Abstract
Leaves of many plant species open during the day and fold at night. Diurnal leaf movement, named nyctinasty, has been of great interest to researchers since Darwin's time. Nyctinastic leaf movement is generated by the pulvinus, which is a specialized motor organ located at the base of leaf and leaflet. The molecular basis and functional reason behind nyctinasty are unknown.
In a forward screening of a retrotransposon-tagged mutant population of Medicago truncatula, four petiolule-like pulvinus (plp) mutant lines with defects in leaf movement were identified and characterized.
Loss of function of PLP results in the change of pulvini to petiolules. PLP is specifically expressed in the pulvinus, as demonstrated by quantitative reverse-transcription polymerase chain reaction analysis, expression analysis of a PLP promoter-β-glucuronidase construct in transgenic plants and in situ hybridization. Microarray analysis revealed that the expression levels of many genes were altered in the mutant during the day and at night. Crosses between the plp mutant and several leaf pattern mutants showed that the developmental mechanisms of pulvini and leaf patterns are likely independent.
Our results demonstrated that PLP plays a crucial role in the determination of pulvinus development. Leaf movement generated by pulvini may have an impact on plant vegetative growth.
Keywords: leaf movement, Medicago truncatula, nyctinasty, PETIOLULE-LIKE PULVINUS, pulvinus
Introduction
The metabolism, physiology and behavior of plants are orchestrated by diurnal rhythms generated by the rising and setting of the sun (McClung, 2006; de Montaigu et al., 2010). The ability to anticipate such circadian rhythms gives plants fitness advantages, such as enhanced chlorophyll content and improved photosynthesis (Dodd et al., 2005). Unlike animals, plants are unable to move on the ground. However, their leaves can respond to environmental stimulations such as light, temperature, touching and chemical substances by visible movement (McClung, 2006; Ueda & Nakamura, 2007).
Many plants fold their leaves at night. Such nyctinastic movements are common in the legume family (Leguminoseae) and the wood sorrel family (Oxalidaceae). Nyctinastic leaf movements are regulated by a circadian clock with a cycle of c. 24 h (Ueda & Nakamura, 2007). Although the phenomenon was observed in ancient times, it was Darwin who studied and illustrated leaf movements in various plants including Medicago (Darwin, 1880). Videos of such movement can now be easily found on the internet, and people are still fascinated with it. Despite the long history of observing and studying this interesting phenomenon, the molecular basis and functional reason behind nyctinasty are still unknown.
Structurally, nyctinastic movement is mediated by the pulvinus, which is a specialized motor organ located at the base of leaves and leaflets. Two functionally different tissues, the adaxial flexor and the abaxial extensor, constitute the pulvinus (Uehlein & Kaldenhoff, 2008). The oscillations in leaf movement are generated by rhythmic swelling and shrinking of the motor cells of pulvini. Therefore, the pulvinus plays a key role in leaf movement.
Several mutants of legume species with defects in pulvini have been reported, such as the apulvinic mutant in Pisum sativum (Marx, 1987) and the sleepless mutant in Lotus japonicus (Kawaguchi, 2003). These mutants lacked pulvini and formed petiole-like structures at the base of leaflets. However, the gene responsible for pulvinus development is unknown. In addition, it is unclear whether such leaf movement may confer any advantage for plant growth.
Medicago truncatula is a model legume species that undergoes nyctinastic movement by folding its leaflets from horizontal to vertical. In recent years, a Tnt1 retrotransposon-tagged M. truncatula mutant population was generated (Tadege et al., 2008) and a number of mutants affecting leaf patterning and development were identified and characterized. It has been documented that multiple genes are involved in leaf pattern identification in M. truncatula, including PALMATE-LIKE PENTAFOLIATA1 (PALM1), SINGLE LEAFLET1 (SGL1) and Mt NO APICAL MERISTEM (MtNAM) (Wang et al., 2008; Chen et al., 2010; Cheng et al., 2012). The palm1, sgl1 and mtnam mutants exhibit pentafoliate leaves, simple leaves and fused leaves, respectively. In addition, compound leaf formation requires the definition of leaflet boundaries. LATERALORGAN BOUNDARIES (LOB) is a gene that is expressed specifically in organ boundaries. In Arabidopsis, however, the lob mutant does not display obvious phenotype as a result of functional redundancy of other gene members in this family (Shuai et al., 2002). The role of LOB in compound-leafed species remains undetermined.
In this study, mutants with defects in leaf movement were isolated from the Tnt1 retrotransposon-tagged mutant population of M. truncatula. Loss of function in PETIOLULE-LIKE PULVINUS (PLP) leads to the change of pulvini to petiolules. The gene was cloned and molecular analysis showed that PLP is a unique LOB domain protein. The specific spatial and temporal expression pattern of PLP suggests that PLP plays a key role in the development of pulvini. Global transcript profiling analysis revealed that loss of function of PLP affects the expression of a large number of genes during both day and night. Comparison of biomass yield between wild-type and plp mutants indicates that leaf movement may have an impact on plant vegetative growth.
Materials and Methods
Plant material and growth conditions
Medicago truncatula Gaertn. Ecotype R108 was used as the wild type for all the experiments described in this study. Plants were grown in Metro-Mix 830 soil mix at 22°C day/20°C night temperature, 16-h day/8-h night photoperiod and 70–80% relative humidity. The four plp alleles identified were: NF2623 (plp-1, ecotype R108; a Tnt1 inserted between 178 bp and 179 bp), NF0571 (plp-2, ecotype R108; a Tnt1 inserted between 130 bp and 131 bp), F9359-LTR4 (plp-3, ecotype A17; a Tnt1 inserted between 36 bp and 37 bp of PLP) and NF5514 (plp-4, ecotype R108; a Mere1 inserted between 246 bp and 247 bp).
Molecular cloning of PLP and vector construction
The coding sequence of PLP was amplified by polymerase chain reaction (PCR) from cDNA of wild-type M. truncatula and deposited in GenBank (JN412594). The primers used for cloning of PLP were: PLP-CDS-F, CACCATGGCATCATCAAGCTCATAC; and PLP-CDS-R, TCACAAATTACCTCCTCCTACA. To functionally complement the plp mutant, a 2414 bp PLP promoter sequence plus a 1611 bp PLP genomic DNA sequence were PCR-amplified and cloned into the pENTR™/D-TOPO cloning vector (Invitrogen, Chicago, IL, USA). Then the fragment was transferred into the pEarleyGate 301 vector (Earley et al., 2006) by attL × attR recombination reactions (Invitrogen). The following primers were used to amplify the PLP promoter and the genomic DNA sequences: PLP-Com-F: CACCGTTAAAGTGTATCATAAGGGAG and PLP-Com-R:TCACTGCTCGTTTTCCTGTTAG.
β-Glucuronidase (GUS) staining and microscopy
Fully expanded leaves were collected for GUS staining. The GUS activity was histochemically detected as previously described (Jefferson et al., 1987). For scanning electron microscopy, tissue samples were fixed in 3% (v/v) glutaraldehyde and then dehydrated in a series of ethanol. The samples were observed using Hitachi TM-3000 SEM at an accelerating voltage of 15 kV. For fluorescent imaging, a Leica TCS SP2 AOBS confocal laser scanning microscope was used. The 488-nm line of an argon laser was chosen for the green fluorescent protein (GFP) signal, and emission was detected at 150 nm.
Plant transformation
The final binary vectors were transferred into the disarmed Agrobacterium tumefaciens strain EHA105 using the freezing/heat shock method. Leaves of wild-type and plp-1 were transformed with EHA105 harboring various vectors (Cosson et al., 2006).
RNA extraction, RT-PCR, qRT-PCR and microarray analysis
Leaves of 4-wk-old wild-type and plp mutant plants were collected for RNA isolation. Total RNA was isolated using Trizol Reagent (Invitrogen). One microgram of RNA was reverse transcribed with SuperscriptIII (Invitrogen) following the manufacturer's instructions. The cDNA was used as a template for reverse transcription PCR (RT-PCR) and quantitative RT-PCR (qRT-PCR). Primers used for quantifying the expression level of PLP were PLP-qPCR-F (ATCACAAACAGCGCAGGAGAA) and PLP-qPCR-R(GGCTGCACATGGTGAATTGTAT). For microarray analysis, total RNA was extracted from three biological replicates of fully expanded leaves of 4-wk-old plp-1 mutant and wild-type-like plants in a segregating F2 population. RNA purification, probe labeling, hybridization, and scanning for microarray analysis were conducted as previously described (Zhou et al., 2011a). Functional enrichments were calculated with PageMan (Usadel et al., 2006) and visualized with mapman (Thimm et al., 2004).
In situ hybridization analysis
A fragment of 510 bp PLP coding sequence (CDS) was amplified by PCR using primers PLP-is-F: TTGCACCATACTTTCCACCGGA and PLP-is-R: ACTTCTTCTGTCACCAGTGCCT. The PCR product was labeled with Digoxigenin (Digoxigenin-11-UTP, Roche Diagnostics). RNA in situ hybridization was performed on shoot apices or inflorescence apices of 4-wk-old wild-type plants as previously described (Zhou et al., 2011b).
Phylogenetic tree
Full-length protein sequences of PLP and other lateral organ boundary (LOB) members were used for phylogenetic analysis. Alignments were performed using jalview with default parameters (Waterhouse et al., 2009). Phylogenetic tree was constructed by the mega4 program (Tamura et al., #b300) with 1000 bootstrap replications.
Results
PLP is required for pulvinus development
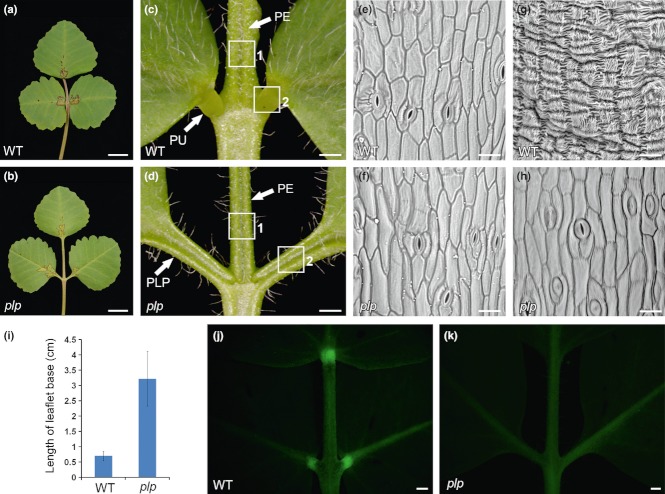
The adult leaves of M. truncatula are in trifoliate form consisting of one terminal leaflet and two lateral leaflets. Each leaflet has a pulvinus that is responsible for leaflet movement. To identify mutants with defects in leaf movement, a large number (c. 10 000) of Tnt1 retrotransposon-tagged M. truncatula lines were screened. Four independent mutant lines with obvious defects in leaf movement were identified. In contrast to the wild-type, the pulvini developed in the mutants were changed to petiolule-like structures (Fig. 1a–d). Therefore, the mutation was named petiolule-like pulvinus (plp) and the allelic lines were designated plp-1, plp-2, plp-3, and plp-4.
Fig. 1.
The petiolule-like pulvinus (plp) mutant of Medicago truncatula shows developmental defects in pulvini. (a,b) Adult leaf of wild type and plp mutant. (c,d) Close-up view of leaflet base in wild-type and plp mutant. (e,f) Scanning electron micrograph of epidermal cells in petiolule of wild type (box 1 in (c)) and plp mutant (box 1 in (d)). (g,h) Scanning electron micrograph of epidermal cells in pulvinus of wild type (box 2 in (c)) and petiolule-like pulvinus of plp mutant (box 2 in (d)). (i) Length of lateral leaflet base of wild type and plp mutant. Values are means ± SD (n = 20). (j,k) DR5rev:green fluorescent protein (GFP) expression in the leaf (abaxial side) of wild type and plp mutant. PE, petiolule; PU, pulvinus; PLP, petiolule-like pulvinus; Bars, (a,b) 5 mm; (c,d,j,k) 500 μm; (e–h) 20 μm.
Scanning electron microscopy (SEM) analysis was performed to characterize the defects of the plp mutants. While the epidermal cells of petiolules in wild-type and mutants showed similar shape (Fig. 1e,f), the shape of the epidermal cells of pulvini in wild-type and mutants were different (Fig. 1g,h). The pulvini of the wild-type consisted of specific motor cells (Fig. 1g). However, cells of petiolule-like pulvini in plp mutants were elongated and the stomata were observable, similar to the epidermal cells of petiolule (Fig. 1h). Accompanying the changes in pulvini was an increase in the leaflet base length in the plp mutants (Fig. 1i). These observations indicate that loss of function in PLP leads to defects in the determination of pulvinus cell fate, resulting in the development of petiolule-like pulvinus in the plp mutants. A previous study showed that auxin plays important roles in various organ development in M. truncatula (Zhou et al., 2011b). To test if auxin accumulation is associated with pulvinus development, the DR5rev:GFP auxin response reporter was introduced into the wild-type and plp-1 mutant, respectively. A GFP signal was detected specifically in the pulvini, but not in petiolules of wild type (Fig. 1j). In contrast, no obvious auxin accumulation was observed at the same location in the plp mutants (Fig. 1k). This observation further suggests that the pulvini lost their developmental characteristics and were changed to petiolules in the plp mutants.
PLP encodes a LOB domain protein
To identify the gene responsible for the mutant phenotype, thermal asymmetric interlaced-PCR (TAIL-PCR) was performed to recover the flanking sequences from plp-1. In total, 16 flanking sequences were identified in plp-1, and one of the sequences was associated with the mutant phenotype. A full-length genomic sequence was obtained after using the flanking sequence to search against the M. truncatula genome from the National Center for Biotechnology Information. The 579-nucleotide full-length CDS of this gene was cloned by RT-PCR. Alignment between the cDNA and genomic sequences revealed that PLP contains one exon (Fig. 2a). The Tnt1 insertion in plp-1 was located in the exon. Further sequence analysis revealed that the other three mutants carried either a tobacco Tnt1 retrotransposon or a M. truncatula Mere1 retrotransposon (Rakocevic et al., 2009) in the exon of PLP. The results were confirmed by genomic PCR analysis (Fig. 2b). Reverse transcription-PCR was performed and no transcripts of PLP were detected in the four alleles (Fig. 2b). To further confirm that the mutant phenotype was caused by disruption of this gene, a construct carrying a 4.2-kb genomic sequence, including a 2.5-kb promoter region and the PLP open reading frame, was introduced into homozygous plp-1 mutant plants. Phenotypic observation showed that the defects in pulvini were fully rescued (Fig. 2c).
Fig. 2.
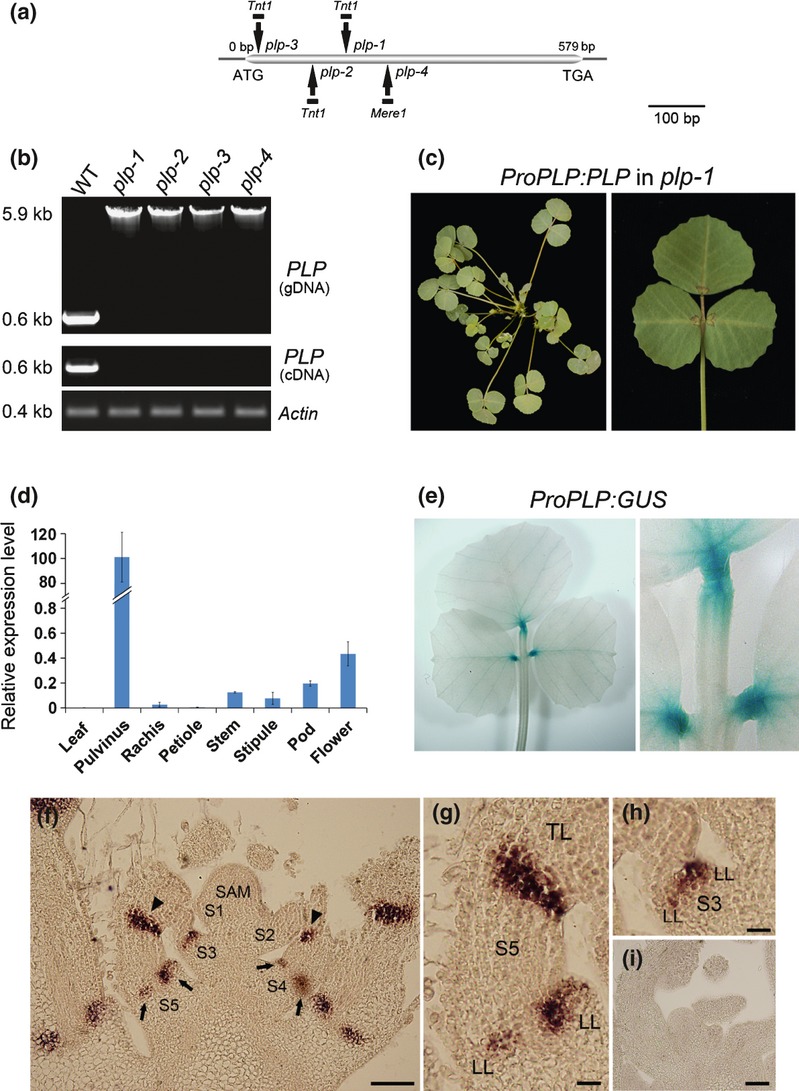
Molecular cloning and expression pattern of PLP in Medicago truncatula. (a) Schematic representation of the gene structure of PLP. The positions of ATG start and TGA stop codons are shown. Vertical arrows mark the location of Tnt1 or Mere1 retrotransposons in plp alleles. (b) PCR and RT-PCR amplification of PLP from the wild-type (WT) and plp mutants. A single insertion (c. 5.3 kb) was detected in each mutant line. PLP expression was not detected in the mutants. Actin was used as a loading control. Three technical replicates were performed. (c) Genetic complementation of the plp mutant. A representative plp-1 line transformed with the PLP genomic sequence (ProPLP : PLP) showed normal wild-type-like leaves and pulvini. (d) Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis of the expression pattern of PLP. Values are means ± SD (n = 3). (e) β-Glucuronidase (GUS) staining of trifoliates of transgenic M. truncatula plants carrying the PLP Promoter-GUS construct. (f–i) In situ hybridization analysis of PLP mRNA in vegetative apices of the wild type. Longitudinal section of the shoot apical meristem (SAM) is shown in (f). Arrows point to lateral leaflet base. Arrowheads point to terminal leaflet base. The close-up views of S5 and S3 are shown in (g) and (h), respectively. (i) In situ hybridization using the sense probe as control. S1–S5, stage1 to stage5; TL, terminal leaflet primordia; LL, lateral leaflet primordia. Bars, (f,i) 50 μm; (g,h) 10 μm.
Sequence alignment was performed between PLP and its putative orthologs from Glycine max, Vitis vinifera, Lotus japonicus, Zea mays, Triticum aestivum and Arabidopsis thaliana. The results showed that PLP contains a highly conserved LOB domain (see the Supporting Information, Fig. S1). Phylogenetic analysis with 42 members of the LOB domain gene family from Arabidopsis revealed that PLP was evolutionarily closer to ASL4/LOB (At5g63090) and showed 75% identity with ASL4/LOB (Fig. S2).
PLP specifically expresses in pulvini
The expression pattern of PLP from the M. truncatula Gene Expression Atlas showed a relatively high expression level of PLP in vegetative buds (Fig. S3). To compare the expression level of PLP in pulvini and other tissues, qRT-PCR was performed. The data revealed that PLP was highly expressed in the pulvinus compared with other plant organs (Fig. 2d). To confirm this specific expression pattern, a PLP promoter driven β-glucuronidase (GUS) reporter gene was introduced into wild-type plants. The expression of GUS was mainly detected in the pulvini of transgenic plants (Fig. 2e). These results suggest that PLP plays a highly specific role in the development of pulvinus.
In M. truncatula, it has been shown that lateral leaflet primordia and the terminal leaflet primordium develop at different stages (Wang et al., 2008). A pair of lateral leaflet primordia (LL) is developed first at the proximal end of the common leaf primordium, followed by the development of the terminal leaflet primordium (Wang et al., 2008). The development of both terminal and lateral leaflets is associated with the formation of pulvini. To explore the spatial and temporal localizations of PLP during the development of pulvini, RNA in situ hybridization was performed in the wild type (Fig. 2f–i). PLP mRNA was first detected in the leaf primordium at stage 3 (S3) in which lateral leaflet primordia were developing (Fig. 2h). At stage 4 (S4), three strong hybridization signals were detected in leaf primordium where the pulvini were formed. Two of them were associated with the lateral leaflets (Fig. 2f, arrows) and one was associated with the terminal leaflet (Fig. 2f, arrowheads). This elaborate expression pattern was continually exhibited in the older leaf primordia (Fig. 2g). The specific spatial and temporal expression pattern of PLP further demonstrates that PLP is tightly associated with the development of pulvini.
Genetic interactions between plp and leaf pattern mutants
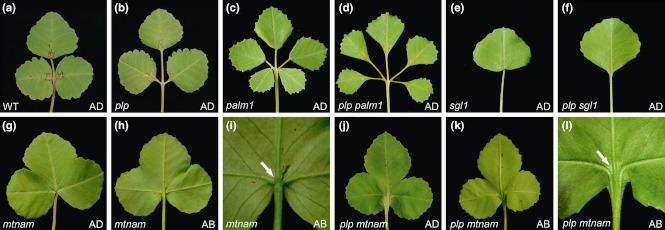
Defects in the pulvini of plp mutants suggest that PLP is required for the proper formation of leaves. To investigate the possible role of PLP in leaf patterning, the plp mutant was crossed with the leaf pattern mutants palm1, sgl1 and mtnam. Compared with the phenotype of wild type and single mutants, the plp palm1 double mutant exhibited pentafoliolate leaves with petiolule-like pulvini (Fig. 3a–d). The plp sgl1 double mutant had simple leaves, resembling those of sgl1 single mutant. The pulvini of plp sgl1 were similar to those in the plp single mutant (Fig. 3e–f). The weak allele of mtnam was used for generating a double mutant because the strong mtnam allele was seedling-lethal. In mtnam, the midveins of lateral leaflets were fused with the midvein of the terminal leaflet (Fig. 3g–i). The plp mtnam double mutant also showed fused leaflets. However, elongation of the proximal end of midveins was observed, indicating that the pulvini were also changed to petiolules in the double mutant. In addition, clustered pulvini were observed in mtnam (Fig. 3i, arrow) while separated petiolule-like pulvini were found in plp mtnam (Fig. 3l, arrow), indicating that MtNAM and PLP may play different roles in organ separation and boundary identification. These observations suggest that the developmental mechanism of pulvini determination and leaf pattern are probably independent. The findings further imply that the pulvini are tightly associated with leaflet formation irrespective of changes in leaflet number and pattern.
Fig. 3.
Genetic interactions between plp and different leaf pattern mutants. Adult leaves of wild-type Medicago truncatula and the mutants are shown. (a) Wild type; (b) plp mutant; (c) palm1 mutant; (d) plp palm1 double mutant; (e) sgl1 mutant; (f) plp sgl1 double mutant; (g,h) mtnam mutant; (i) close-up view of leaf base of the mtnam mutant, the arrow points to the clustered pulvini; (j,k) plp mtnam double mutant; (l) close-up view of leaf base in the plp mtnam double mutant, the arrow points to the separated petiolule-like pulvini. AD, adaxial side; AB, abaxial side.
Global changes in gene expression in plp mutant
The leaflets of M. truncatula exhibit nyctinastic movement with circadian rhythms. They are usually horizontal (open) during the day and vertical (folded or closed) at night (Fig. 4a,b). The closed leaflets start to move becoming horizontal at dawn (06:00 h, Zeitgeber time 0, ZT0). The opened leaflets start to move vertically at 19:00 h (ZT13) and continue the movement until c. 21:00 h (ZT15) to close completely. In plp mutants, the leaflets lost their usual dynamic movement because of the altered structure of pulvini (Fig. 4c,d). To investigate molecular response to PLP associated leaf movement, gene transcript levels in wild type and plp-1 mutant were measured using AffymetrixMedicagoGenechips (Affymetrix, CA, USA). Fully expanded leaves were collected from plants at noon (ZT6) and at midnight (ZT18) and used for RNA extraction and microarray analysis.
Fig. 4.
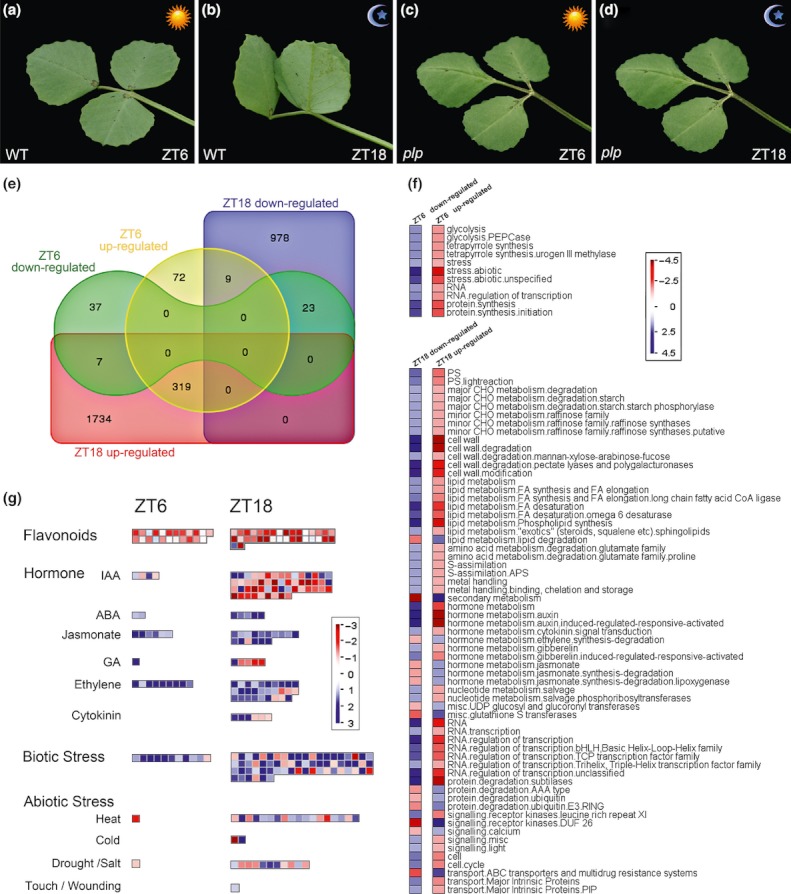
Leaf movement and differential expression of metabolic genes in Medicago truncatula wild-type and plp mutant leaves at ZT6 (noon) and ZT18 (midnight). (a,b) Leaf movement of wild type (WT) at ZT6 and ZT18. (c,d) The plp mutant lost its usual dynamic movement of leaflets because of the altered structure of pulvinus. (e) Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) of total numbers of upregulated and downregulated genes in the plp mutant at ZT6 and ZT18. (f) PageMan functional enrichments of differentially expressed genes. Red, underrepresented functional categories; blue, overrepresented functional categories; (g) Selected significantly differentially expressed genes in flavonoids and hormone metabolism, as well as in biotic and abiotic stress. Visualization of these genes with MapMan 3.5.1R2 using log2-transformed ratios of the mutant divided by wild-type. Red, up-regulated genes; blue, down-regulated genes.
Compared with the wild type, the expression levels of 467 genes at ZT6 and of 3070 genes at ZT18 were altered by at least twofold in the plp mutant (see the Supporting Information, Tables S1, S2). Among 400 upregulated and 67 downregulated genes in plp mutants at ZT6, only nine and seven genes were oppositely regulated at ZT18, respectively, suggesting that abolished leaf movement associated with PLP affects gene expression in a consistent manner during both day and night (Fig. 4e). To characterize the possible functions of differentially expressed genes, functional enrichment was analysed by PageMan (Usadel et al., 2006). The results revealed that upregulated genes at ZT18 were enriched in lipid metabolism, secondary metabolism, hormone metabolism, protein degradation and transport. The upregulated genes at ZT6 were under-represented in all functional categories. However, most of the downregulated genes at both ZT6 and ZT18 were over-represented. They were enriched for genes involved in photosynthesis, stress, protein synthesis, cell wall degradation, lipid metabolism, hormone metabolism and RNA transcription. The data suggests that broad effects in multiple biological processes were induced by PLP associated leaf movement.
To further illustrate the effects of PLP-associated leaf movement on plant growth and development, differentially expressed genes at ZT6 and ZT18 were mapped onto MapMan (Fig. S4, metabolism overview; Tables S3, S4) (Thimm et al., 2004). The data showed that changes in the amount of gene transcript were observed in most metabolic processes. Among these changes, genes involved in secondary metabolism of terpenes, flavonoids and phenylpropanoids/phenolics were strongly upregulated at ZT18. It is noted that the flavonoid-related genes are induced at both ZT6 and ZT18, suggesting that flavonoid metabolism was affected in plp mutants both during the day and at night (Fig. 4g). Further analysis showed that genes involved in hormone metabolism were changed dramatically at ZT18 compared with those at ZT6 (Fig. 4g). In addition, the expression levels of both biotic and abiotic stress-related genes were changed at both time-points, especially at ZT18. These observations imply that loss of function of PLP probably has negative effects on their growth. To verify this hypothesis, the vegetative growth of 3-wk-old plants of wild type and plp mutants was analysed. The projected leaf area of 48 plants from wild-type and plp mutant was calculated (Fig. S5a,b). The data showed that wild-type plants had 15% more projected leaf area than that of the mutants (Fig. S5c). Furthermore, biomass yield of the aboveground parts of wild-type plants was significantly higher than the yield of mutant plants (Fig. S5d).
Discussion
In this study, four independent PLP alleles were found. These alleles showed the same defects in the determination of developmental identity of pulvini. PLP is identified as a member of the plant-specific LOB domain gene family in M. truncatula. In Arabidopsis, the LOB gene is expressed at the base of all lateral organs. The expression pattern indicates that it may play a crucial role in defining organ boundaries (Shuai et al., 2002; Majer & Hochholdinger, 2011). In M. truncatula, pulvini are formed when the leaflet primordia emerge and localize at the joint between leaflet and petiole. Therefore, the formation of pulvini defines of boundaries between leaflet and petiole. In compound-leafed species, loss of function in boundary formation-related genes resulted in fused leaves (Berger et al., 2009; Cheng et al., 2012). Unlike those genes, PLP defines the specific boundaries between leaflet and petiole by playing a unique role in the development of pulvini, as revealed by the following lines of evidence. First, the pulvinous differentiation program did not proceed because of the absence of PLP. The pulvini in the plp mutants were changed to petiolules with elongated epidermal cells and increased organ length. In addition, auxin accumulation was specifically observed in the pulvini of adult leaves in wild type, while no such auxin accumulation was detected in the plp mutants, further confirming that the unique structure of pulvini was altered in the mutant. Second, PLP was highly expressed in the pulvini, supported by the qRT-PCR data and the analysis of transgenics carrying the PLP promoter–GUS reporter construct. Furthermore, the developmental link between PLP expression and pulvini emergence was revealed by in situ hybridization. Third, genetic interactions between plp and leaf pattern mutants sgl1 and palm1 indicate that PLP is not involved in the determination of leaf pattern but is associated with leaflet formation. The weak allele of mtnam was used to produce the double mutant with plp. Fused leaflets but separated petiolule-like pulvini in the double mutant suggest that MtNAM and PLP may regulate the formation of different boundaries. In this proposed scenario, MtNAM plays a conserved role in boundary identification between leaflets (Blein et al., 2008; Cheng et al., 2012) while PLP specifically elaborates boundary formation between leaflet and petiole through activation of the pulvinous differentiation program. Such a program is required for the identification of pulvini where compact and convoluted motor cells are produced. The loss of cell fate identity in the petiolule-like pulvinus of plp led to the development of elongated cells similar to those in rachis. Therefore, the formation of elongated petiolules in the plp mutant results from the conversion of convoluted motor cells to regular rachis cells. It has been reported that conserved gene modules function in a context-dependent manner (Efroni et al., 2010; Hay & Tsiantis, 2010). Petiolules are present in most compound leafed species, but pulvini only exist in some species with nyctinastic movement. This phenomenon suggests that pulvini development is the result of context-specific effects and that the conserved LOB genes are differentially deployed among species, resulting in morphological diversity. We propose that the formation of boundaries regulated by PLP is required for plant species that are capable of displaying nyctinastic leaf movement.
Leaf movement may have particular functional significance for plant growth by optimizing light intensity on leaf surface during the day. It has been suggested that nyctinastic leaf movement could be a type of self-adjusting mechanism in response to environmental changes. In order to better understand the biological effects of leaf movement, we analysed global changes of gene expression in plp-1 mutant during the day and at night. The microarray experiments revealed that transcriptional changes occur at both ZT6 and ZT18. Surprisingly, the number of upregulated or downregulated genes at ZT18 was about seven times more than that at ZT6, suggesting that loss of function of PLP has a large impact on gene expression at night. It seems that a dynamic gene regulatory mechanism exists even after the closing of leaflets and without light stimulation. Further research is needed to explore this area.
Functional enrichment analysis revealed overrepresentation of genes at ZT18 involved in multiple biological processes, suggesting that loss of function of PLP has broad effects on plants. Previous reports showed that flavonoid accumulation is associated with the biotic or abiotic stress conditions (Christie et al., 1994; Dixon & Paiva, 1995; Vanderauwera et al., 2005; Hernandez & Van Breusegem, 2010; Kang et al., 2011). In addition, flavonoids also protect leaves from light stress and UV-B radiation (Gould et al., 2000; Jordan, 2002; Deavours & Dixon, 2005). The upregulation of genes involved in flavonoid metabolism in the plp mutant indicate that the mutant plants may undergo certain stresses. This hypothesis is supported by the expression changes of genes involved in biotic stress and abiotic stress. The expression level of hormone metabolism-related genes are also altered in mutants. The comprehensive changes in hormone metabolism are probably the result of crosstalk between plant hormones in response to biotic and abiotic stresses (Spoel & Dong, 2008; Wolters & Jurgens, 2009; Robert-Seilaniantz et al., 2011). Consistent with these findings, wild-type plants showed a larger leaf area and higher biomass yield than those of the mutants, suggesting that nyctinastic leaf movement associated with PLP may have an important impact on plant vegetative growth.
In summary, we identified plp mutants with defects in leaf movement. The specific function of PLP was characterized in detail. PLP encodes a unique LOB domain protein and plays a crucial role in the determination of pulvinus development. Nyctinastic leaf movement associated with PLP greatly impacts on gene expression and may benefit plant growth and development.
Acknowledgments
We thank Amy Mason and Steven Tudor for critical reading of the manuscript and Kirankumar Mysore, Million Tadege, Pascal Ratet and René Geurts for providing the Tnt1 mutants. This work was supported by the Samuel Roberts Noble Foundation, the National Science Foundation (Grant No. EPS-0814361) and the BioEnergy Science Center. The BioEnergy Science Center is a US Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1 Alignment of amino acid sequences of PLP and its putative orthologs.
Fig. S2 Phylogenetic tree of PETIOLULE-LIKE PULVINUS (PLP) and members of the lateral organ boundary (LOB) domain gene family from Arabidopsis.
Fig. S3 Expression profiling of PLP transcripts.
Fig. S4 Differential expression of metabolic genes in the plp mutant.
Fig. S5 Vegetative growth of wild-type and plp mutant plants.
Table S1 List of downregulated and upregulated genes/probe sets in the plp mutant at ZT6
Table S2 List of downregulated and upregulated genes/probe sets in the plp mutant at ZT18
Table S3 List of genes involved in flavonoid metabolism, hormone metabolism and stress response at ZT6
Table S4 List of genes involved in flavonoid metabolism, hormone metabolism and stress response at ZT18
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development. 2009;136:823–832. doi: 10.1242/dev.031625. [DOI] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P. A conserved molecular framework for compound leaf development. Science. 2008;322:1835–1839. doi: 10.1126/science.1166168. [DOI] [PubMed] [Google Scholar]
- Chen J, Yu J, Ge L, Wang H, Berbel A, Liu Y, Chen Y, Li G, Tadege M, Wen J, et al. Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proceedings of the National Academy of Sciences, USA. 2010;107:10754–10759. doi: 10.1073/pnas.1003954107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Peng J, Ma J, Tang Y, Chen R, Mysore KS, Wen J. NO APICAL MERISTEMMtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula. New Phytologist. 2012;195:71–84. doi: 10.1111/j.1469-8137.2012.04147.x. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Alfenito MR, Walbot V. Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta. 1994;194:541–549. [Google Scholar]
- Cosson V, Durand P, d'Erfurth I, Kondorosi A, Ratet P. Medicago truncatula transformation using leaf explants. Methods Molecular Biology. 2006;343:115–127. doi: 10.1385/1-59745-130-4:115. [DOI] [PubMed] [Google Scholar]
- Darwin C. The power of movement in plants. London, UK: J. Murray; 1880. [Google Scholar]
- Deavours BE, Dixon RA. Metabolic engineering of isoflavonoid biosynthesis in alfalfa. Plant Physiology. 2005;138:2245–2259. doi: 10.1104/pp.105.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant Journal. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Efroni I, Eshed Y, Lifschitz E. Morphogenesis of simple and compound leaves: a critical review. Plant Cell. 2010;22:1019–1032. doi: 10.1105/tpc.109.073601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KS, Markham KR, Smith RH, Goris JJ. Functional role of anthocyanins in the leaves of Quintinia serrata A. Cunn. Journal of Experimental Botany. 2000;51:1107–1115. doi: 10.1093/jexbot/51.347.1107. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. KNOX genes: versatile regulators of plant development and diversity. Development. 2010;137:3153–3165. doi: 10.1242/dev.030049. [DOI] [PubMed] [Google Scholar]
- Hernandez I, Van Breusegem F. Opinion on the possible role of flavonoids as energy escape valves: novel tools for nature's Swiss army knife? Plant Science. 2010;179:297–301. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BR. Molecular response of plant cells to UV-B stress. Functional Plant Biology. 2002;29:909–916. doi: 10.1071/FP02062. [DOI] [PubMed] [Google Scholar]
- Kang Y, Han Y, Torres-Jerez I, Wang M, Tang Y, Monteros M, Udvardi M. System responses to long-term drought and re-watering of two contrasting alfalfa varieties. Plant Journal. 2011;68:871–889. doi: 10.1111/j.1365-313X.2011.04738.x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M. SLEEPLESS, a gene conferring nyctinastic movement in legume. Journal of Plant Research. 2003;116:151–154. doi: 10.1007/s10265-003-0079-5. [DOI] [PubMed] [Google Scholar]
- Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends in Plant Science. 2011;16:47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Marx GA. A suite of mutants that modify pattern formation in pea leaves. Plant Molecular Biology Reporter. 1987;5:311–335. [Google Scholar]
- McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Montaigu A, Toth R, Coupland G. Plant development goes like clockwork. Trends in Genetics. 2010;26:296–306. doi: 10.1016/j.tig.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Rakocevic A, Mondy S, Tirichine L, Cosson V, Brocard L, Iantcheva A, Cayrel A, Devier B, Abu El-Heba GA, Ratet P. MERE1, a low-copy-number copia-type retroelement in Medicago truncatula active during tissue culture. Plant Physiology. 2009;151:1250–1263. doi: 10.1104/pp.109.138024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annual Review of Phytopathology. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Pena CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiology. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host & Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant Journal. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Ueda M, Nakamura Y. Chemical basis of plant leaf movement. Plant & Cell Physiology. 2007;48:900–907. doi: 10.1093/pcp/pcm060. [DOI] [PubMed] [Google Scholar]
- Uehlein N, Kaldenhoff R. Aquaporins and plant leaf movements. Annals of Botany. 2008;101:1–4. doi: 10.1093/aob/mcm278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Blasing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inze D, Van Breusegem F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiology. 2005;139:806–821. doi: 10.1104/pp.105.065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chen J, Wen J, Tadege M, Li G, Liu Y, Mysore KS, Ratet P, Chen R. Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiology. 2008;146:1759–1772. doi: 10.1104/pp.108.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters H, Jurgens G. Survival of the flexible: hormonal growth control and adaptation in plant development. Nature Reviews Genetics. 2009;10:305–317. doi: 10.1038/nrg2558. [DOI] [PubMed] [Google Scholar]
- Zhou C, Han L, Hou C, Metelli A, Qi L, Tadege M, Mysore KS, Wang ZY. Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. Plant Cell. 2011b;23:2106–2124. doi: 10.1105/tpc.111.085464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Pislariu C, Nakashima J, Fu C, Jiang Q, Quan L, Blancaflor EB, Tang Y, Bouton JH, et al. From model to crop: functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiology. 2011a;157:1483–1496. doi: 10.1104/pp.111.185140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.