperideae and Trib. Matthioleae, six SNPs in Trib. Arabideae and four polymorphic sites in Trib. Alysseae. It should be noted that there are distinctly different in mutation sites within atp6 gene among different genus or species of the same tribes. ORG- EcoTILLING images of seven samples (Figure 6) revealed that 13 nucleotide mutations were detected in the atp6 locus in samples A67, A76, A83 and A90, while samples A18, A24 and A32 lacking cleaved positions. Therefore, it was clearly shown that ORG- EcoTILLING could be applied to the detection of mutations in a mitochondrial gene.
Table 5. Band analysis of atp6 gene in the family of Brassicaceae identified by ORG- EcoTILLING.
| MW (bp) IRD700 | |||||||||||||||||
| Mutated sample code | 134 | 140 | 240 | 296 | 305 | 314 | 338 | 351 | 368 | 465 | 512 | 533 | 625 | 637 | 647 | 653 | 663 |
| A56 | + | + | |||||||||||||||
| A58 | + | + | + | ||||||||||||||
| A60 | + | + | + | + | + | ||||||||||||
| A61 | + | + | + | + | + | ||||||||||||
| A67 | + | + | + | + | + | + | + | ||||||||||
| A68 | + | + | |||||||||||||||
| A70 | + | ||||||||||||||||
| A71 | + | + | + | ||||||||||||||
| A72 | + | ||||||||||||||||
| A73 | + | + | + | + | |||||||||||||
| A75 | + | + | + | + | + | ||||||||||||
| A76 | + | + | + | + | + | + | + | ||||||||||
| A79 | + | + | + | ||||||||||||||
| A82 | + | + | + | ||||||||||||||
| A83 | + | + | + | + | + | + | |||||||||||
| A87 | + | + | + | ||||||||||||||
| A88 | + | + | + | + | |||||||||||||
| A90 | + | + | + | + | + | + | |||||||||||
“+” indicated that there was an intense band visualized by denaturing polyacrylamide gel electrophoresis with the LI-COR DNA analyzer.
Figure 6. Images of ORG-EcoTILLING gels of CEL I-digested products of a PCR fragment of atp6 gene obtained from each of the two fluorescent channels of the LI-COR 4300 DNA Analyzer.
The intense bands (circled in different color geometric figures of IRD 700 channel and corresponding same color ones of IRD 800 channel) came from the product of CEL I-cleaved heteroduplexes. The sample in each lane from left to right was A24, A18, A32, A67, A76, A83 and A90, respectively, and sample A24 acted as the reference sample of atp6 gene for ORG-EcoTILLING. Therefore, it was obvious that 13 polymorphism sites were detected in atp6 gene among these seven samples.
Validation of the accuracy of ORG-EcoTILLING for the identification of cytoplasmic DNA Polymorphism
We validated the accuracy of ORG-EcoTILLING by Sanger DNA sequencing. At the species level for the accD, matK and rbcL genes in 96 samples of B. napus, B. rapa and B. oleracea, mutation points were detected in 26, 26, and 19 samples, respectively. However, 70, 70 and 77 samples of B. napus presented no mutation points in the corresponding genes, accD, matK and rbcL by ORG-EcoTILLING. According to the Sanger sequencing results, 116, 89 and 19 DNA polymorphisms were detected in accD, matK and rbcL genes, respectively, and the results of ORG-EcoTILLING were entirely consistent with those of DNA sequencing results (Table 6). In addition, we calculated that the accD gene in B. napus, B. rapa and B. oleracea had approximately one mutation per 1.22 kb, compared with approximately one mutation per 1.70 kB and 7.28 kB in the matK and rbcL genes, respectively (Table 6). We then analyzed SNP classes based on the results of DNA sequencing. Two SNP classes of deletion and transition mutations were detected in the three genes, with the deletion type only presented in the accD gene (in the form of six nucleotide base pair deletions). However, the majority of the transitions detected were C to A in the accD gene; G to T, T to G and T to A in the matK gene; and only G to A transitions in the rbcL gene. All sequence variants detected by DNA sequencing were also detected by ORG-EcoTILLING. Therefore, the detection rate of ORG- EcoTILLING was 100%.
Table 6. SNP mutation types and band analysis of B. napus varieties identified by ORG- EcoTILLING and DNA sequencing.
| Sample code | accD mutation position | matK mutation position | rbcL mutation position | |||||||||||||
| 216 (G/T)* | 335 (T/A) | 560–565 (AAAGTG) | 644 (A/C) | 797 (C/T) | 858 (G/A) | 1058 (G/A) | 1177 (T/G) | 1275 (T/C) | 201 (A/G) | 822 (T/A) | 889 (T/G) | 934 (G/A) | 1031–1032 (TT/AA) | 1132 (G/T) | 66 (G/A) | |
| B11 | + | + | + | + | + | + | ||||||||||
| B12 | + | + | + | + | + | |||||||||||
| B14 | + | + | + | + | + | + | + | + | ||||||||
| B25 | + | + | + | |||||||||||||
| B27 | + | + | + | + | + | + | ||||||||||
| B29 | + | + | + | + | + | + | ||||||||||
| B32 | + | + | + | + | + | |||||||||||
| B45 | + | + | + | + | + | |||||||||||
| B51 | + | + | + | + | + | + | ||||||||||
| B60 | + | + | + | + | + | + | + | + | ||||||||
| B64 | + | + | + | + | + | |||||||||||
| B65 | + | + | + | + | + | |||||||||||
| B66 | + | + | + | + | + | |||||||||||
| B68 | + | + | + | + | ||||||||||||
| B69 | + | + | + | + | ||||||||||||
| B73 | + | + | + | + | + | |||||||||||
| B74 | + | + | + | + | + | |||||||||||
| B76 | + | + | + | + | + | |||||||||||
| B79 | + | + | + | + | ||||||||||||
| B81 | + | + | + | + | + | |||||||||||
| B90 | + | + | + | + | + | + | + | + | ||||||||
| B91 | + | + | + | + | + | + | ||||||||||
| B92 | + | + | + | + | + | + | ||||||||||
| B93 | + | + | + | + | + | + | ||||||||||
| B94 | + | + | + | + | + | + | + | + | + | + | ||||||
| B95 | + | + | + | + | + | + | + | + | + | + | ||||||
| B96 | + | + | + | + | + | + | + | + | + | + | ||||||
| 3 | 3 | 72 | 23 | 3 | 3 | 3 | 3 | 3 | 3 | 26 | 23 | 4 | 6 | 27 | 19 | |
| Total | 116 | 89 | 19 | |||||||||||||
| sequencing | 116 | 89 | 19 | |||||||||||||
| Detection rate | 100% | 100% | 100% | |||||||||||||
| Total size | 141120 bp | 151200 bp | 138240 bp | |||||||||||||
| Frequency | 1/1.22 kb | 1/1.70 kb | 1/7.28 kb | |||||||||||||
Note: The sample codes were corresponding with Table 2, and mutation position was located from initiation site (ATG) of a gene. The length of accD, matK and rbcL were 1470 bp, 1575 bp and 1440 bp, respectively. “+” indicated that there was an intense band visualized by denaturing polyacrylamide gel electrophoresis with the LI-COR DNA analyzer.
The base pairs changes in round bracket were detected by DNA sequencing and G to T change, while “AAAGTG” meant deletion of base pairs. The reference sample of accD, matK and rbcL were B26, B26 and B85, respectively.
At the intertribal, intergeneric and interspecific levels, 40 and 13 mutation positions were detected by ORG-EcoTILLING from the total of 13 samples screened for the accD and atp6 gene fragments, respectively (Figure 5 and 6). Although we identified only one type of mutation (base substitution) in the accD and atp6 genes, there were abundant SNP types in these mutations (Table 7, Figure S2 and S3). There were 37, 15, 27 and 29 polymorphism sites (a total of 108 polymorphic sites)in accD gene among sample A65, A67, A68 and A69, respectively, and only 29, 9, 15 and 16 mutation sites (a total of 69 polymorphic sites) were discovered in these corresponding samples by ORG-EcoTILLING. Therefore, the detection rate within this gene was 64%. Two particular discrepancies primarily contributed to the low detection rate of ORG-EcoTILLING. Within the first 80 bp only one out of 13 polymorphic sites was detected. Additionally, between nucleotides 354 and 380 within accD gene in A65, A67, A68 and A69, there were 14 polymorphism sites within a range of 27 bases (Table 7, Figure S2), but only two of these were detected compared with the results of DNA sequencing. In contrast, all 13 polymorphic sites representing different types of SNPs within atp6 gene fragments were detected using ORG-EcoTILLING (Table 7), thus the detection rate within this gene was 100%.
Table 7. Comparison of efficiency in different types of SNPs in accD and atp6 gene detected by ORG-EcoTILLING and DNA sequencing.
| positions of polymorphisms | SNPs detected by sequencing | SNPS detected by ORG-EcoTILLING | Detection rate (%) | |||||||||
| A64* | A65 | A67 | A68 | A69 | A64* | A65 | A67 | A68 | A69 | |||
| accD gene | 56 | C | T | T | T | T | NP | − | − | − | − | 0 |
| 63 | A | A | A | G | G | NP | NP | NP | − | − | 0 | |
| 73 | G | G | G | A | G | NP | NP | NP | + | NP | 100 | |
| 74 | T | C | C | C | C | NP | − | − | − | − | 0 | |
| 81 | T | T | T | G | G | NP | NP | NP | − | − | 0 | |
| 86 | G | A | G | G | G | NP | + | NP | NP | NP | 100 | |
| 87 | A | C | A | A | A | NP | + | NP | NP | NP | 100 | |
| 88 | T | A | T | T | T | NP | + | NP | NP | NP | 100 | |
| 100 | G | T | G | G | G | NP | + | NP | NP | NP | 100 | |
| 109 | G | G | G | A | A | NP | NP | NP | − | − | 0 | |
| 119 | A | G | G | G | G | NP | + | + | + | + | 100 | |
| 127 | A | G | A | A | A | NP | + | NP | NP | NP | 100 | |
| 134 | C | A | C | C | C | NP | + | NP | NP | NP | 100 | |
| 135 | G | T | G | G | G | NP | + | NP | NP | NP | 100 | |
| 144 | C | A | C | A | A | NP | + | NP | + | + | 100 | |
| 151 | A | C | A | A | A | NP | + | NP | NP | NP | 100 | |
| 152 | C | T | C | C | C | NP | + | NP | NP | NP | 100 | |
| 187 | G | T | G | G | G | NP | + | NP | NP | NP | 100 | |
| 191 | A | G | A | A | A | NP | + | NP | NP | NP | 100 | |
| 207 | A | A | A | G | G | NP | NP | NP | + | + | 100 | |
| 225 | C | C | C | G | G | NP | NP | NP | + | + | 100 | |
| 227 | A | A | A | A | G | NP | NP | NP | NP | + | 100 | |
| 250 | G | G | G | C | C | NP | NP | NP | + | + | 100 | |
| 251 | T | T | T | C | C | NP | NP | NP | + | + | 100 | |
| 260 | A | G | G | G | G | NP | + | + | + | + | 100 | |
| 266 | C | T | C | T | T | NP | − | NP | − | − | 0 | |
| 283 | C | G | C | C | C | NP | + | NP | NP | NP | 100 | |
| 305 | T | C | T | T | T | NP | + | NP | NP | NP | 100 | |
| 339 | C | A | C | C | C | NP | + | NP | NP | NP | 100 | |
| 345 | A | C | A | A | A | NP | + | NP | NP | NP | 100 | |
| 346 | A | G | G | G | G | NP | + | + | + | + | 100 | |
| 354 | C | A | C | C | T | NP | − | NP | NP | − | 0 | |
| 365 | C | A | T | G | G | NP | − | − | − | − | 0 | |
| 368 | T | T | T | C | C | NP | NP | NP | − | − | 0 | |
| 372 | G | G | G | A | A | NP | NP | NP | + | + | 100 | |
| 380 | G | T | T | T | T | NP | − | − | − | − | 0 | |
| 388 | C | T | C | A | A | NP | + | NP | + | + | 100 | |
| 389 | T | G | G | G | G | NP | + | + | + | + | 100 | |
| 395 | A | A | A | G | G | NP | NP | NP | − | − | 0 | |
| 408 | G | G | G | A | A | NP | NP | NP | − | − | 0 | |
| 416 | A | C | A | A | A | NP | + | NP | NP | NP | 100 | |
| 449 | C | C | C | T | T | NP | NP | NP | − | − | 0 | |
| 450 | C | A | C | C | C | NP | − | NP | NP | NP | 0 | |
| 468 | C | A | A | C | C | NP | + | + | NP | NP | 100 | |
| 469 | G | A | A | A | A | NP | + | + | + | + | 100 | |
| 470 | G | T | G | G | G | NP | + | NP | NP | NP | 100 | |
| 474 | A | C | T | A | A | NP | + | + | NP | NP | 100 | |
| 522 | A | G | A | A | A | NP | − | NP | NP | NP | 0 | |
| 543 | T | C | C | T | T | NP | + | + | NP | NP | 100 | |
| 704 | A | G | G | G | G | NP | + | + | + | + | 100 | |
| 707 | G | G | C | G | G | NP | NP | − | NP | NP | 0 | |
| 713 | T | T | T | A | A | NP | NP | NP | + | + | 100 | |
| 746 | C | C | T | C | C | NP | NP | − | NP | NP | 0 | |
| 755 | T | T | T | T | C | NP | NP | NP | NP | + | 100 | |
| 781 | G | T | G | G | G | NP | + | NP | NP | NP | 100 | |
| Total mutation sites | 108 69 | |||||||||||
| Average Detection rate (%) | 64 | |||||||||||
| atp6 gene | A24* | A67 | A76 | A83 | A90 | A24* | A67 | A76 | A83 | A90 | ||
| 134 | G | T | T | G | G | NP | + | + | NP | NP | 100 | |
| 240 | G | C | C | C | G | NP | + | + | + | NP | 100 | |
| 296 | T | T | T | C | T | NP | NP | NP | + | NP | 100 | |
| 305 | T | C | C | T | T | NP | + | + | NP | NP | 100 | |
| 314 | T | G | G | G | T | NP | + | + | + | NP | 100 | |
| 338 | A | A | A | A | G | NP | NP | NP | NP | + | 100 | |
| 351 | C | G | G | G | C | NP | + | + | + | NP | 100 | |
| 512 | T | T | T | T | C | NP | NP | NP | NP | + | 100 | |
| 533 | T | T | T | T | C | NP | NP | NP | NP | + | 100 | |
| 625 | T | C | C | C | C | NP | + | + | + | + | 100 | |
| 637 | G | G | G | G | A | NP | NP | NP | NP | + | 100 | |
| 647 | A | A | A | A | T | NP | NP | NP | NP | + | 100 | |
| 653 | C | A | A | A | C | NP | + | + | + | NP | 100 | |
| Average Detection rate (%) | 100 | |||||||||||
indicated that the reference sample was A64 and A24 in accD gene and atp6 gene, respectively. “+” indicated that there was an intense band visualized by ORG-EcoTIILLING. “−” indicated that there was no intense band detected by ORG-EcoTIILLING. “NP” indicated no polymorphism.
Very interestingly, we observed that the same polymorphic position detected by ORG-EcoTILLING revealed distinct transitions in different samples for the accD gene. For example, based on sequencing results, at position 365, base C mutated to base A, T and G among these samples, and for position 474, base A mutated to base C and T (Table 7, Figure S2). The latter was detected by ORG-EcoTILLING, while the former not.
Since this study involved quite distantly related accessions, it was necessary to verify whether the same polymorphic position detected by ORG-EcoTILLING represented identical transversion events within these samples. We therefore designed an experiment to validate the difference for the same polymorphic position discovered by ORG-EcoTILLING among species or higher taxa. Based on the results of ORG-EcoTILLING in the matK-2 fragment of Brassica species in Figure 3, we selected B76 mutated sample as the reference to mix with B26 (as the reference sample in Figure 3) and 19 mutated samples (also including B76, Figure 3), respectively. The result showed that there was no intense band in samples of B51, B64, B65, B66, B68, B69, B73, B74, B76, B79, B81, B91, B92 and B93, because these mutated samples had the same polymorphisms and positions as the reference sample B76. However, three polymorphic positions were discovered in the lane of B26, identical to those of the reference sample B76 (Figure 3). Interestingly, the lanes for B60, B90, B94, B95 and B96 all showed one polymorphic position (Figure 7a), primarily due to the fact that these samples held the special polymorphism which the reference sample B76 did not share with (Figure 3). Thus, the result clearly demonstrated that the same polymorphic position detected by ORG-EcoTILLING revealed the same polymorphism between Brassica intraspecies.
Figure 7. Gel image showing the result of validation in the difference of the same polymorphic position discovered by ORG-EcoTILLING among Brassica species or Cruciferae.
(a) Based on the results of ORG-EcoTILLING in matK-2 fragment of Brassica intraspecies in Figure 3, 19 mutated samples and no mutation sample B26 (as the reference in Figure 3) were selected to mix with B76 (as the reference), respectively. The sample in each lane from left to right in the channel of IRD700 or IRD800 was B26, B51, B60, B64, B65, B66, B68, B69, B73, B74, B76, B79, B81, B90, B91, B92, B93, B94, B95 and B96. CEL I-cleaved heteroduplexes appeared as intense bands (circled in different color geometric figures of IRD 700 channel and corresponding same color ones of IRD 800 channel). (b) ORG-EcoTILLING gel image of validation in polymorphism site 474 of accD gene (Figure S1, Table 7) by using one of the three samples of A64, A65 and A67 as reference with the others. In the channel of IRD700 or IRD800, the lane of 1, 2, and 3; 4, 5 and 6; 7, 8 and 9 were sample A64, A65 and A67, respectively, but the reference of 1, 2, and 3 was A64; the reference of 4, 5 and 6 was A65, and the reference of 7, 8 and 9 was A67. It was obvious that there was different polymorphism in the same position 474 because polymorphism appeared when 65 mixed with A67 (blue arrow showed polymorphism band).
Plants from different genera or tribe shared polymorphisms in accD and/or atp6 whereas they did not share with other accessions of the same species. For example, it was obviously not the case for position 474 in accD between samples A65 and A67 (Figure S2). Therefore, we designed an ORG-EcoTILLING experiment to test them by using one of the three samples of A64, A65 and A67 as reference with the others. The result revealed the presence of polymorphisms when A65 was mixed with A67, suggesting that distinct polymorphisms were located at the same position (Figure 7b).
Phylogenetic relationships of chloroplast genes among three Brassica species
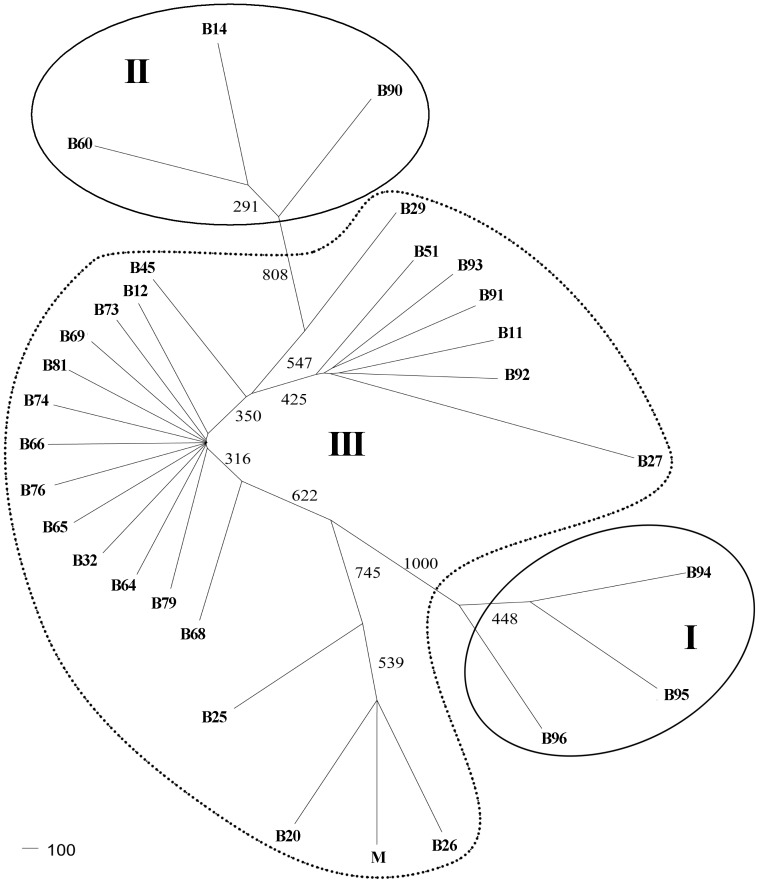
In order to exploit the phylogenetic relationship among the different Brassica species based on the polymorphism results by ORG-EcoTILLING, a phylogenetic tree was constructed using 288 chloroplast genes sequences. Ninety DNA sequences representing each of accD, matK and rbcL in B. napus, and three sequences of each of three chloroplast genes in B. oleracea and B. rapa were analysed. Based on the results no mutation was found to have an identical DNA sequence as that of the reference sample, and this greatly reduced the samples available for DNA sequencing. For example, 27 samples were sequenced for accD and matK (including the reference), and 20 samples for rbcL (including the reference). Therefore, it was possible for us to analyze DNA polymorphisms in a large number of samples, merely through the strategy of combining ORG-EcoTILLING results with existing sequences data. The results revealed three distinct classes (I, II and III) within the genus (Figure 8). The three B. oleracea accessions were distinctly diverged from both B. napus and B. rapa accessions, and the majority of B. napus clustered with three accessions of B. rapa. However, the three accessions of B. napus, B14, B60 and B90 clustered in a clade (II), distinct from other accessions.
Figure 8. Phylogenetic tree was constructed based on the integrating data of the three chloroplast genes (accD, matK and rbcL) in Brassica species.
A total of accD, matK and rbcL sequences of 90 accessions of B. napus and each of 3 accessions of B. rapa and B. oleracea were collected for analysis. The tree was constructed by the neighbour-joining method and bootstrap values above 750 from 1000 resamplings were shown for each node. The code in the tree corresponded to the locality code of Table 2. M represented 69 no mutation samples in B. napus. The bar corresponded with 100 substitutions per site.
In conclusion, our results demonstrate that there are obvious differences in the detection rate of cytoplasmic DNA polymorphisms by ORG-EcoTILLING within the Brassicaceae family. At Brassica interspecific and intraspecific levels, 100% of mutations were detected in SNPs and INDELs in the accD, matK and rbcL genes. However, this method could not detect all mutation events in the accD gene at the genus level in the taxa investigated. In contrast, the detection rate within the atp6 gene was 100% accurate among the Brassicaceae family. Moreover, the information from ORG-EcoTILLING revealed genetic variations within the species of B. napus.
Discussion
We have demonstrated that a CEL I-based heteroduplex cleavage strategy, which was originally developed for TILLING and EcoTILLING [48], [49], [63], can be successfully applied to the discovery of chloroplast and mitochondrial DNA polymorphisms, especially at the inter- and intraspecific levels.
TILLING and EcoTILLING have been proven to be inexpensive and efficient technologies for the discovery of DNA polymorphisms within specific genes. By applying these methods, much progress has been achieved. However, all existing studies have focused on detecting genetic variation within nuclear genomes of different eukaryotic organisms [44]–[47], [64]–[67]. The results of the present study suggested that the development of ORG-EcoTILLING will provide a high-throughput and cost-effective alternative to the current state-of-the-art techniques for studying chloroplast and mitochondrial genomes. Application of new methods in phylogenetic and ecological studies usually leads to important progress in a range of fields. For example, using chloroplast microsatellites, much progress has been achieved in recent decades in investigations of population genetics, crop plant evolution and domestication and phylogenetics [14]. As ORG-EcoTILLING has several important advantages over chloroplast microsatellites, we can expect widespread application of this new tool in chloroplast and mitochondrial genome studies, with more informative outcomes.
Compared with existing techniques, this method requires only a minimal amount of plant tissue or DNA and avoids the laborious experimental procedures associated with cpDNA and mtDNA purification, DNA enzymatic digestion and Southern hybridization. The use of genomic DNA is one of the key advantages of the successful application of chloroplast microsatellites. We can apply this method to survey natural variation in a region of interest accurately and affordably, which is extremely important for chloroplast and mitochondrial genome research because these genomes share the common characteristic of slow nucleotide substitution rates. If more regions are assessed, the information content will rise accordingly. Point mutations are unevenly distributed in plant chloroplast genomes. Magee et al. (2010) found a region of chloroplast DNA in plants related to the sweet pea (Lathyrus) in which the local point mutation rate is at least 20 times higher than elsewhere in the same molecule [68]. In this study, it was found that the accD, matK and rbcL genes present different point mutation rates among different regions in Brassica species (Table 7), which suggests that more regions need to be studied. ORG-EcoTILLING can efficiently detect rare SNPs and INDELs in specific chloroplast genes with high throughput (including coding regions and non-coding regions). Using a Li-COR gel analyzer, 96 samples can be screened in a single gel run using 1∶1 reference and query pooled samples, and the efficiency is much higher than that of DNA sequencing.
Under the experimental conditions used here, the endonuclease CEL I cuts with partial efficiency, allowing the detection of multiple mismatches in a DNA duplex [48], [69]. We identified both SNPs and small INDELs in chloroplast and mitochondrial gene fragments over a roughly 800-bp window (1 kb-2×100 bp, the terminal noisy regions), similar to EcoTILLING for nuclear genes [48]. However, the degree of detection accuracy varies at different taxon levels. For example, at inter- and intraspecific levels, 10, six and one mutation sites were discovered by ORG-EcoTILLING in the accD, matK and rbcL genes, respectively, and the detection rates of these chloroplast genes were all 100% (Table 6), which corresponded with the results of DNA sequencing. Furthermore, at higher taxonomic levels (among tribes and genera in Brassicaceae), the total detection rate of the chloroplast gene accD was only 64%, whereas it was 100% in the mitochondrial gene atp6 (Table 7).
We found that as the number of mutations detected per fragment increases, the scoring and tracking of cleaved fragments becomes more difficult. The lower accuracy at high taxonomic levels could be attributed to higher numbers of polymorphisms occurring among distant samples at intertribal or intergeneric levels, as in the case of the accD gene, from nucleotide 354 to 380 of the accD gene in A65, A67, A68 and 69, there were 14 polymorphism sites within a range of 27 bases, but only two of these were detected compared with the results of DNA sequencing (Table 7). Therefore, overestimating the number of mismatches decreased the resolution and signal intensity in the CEL I analysis, which was also found in the case of the PIF2-2 haplotype [48]. Moreover, the accuracy of this method may be reduced in extreme situations such as where multiple mutations occur in a very short region. In this study, only one of 13 polymorphic sites in the accD gene was discovered within the first 80 bp (Table 7).
Recently, increasingly complete chloroplast and mitochondrial genome sequences of many species of animals, plants and microbes have been obtained by applying next-generation DNA sequencing technologies [56], [70]–[72]. These complete genome sequences provide important references, but they are still not sufficient for conducting phylogenetic analysis and functional studies of chloroplast and mitochondrial genes, in which more information for individuals in populations is needed. The increased number of polymorphisms screened in organelle DNA is particularly advantageous for evolutionary and ecological studies because the level of DNA polymorphism at inter- and intraspecific levels is generally very low, and our results indicate that ORG-EcoTILLING is ideally suited for the high-throughput, accurate discovery of rare chloroplast and mitochondrial DNA polymorphisms. Therefore, ORG- EcoTILLING is a good alternative for assessment of organelle genome variation.
The most important and attractive application of this method is to conduct genome-wide assessments of chloroplast and mitochondrial DNA diversity within a population at the inter- and intraspecific level, which is particularly important for providing a better understanding of the evolution of organelle genomes and the evolution and ecology of species of interest. By using a gene overlapping strategy, we will be able to construct haplotype maps of chloroplast and mitochondrial genomes efficiently. At present, only a few haplotype maps of the nuclear genomes of humans, Arabidopsis and maize have been constructed [73]–[75]. Unlike the nuclear genome, because the polymorphism level of organelle genomes are much lower, and the recombination is much rarer, it is unnecessary to sequence genome of all individuals of a species for the purpose of haplotype map construction. A more efficient way to do this is to use the ORG-EcoTILLING strategy. The size of the chloroplast genome in plants is 120–200 kb [11]. A single EcoTILLING run generally covers a 1-kb target region; considering overlap and terminal noisy regions. With approximately 200 runs, we can expect to obtain genome-wide coverage of 96 samples and observe the full spectrum of variation. Organelle haplotype maps will provide global documented information on inter- and intraspecific variations, which will greatly facilitate studies of the evolution, domestication, population genetics and gene structure and organization.
The data from ORG-EcoTILLING could be applied to analyze phylogenetic relationships. Our study demonstrated that the three B. oleracea accessions were distinctly diverged from both B. napus and B. rapa accessions, while the majority of B. napus clustered with three accessions of B. rapa. The probable reason is that most accessions of B. napus originated from the cytoplasm of B. rapa, suggesting that B. rapa was a much more likely maternal progenitor for B. napus than B. oleracea, which was consistent with previous reports [76], [77]. Moreover, genetic variations were also observed within the B. napus, such as B14, B60 and B90, which were diverged from other accessions (Figure 8), most likely due to the multiple origins or evolution in the relatively recent domestication and modern breeding B. napus.
Although this study focused on plant chloroplast and mitochondrial DNA polymorphisms, the method developed here is also likely to be applicable to human and animal mitochondrial genomes. The high-throughput 454 method was recently used to sequence the complete human mtDNA genome of 109 individuals from the Philippines, and ∼55-fold coverage was achieved, generating <1% missing data per sequence [78]. If ORG-EcoTILLING is proven to be applicable to human and animal mitochondrial genomes, as the size of human and animal mitochondrial genomes is approximately 16 kb [79], additional genome-wide investigations of mitochondrial genomes will become practical not only in humans, but also in many animal species.
There are some limitations to ORG-EcoTILLING. For example, the method does not directly generate sequence information about the detected mutations, and one representative sample among many individuals sharing identical DNA haplotypes must be sequenced to acquire sequence information for the subset. In this case, complementary DNA sequencing would be needed for the extraordinary region.
The rapid development of NGSts promises the possibility of global studies of chloroplast and mitochondrial genomes at population levels. However, ORG- EcoTILLING is ideally suited to screening large numbers to survey populations and diversity collections in chloroplast and mitochondrial genomes, thus reducing samples for which sequencing would provide more detailed information. Therefore, ORG-EcoTILLING represents an alternative new tool for high-throughput, high-resolution analysis of organelle genomes.
Supporting Information
A schematic representation of the position of the primers on accD , matK , rbcL and atp6 genes. Two pairs of primers were designed in accD, matK and rbcL of 96 Brassica species, whereas only one pair of labeled primer was designed in accD and atp6 genes of 91 accessions from Cruciferae. Black boxes showed the genes, and detailed information on each primer was shown in Table 3.
(TIF)
Alignment of representative DNA sequences showing polymorphism in accD gene region regarding INDELs and substitutions. The sequence in each row is a representative sequence, and the red line showed the sequence of the reference sample A64. The numbers in parentheses show the number of samples in Brassicaceae. Blue arrows above the alignment indicated the position of mutation. The position showed in the sequence map began from initial primers attached to M13 adaptors.
(TIF)
Alignment of representative DNA sequences showing polymorphism in atp6 gene region regarding INDELs and substitutions. The sequence in each row is a representative sequence, and the red line showed the sequence of the reference sample A24. The numbers in parentheses show the number of samples in Brassicaceae. Blue arrows above the alignment indicated the position of mutation. The result of DNA sequencing was DNA antisense strand of atp6 gene.
(TIF)
Analysis of ORG-EcoTILLING information in 700 and 800 channel from accD , matK and rbcL gene.
(XLS)
Band analysis of accD gene in 91 Brassicaceae samples identified by ORG-EcoTILLING.
(XLS)
Acknowledgments
We are very grateful to Professor Graham King and Dr. Cathy Nock for the constructive suggestions in revising our manuscript. We also thank the reviewer's valuable comments.
Funding Statement
This work was supported by the China Postdoctoral Science Foundation (20100470417), the Chinese National Basic Research and Development Program (2011CB109302), and the National Natural Science Foundation of China (31100911). The website is http://www.nsfc.gov.cn/Portal0/default152.htm. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Azam A, Paul J, Sehgal D, Prasad J, Bhattacharya S, et al. (1996) Identification of novel genes from Entamoeba histolytica by expressed sequence tag analysis. Gene 181: 113–116. [DOI] [PubMed] [Google Scholar]
- 2. Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of Agenetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32: 314–331. [PMC free article] [PubMed] [Google Scholar]
- 3. Féral J (2002) How useful are the genetic markers in attempts to understand and manage marine biodiversity? J Exp Mar Biol Ecol 268: 121–145. [Google Scholar]
- 4. Liu Z, Cordes J (2004) DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238: 1–37. [Google Scholar]
- 5.Ryman N, Utter F (1987) Population genetics and fishery management. Seattle: University of Washington Press.
- 6. Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic acids Res 18: 7213–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Palmer JD, Herbon LA (1988) Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J Mol Evol 28: 87–97. [DOI] [PubMed] [Google Scholar]
- 8.Palmer JD (1985) Evolution of chloroplast and mitochondrial DNA in plants and algae. in Molecular Evolutionary Genetics. New York: Plenum Press, pp. 131–240.
- 9. Ward BL, Anderson RS, Bendich AJ (1981) The mitochondrial genome is large and variable in a family of plants (Cucurbitaceae). Cell 25: 793–803. [DOI] [PubMed] [Google Scholar]
- 10.Brown W (1983) Evolution of animal mitochondrial DNA. In Evolution of genes and proteins (eds. Nei M, Koehn RK), Sinauer, Sunderland MA.pp. 62–88.
- 11. Palmer JD (1985) Comparative organization of chloroplast genomes. Annu Rev Genet 19: 325–354. [DOI] [PubMed] [Google Scholar]
- 12. Olmstead RG, Palmer JD (1994) Chloroplast DNA systematics: a review of methods and data analysis. Am J Bot 81: 1205–1224. [Google Scholar]
- 13. Galtier N, Nabholz B, Glemin S, Hurst G (2009) Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol 18: 4541–4550. [DOI] [PubMed] [Google Scholar]
- 14. Provan J, Powell W, Hollingsworth P (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16: 142–147. [DOI] [PubMed] [Google Scholar]
- 15. Birky C (1995) Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci 92: 11331–11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dumolin S, Demesure B, Petit R (1995) Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor Appl Genet 91: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 17. Radetzky R (1990) Analysis of mitochondrial DNA and its inheritance in Populus. Curr Genet 18: 429–434. [Google Scholar]
- 18. Rajora O, Dancik B (1992) Chloroplast DNA inheritance in Populus. Theor Appl Genet 84: 280–285. [DOI] [PubMed] [Google Scholar]
- 19. Hodkinson T, Chase M, Takahashi C, Leitch I, Bennett M, et al. (2002) The use of DNAsequencing (ITS and trnL-F), AFLP, and fluorescent in situ hybridization to study allopolyploid Miscanthus (Poaceae). Am J Bot 89: 279–286. [DOI] [PubMed] [Google Scholar]
- 20. Halldén C, Gertsson B, Säll T, Lind-Hallden C (1993) Characterization of organellar DNA in alloplasmic lines of Brassica napus L. Plant Breeding 111: 185–191. [Google Scholar]
- 21. Cronn R, Liston A, Parks M, Gernandt D, Shen R, et al. (2008) Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic acids Res 36: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rieseberg L, Soltis D (1991) Phylogenetic consequences of cytoplasmic gene flow in plants. Evol Trends Plants 5: 65–84. [Google Scholar]
- 23. Byrne M, Moran G, Tibbits W (1993) Restriction map and maternal inheritance of chloroplast DNAin Eucalyptus nitens. J Heredity 84: 218. [Google Scholar]
- 24. Chatterjee A, Faust CJ, Herman GE (1993) Genetic and physical mapping of the biglycan gene on the mouse X chromosome. Mammalian Genome 4: 33–36. [DOI] [PubMed] [Google Scholar]
- 25. Vedel F, Quetier F, Dosba F, Doussinault G (1978) Study of wheat phylogeny by coRI analysis of chloroplastic and mitochondrial DNAs. Plant Sci Letters 13: 97–102. [Google Scholar]
- 26. Fernandes CA, Ginja C, Pereira I, Tenreiro R, Bruford MW, et al. (2008) Species-specific mitochondrial DNA markers for identification of non-invasive samples from sympatric carnivores in the Iberian Peninsula. Conserv Genet 9: 681–690. [Google Scholar]
- 27. Malisa A, Gwakisa P, Balthazary S, Wasser S, Mutayoba B (2006) The potential of mitochondrial DNA markers and polymerase chain reaction-restriction fragment length polymorphism for domestic and wild species identification. Afri J Biotechnol 5: 1588–1593. [Google Scholar]
- 28. Wu F, Zhang Z, Dai H, Zhang Y, Chang L (2008) Genetic relationship of some Crataegus spp. (Hawthorn) revealed by chloroplast DNAPCR-RFLP. J Biotechnol 136: S103. [Google Scholar]
- 29. Li YP, Yang BS, Wang H, Xia RX, Wang L, et al. (2009) Mitochondrial DNAanalysis reveals a low nucleotide diversity of Caligula japonica in China. Afri. J Biotechnol 8: 2707–2712. [Google Scholar]
- 30. Ursin VM, Becker CK, Shewmaker CK (1993) Cloning and nucleotide sequence of a tobacco chloroplast translational elongation factor, EF-Tu. Plant Physiol 101: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cubas P, Pardo C, Tahiri H (2005) Genetic variation and relationships among Ulex (Fabaceae) species in southern Spain and northern Morocco assessed by chloroplast microsatellite (cpSSR) markers. Am J Bot 92: 2031. [DOI] [PubMed] [Google Scholar]
- 32. Jakobsson M, Säll T, Lind-Hallden C, Halldén C (2007) Evolution of chloroplast mononucleotide microsatellites in Arabidopsis thaliana. Theor Appl Genet 114: 223–235. [DOI] [PubMed] [Google Scholar]
- 33. Chase M, Soltis D, Olmstead R, Morgan D, Les D, et al. (1993) Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann Mo Bot Gard 80: 528–580. [Google Scholar]
- 34. Steele K, Vilgalys R (1994) Phylogenetic analyses of Polemoniaceae using nucleotide sequences of the plastid gene matK. Syst Bot 19: 126–142. [Google Scholar]
- 35. Nagano Y, Matsuno R, Sasaki Y (1991) Sequence and transcriptional analysis of the gene cluster trnQ-zfpA-psaI-ORF231-petA in pea chloroplasts. Curr Genet 20: 431–436. [DOI] [PubMed] [Google Scholar]
- 36. Fukami H, Omori M, Hatta M (2000) Phylogenetic relationships in the coral family Acroporidae, reassessed by inference from mitochondrial genes. Zool Sci 17: 689–696. [DOI] [PubMed] [Google Scholar]
- 37. Ratnasingham S, Hebert PDN (2007) BOLD: The Barcode of Life DatA System (http://www.barcodinglife.org). Mol Ecol Notes 7: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11: 31–46. [DOI] [PubMed] [Google Scholar]
- 39. Kristensen V, Kelefiotis D, Kristensen T, Borresen-Dale A (2001) High-throughput methods for detection of genetic variation. Biotechniques 30: 318–333. [DOI] [PubMed] [Google Scholar]
- 40. Tsuchihashi Z, Dracopoli N (2002) Progress in high throughput SNP genotyping methods. Pharmacogenomics J 2: 103–110. [DOI] [PubMed] [Google Scholar]
- 41. Goremykin V, Hirsch-Ernst K, Wölfl S, Hellwig F (2004) The chloroplast genome of Nymphaea alba: whole-genome analyses and the problem of identifying the most basal angiosperm. Mol Biol Evol 21: 1445–1454. [DOI] [PubMed] [Google Scholar]
- 42. Soltis D, Albert V, Savolainen V, Codomo CA, Enns LC, et al. (2004) Genome-scale data, angiosperm relationships, and ‘ending incongruence’: a cautionary tale in phylogenetics. Trends Plant Sci 9: 477–483. [DOI] [PubMed] [Google Scholar]
- 43. Gilchrist E, Haughn G (2005) TILLING without A plough: A new method with applications for reverse genetics. Curr Opin Plant Biol 8: 211–215. [DOI] [PubMed] [Google Scholar]
- 44. Henikoff S, Comai L (2003) Single-Nucleotide Mutations for Plant Functional Genomics. Annu Rev Plant Biol 54: 375–401. [DOI] [PubMed] [Google Scholar]
- 45. Henikoff S, Till BJ, Comai L (2004) TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol 135: 630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Slade A, Knauf V (2005) TILLING moves beyond functional genomics into crop improvement. Transgenic Res 14: 109–115. [DOI] [PubMed] [Google Scholar]
- 47. Stemple D (2004) TILLING-a high-throughput harvest for functional genomics. Nat Rev Genet 5: 145–150. [DOI] [PubMed] [Google Scholar]
- 48. Comai L, Young K, Till B, Reynolds S, Greene E, et al. (2004) Efficient discovery of DNA polymorphisms in natural populations by EcoTILLING. Plant J 37: 778–786. [DOI] [PubMed] [Google Scholar]
- 49. Colbert T, Till B, Tompa R, Reynolds S, Steine M, et al. (2001) High-throughput screening for induced point mutations. Plant Physiol 126: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McCallum C, Comai L, Greene E, Henikoff S (2000) Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol 123: 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsai H, Howell T, Nitcher R, Missirian V, Watson B, et al. (2011) Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol 156: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Appel O, Al-Shehbaz I (2003) Cruciferae. In The families and genera of vascular plants (Kubitzki K, Bayer C. eds). Heidelberg: Springer-Verlag Berlin, pp. 75–174.
- 53. Baum D, Yoon H, Oldham R (2005) Molecular evolution of the transcription factor LEAFY in Brassicaceae. Mol Phylogenet Evol 37: 1–14. [DOI] [PubMed] [Google Scholar]
- 54. Schranz M, Osborn T (2004) De novo variation in life-history traits and responses to growth conditions of resynthesized polyploid Brassica napus (Brassicaceae). Am J Bot 91: 174–183. [DOI] [PubMed] [Google Scholar]
- 55. Song K, Lu P, Tang K, Osborn T (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci 92: 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haughn GW, Gilchrist EJ (2006) TILLING in the Botanical Garden: a reverse genetic technique feasible for all plant species. In: Floriculture, Ornamental and Plant Biotechnology. Teixeira da Silva JA, (ed), Global Science Books, Ltd., London, Vol I, pp. 476–482.
- 57. Murray M, Thompson W (1980) Rapid isolation of high molecular weight plant DNA. Nucleic acids res 8: 4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barkley N, Wang M (2008) Application of TILLING and EcoTILLING as reverse genetic approaches to elucidate the function of genes in plants and animals. Curr Genet 9: 212–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oleykowski CA, Bronson Mullins CR, Godwin AK, Yeung AT (1998) Mutation detection using a novel plant endonuclease. Nucleic Acids Res 26: 4597–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thompson DJ, Higgins DG, Gibson TI (1994) CLUCSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific cap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Felsenstein J (1997) An alternating least-squares approach to inferring phylogenies from pairwise distances. Syst Zool 46: 101–111. [DOI] [PubMed] [Google Scholar]
- 62. Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 63. Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, et al. (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 31: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nieto C, Piron F, Dalmais M, Marco C, Moriones E, et al. (2007) EcoTILLING for the identification of allelic variants of melon eIF4E, A factor that controls virus susceptibility. BMC plant Biol 7: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang G, Imaizumi T, Li W, Saitoh H, Terauchi R, et al. (2008) Self-EcoTILLING to identify single-nucleotide mutations in multigene family. Pestic Biochem Phys 92: 24–29. [Google Scholar]
- 66. Till BJ, Jankowicz-Cieslak J, Sági L, Huynh O, Utsushi H, et al. (2010) Discovery of nucleotide polymorphisms in the Musa gene pool by EcoTILLING. Theor Appl Genet 121: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang N, Shi L, Tian F, Ning H, Wu X, et al. (2010) Assessment of FAE 1 polymorphisms in three Brassica species using EcoTILLING and their association with differences in seed erucic acid contents. BMC plant biol 10: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Magee A, Aspinall S, Rice DW, Cusack BP, Sémon M, et al. (2010) Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res 20: 1700–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yeung A, Hattangadi D, Blakesley L, Nicolas E (2005) Enzymatic mutation detection technologies. Biotechniques 38: 749–758. [DOI] [PubMed] [Google Scholar]
- 70. Robbens S, Derelle E, Ferraz C, Wuyts J, Moreau H, et al. (2007) The complete chloroplast and mitochondrial DNA sequence of Ostreococcus tauri: organelle genomes of the smallest eukaryote are examples of compaction. Mol Biol Evol 24: 956–968. [DOI] [PubMed] [Google Scholar]
- 71. Yang M, Zhang XW, Liu GM, Yin YX, Chen KF, et al. (2010) The Complete Chloroplast Genome Sequence of Date Palm (Phoenix dactylifera L.). PLoS ONE 5: e12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hu ZY, Hua W, Huang SM, Wang HZ (2010) Complete chloroplast genome sequence of rapeseed (Brassica napus L.) and its evolutionary implications. Genet Resour Crop Evol DOI 10. 1007/s10722-010-9626-9. [Google Scholar]
- 73. The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, et al. (2007) Common Sequence Polymorphisms Shaping Genetic Diversity in Arabidopsis thaliana. Science 317: 338–342. [DOI] [PubMed] [Google Scholar]
- 75. Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, et al. (2009) A First-Generation Haplotype Map of Maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- 76. Allender CJ, King GJ (2010) Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Flannery ML, Mitchell FJG, Coyne S, Kavanagh TA, Burke JI, et al. (2006) Plastid genome characterisation in BrassicAand Brassicaseae using Anew set of nine SSRs. Theor Appl Genet 113: 1221–31. [DOI] [PubMed] [Google Scholar]
- 78. Gunnarsdóttir ED, Li M, Bauchet M, Finstermeier K, Stoneking M (2011) High-throughput sequencing of complete human mtDNAgenomes from the Philippines. Genome Res 21: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, et al. (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23: 147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A schematic representation of the position of the primers on accD , matK , rbcL and atp6 genes. Two pairs of primers were designed in accD, matK and rbcL of 96 Brassica species, whereas only one pair of labeled primer was designed in accD and atp6 genes of 91 accessions from Cruciferae. Black boxes showed the genes, and detailed information on each primer was shown in Table 3.
(TIF)
Alignment of representative DNA sequences showing polymorphism in accD gene region regarding INDELs and substitutions. The sequence in each row is a representative sequence, and the red line showed the sequence of the reference sample A64. The numbers in parentheses show the number of samples in Brassicaceae. Blue arrows above the alignment indicated the position of mutation. The position showed in the sequence map began from initial primers attached to M13 adaptors.
(TIF)
Alignment of representative DNA sequences showing polymorphism in atp6 gene region regarding INDELs and substitutions. The sequence in each row is a representative sequence, and the red line showed the sequence of the reference sample A24. The numbers in parentheses show the number of samples in Brassicaceae. Blue arrows above the alignment indicated the position of mutation. The result of DNA sequencing was DNA antisense strand of atp6 gene.
(TIF)
Analysis of ORG-EcoTILLING information in 700 and 800 channel from accD , matK and rbcL gene.
(XLS)
Band analysis of accD gene in 91 Brassicaceae samples identified by ORG-EcoTILLING.
(XLS)