Abstract
A reversibly glycosylated polypeptide from pea (Pisum sativum) is thought to have a role in the biosynthesis of hemicellulosic polysaccharides. We have investigated this hypothesis by isolating a cDNA clone encoding a homolog of Arabidopsis thaliana, Reversibly Glycosylated Polypeptide-1 (AtRGP1), and preparing antibodies against the protein encoded by this gene. Polyclonal antibodies detect homologs in both dicot and monocot species. The patterns of expression and intracellular localization of the protein were examined. AtRGP1 protein and RNA concentration are highest in roots and suspension-cultured cells. Localization of the protein shows it to be mostly soluble but also peripherally associated with membranes. We confirmed that AtRGP1 produced in Escherichia coli could be reversibly glycosylated using UDP-glucose and UDP-galactose as substrates. Possible sites for UDP-sugar binding and glycosylation are discussed. Our results are consistent with a role for this reversibly glycosylated polypeptide in cell wall biosynthesis, although its precise role is still unknown.
The primary cell wall of dicot plants is laid down by young cells prior to the cessation of elongation and secondary wall deposition. Making up to 90% of the cell's dry weight, the extracellular matrix is important for many processes, including morphogenesis, growth, disease resistance, recognition, signaling, digestibility, nutrition, and decay. The composition of the cell wall has been extensively described (Bacic et al., 1988; Levy and Staehelin, 1992; Zablackis et al., 1995), and yet many questions remain unanswered regarding the synthesis and interaction of these components to provide cells with a functional wall (Carpita and Gibeaut, 1993; Carpita et al., 1996).
Heteropolysaccharide biosynthesis can be divided into four steps: (a) chain or backbone initiation, (b) elongation, (c) side-chain addition, and (d) termination and extracellular deposition (Waldron and Brett, 1985). The similarity between various polysaccharide backbones leads to the prediction that the synthesizing machinery would be conserved between them. For example, the backbone of xyloglucan polymers, β-1,4 glucan, can be synthesized independently of or concurrently with side-chain addition (Campbell et al., 1988; White et al., 1993), and this polymer and the chains that make up cellulose are identical. The later addition of side chains to xyloglucan are catalyzed by specific transferases (Kleene and Berger, 1993) such as xylosyltransferase (Campbell et al., 1988), galactosyltransferase, and fucosyltransferase (Faïk et al., 1997), all of which are localized to the Golgi compartment (Brummell et al., 1990; Driouich et al., 1993; Staehelin and Moore, 1995).
The enzymes involved in wall biosynthesis have been recalcitrant to isolation (Carpita et al., 1996; Albersheim et al., 1997). Only recently has the first gene encoding putative cellulose biosynthetic enzymes, celA, been isolated from cotton (Gossypium hirsutum) and rice (Oryza sativa; Pear et al., 1996).
During studies of polysaccharide synthesis in pea (Pisum sativum) Golgi membranes, Dhugga et al. (1991) identified a 41-kD protein doublet that they suggested was involved in polysaccharide synthesis. The authors showed that this protein could be glycosylated by radiolabeled UDP-Glc but that this labeling could be reversibly competed with by unlabeled UDP-Glc, UDP-Xyl, and UDP-Gal, the sugars that make up xyloglucan (Hayashi, 1989). The 41-kD protein was named PsRGP1 (P. sativum Reversibly Glycosylated Polypeptide-1; Dhugga et al., 1997). Furthermore, the conditions that stimulate or inhibit Golgi-localized β-glucan synthase activity are the same conditions that stimulate or inhibit the glycosylation of PsRGP1 (Dhugga et al., 1991). To address the role of this protein in polysaccharide synthesis, the authors purified the polypeptides and obtained the sequences from tryptic peptides (Dhugga and Ray, 1994). Antibodies raised against PsRGP1 showed that it is soluble and localized to the plasma membrane (Dhugga et al., 1991) and Golgi compartment (Dhugga et al., 1997). In addition to its Golgi localization, the steady-state glycosylation of PsRGP1 is approximately 10:7:3 (UDP-Glc:-Xyl:-Gal), which is similar to the typical sugar composition of xyloglucan (1.0:0.75:0.25; Dhugga et al., 1997).
We were interested in studying various aspects of cell wall metabolism, including the synthesis of polysaccharides and their delivery to the cell wall. Studies in pea have shown that a 41-kD protein may be involved in cell wall polysaccharide synthesis, possibly that of xyloglucan (Dhugga et al., 1997). Here we report the characterization of AtRGP1 (Arabidopsis thaliana Reversibly Glycosylated Polypeptide-1), a soluble protein that can also be found weakly associated with membrane fractions, most likely the Golgi fraction. The reversible nature of the glycosylation of this Arabidopsis homolog by the substrates used to make polysaccharides (nucleotide sugars) suggests a possible role for AtRGP1 in polysaccharide biosynthesis.
MATERIALS AND METHODS
Plant and Cell Growth
Arabidopsis thaliana wild-type plants (Columbia ecotype) were grown in soil at 22°C and 80% humidity and with 16 h of light. To obtain large amounts of soil-free root tissue, seeds were germinated in liquid culture medium (1 seed 100 mL−1, 10 g L−1 Suc, 4.3 g L−1 Murashige-Skoog salts [GIBCO-BRL], 0.5 g L−1 Mes, 0.1 g L−1 myo-inositol, 1 mg L−1 thiamine-HCl, and 0.5 mg L−1 nicotinic acid, pH 5.7). Plants grown in liquid culture and maintained under constant agitation (50–60 rpm) and light grew mostly roots.
A. thaliana cell-suspension cultures were obtained from Axelos et al. (1992) and were maintained by diluting a 2-week-old culture 1:5 with fresh suspension-culture medium (3.2 g L−1 Gamborg's B5 medium [Sigma], 20 g L−1 Suc, 2.5 μm 2,4-D, and 0.5 g L−1 Mes, pH 5.7).
Cloning, Sequencing, and Sequence Analysis
Peptide sequences from purified pea (Pisum sativum) proteins (Dhugga and Ray, 1994; Fig. 1) were used to search the EST database (dBEST) comprising the GenBank, EMBL, and DDBJ databases. We used a database search program (Basic BLAST) with the “tblastn” option, which compares a protein query sequence against a nucleotide sequence database dynamically translated in all reading frames (Altschul et al., 1990, http://www.ncbi.nlm.nih.gov/BLAST/). Twenty-eight ESTs have been obtained so far from the original search and subsequent searches (Altschul et al., 1997) using the ESTs themselves to find overlapping ESTs. These ESTs can be grouped based on sequence similarity using a software package (MegAlign module, DNAStar, Genetics Computer Group, Madison, WI).
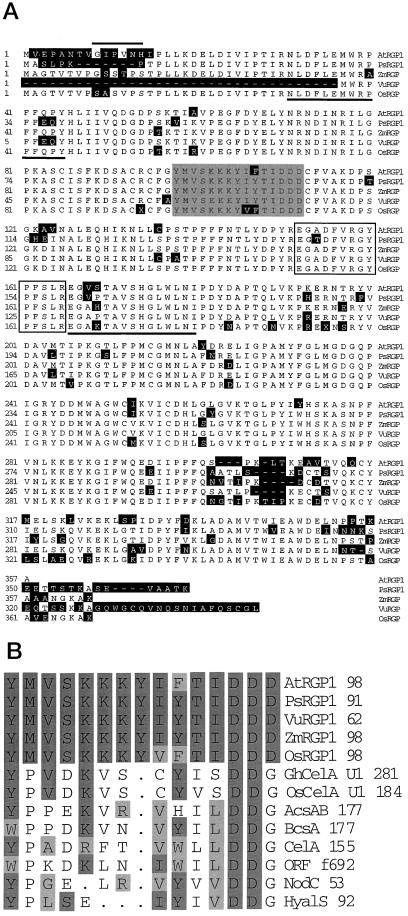
Figure 1.
Predicted protein sequence of AtRGP1 (GenBank accession no. AF013627) and comparison with other RGP proteins. A, Amino acid sequence alignment of AtRGP1, PsRGP1 (U31565), VuRGP (AF005279), ZmRGP (U89897), and OsRGP using the MegAlign module of DNAStar. The black regions represent amino acid differences between AtRGP1 and the other RGPs. The overline highlights a putative N-myristoylation site. The shaded box represents the predicted UDP-Glc-binding domain U1 from cotton CelA. The open box is the site of glycosylation as determined in sweet corn. Underlined are the two pea tryptic peptides used to search the EST database. B, Alignment of UDP-Glc-binding domains from various organisms, as determined by Delmer and Amor (1995), with the predicted binding domain of RGP.
Based on sequence similarity, the following ESTs have been assigned: eight to group I (AtRGP1; T20512, T46245, T42672, T22507, T22943, H37657, N37306, and T44971), five to group II (AtRGP2; T23020, T46745, AA597661, AA650721, and AA650701), and three to group III (AtRGP3; ATTS4214, R30021, and R90614). The rest (ATTS3942, T44394, T45672, ATTS0381, H76915, N65402, N65528, N65622, AA042694, AA650802, AA651536, and AA597661) were too short to be unambiguously assigned to a specific group. Furthermore, similar genomic sequences to AtRGP1 have thus far been identified in only two Arabidopsis chromosomes. A 129-nucleotide stretch at the end of a bacteria artificial chromosome clone (T24B2-Sp6) of chromosome I (http://cbil.humgen.upenn.edu/approximatelyatgc/SPP.html) is 96% identical to AtRGP1, and a 113-nucleotide stretch at the end of a bacteria artificial chromosome clone (T20M8-Sp6) of chromosome II (http://www.tigr.org/tdb/at/atgenome/atgenome.html) is 96% identical to AtRGP1. Full-length cDNA clones (T20512 and T46745) were obtained from the PRL2 Arabidopsis cDNA Library (Newman et al., 1994) kept by the Arabidopsis Biological Research Center at Ohio State University (Columbus). Automated sequencing of the cDNAs was performed by the Plant Biochemistry Facility at Michigan State University (East Lansing).
BLAST searches of the GenBank database using the full-length AtRGP1 cDNA also detected 19 rice ESTs (RICC1486A, RICC2546A, RICR0440A, RICR1510A, RICR2980A, RICS1545A, RICS1750A, RICS1848A, RICS2305A, RICS5091A, C28170, C73320, C27450, C26584, C72510, C26475, C25974, and C71661), four of which (C72510, C26584, RICS5091A, and C27450) can be fused to give a full-length clone (Oryza sativa, OsRGP). Other RGPs identified in this search include those from P. sativum (PsRGP1; U31565), maize (Zea mays, ZmRGP; U89897), and cowpea (Vigna unguiculata, VuRGP; AF005279).
Hydropathy analysis of AtRGP1 was performed using the DNAStar package, which uses the method by Kyte and Doolittle (1982). Protein-sequence analysis of AtRGP1 was also performed with this software package. In addition, the pSORT program (http://psort.nibb.ac.jp/) was used, which compares a given protein sequence against a database of known sequences that mediate protein sorting to various membranes and organelles.
Isolation of RNA and Northern Analysis
Total RNA from Arabidopsis flowers, leaves, roots, and stems was extracted as described previously (Puissant and Houdebine, 1990). Total Arabidopsis RNA (30 μg/lane) was separated on a gel containing 1% agarose and 2% formaldehyde. Electrophoresis was stopped when the dye front had migrated three-quarters of the way down the gel. Sizes were estimated using the sizes of rRNAs as markers. Gels were washed for 20 min in 10× SSC and analyzed as described by Sambrook et al. (1992). Transfer was performed by capillary action onto a nylon membrane (Hybond N, Amersham) overnight. Membranes were washed in 2× SSC for 5 min, cross-linked in a UV-cross-linker, and stored at room temperature between filter papers. Hybridization was overnight at 65°C in northern hybridization buffer (5× SSC, 10× Denhardt's solution [2 mg mL−1 Ficoll, 2 mg mL−1 PVP, 2 mg mL−1 BSA], 0.1 m KPO4, pH 6.8, 100 μg/mL salmon-sperm DNA [freshly boiled], 10% dextran, and 30% deionized formamide) using a random-primed 32P-labeled 1.0-kb EcoRI-BbvI fragment of the AtRGP1 cDNA at 1,000,000 dpm mL−1. Membranes were washed once with 2× SSC and 0.5% SDS, twice with 2× SSC and 0.1% SDS, and once with 0.2× SSC and 0.1% SDS (1× SSC is 0.15 m NaCl and 15 mm sodium nitrate, pH 7.0), each for 30 min at 65°C prior to autoradiographic exposure.
Protoplast Preparation
Twenty grams of cells from a 5- to 8-d-old cell-suspension culture collected on an 80-μm filter was incubated with 50 mL of freshly made protoplasting solution (15.4% [w/v] Suc, 0.32% [w/v] Gamborg's B5 minimal organic, 5 mg mL−1 cellulase [Onozuka R10, Yakutt Honsha, Tokyo], and 1.2 mg mL−1 Macerozyme R10 [Yakutt Honsha]) for 2 h in a rotary shaker at 80 rpm. All manipulations were done at room temperature. The cells were then poured into Babcock centrifuge bottles and centrifuged for 10 min at 1100 rpm in a clinical centrifuge swinging bucket rotor (model HNSII, IEC, Needham Heights, MA). Floated protoplasts were collected, mixed with 20 mL of 0.4 m betaine, 3 mm Mes, and 10 mm CaCl2, pH 5.7, and then pelleted for 5 min at 50g.
Protein Extraction
Four milliliters of cold lysis buffer (20 mm Hepes-KOH, pH 7.0, 13.5% [w/v] Suc, 10 mm potassium acetate, 1 mm DDT, 0.5 mm PMSF, and 1 mm EDTA) was added per milliliter of packed protoplasts and passed through a 25–5/8-gauge needle at 4°C until no unbroken protoplasts could be detected under the microscope. Cell debris were pelleted by centrifugation at 500g for 5 min and this homogenate was called the total protoplast protein. Total plant protein from different tissues (flowers, leaves, roots, stems, and roots grown in liquid culture) was extracted by grinding 1 to 5 g of the given tissue in liquid N2 and mixing with 5 mL of lysis buffer at 4°C. For all plant tissue samples, cell debris were pelleted by centrifugation at 500g for 5 min. This homogenate was called the total tissue protein, where “tissue” specifies the tissue used.
Overexpression of a Fusion Protein in Escherichia coli and Antibody Preparation
The 1.4-kb EcoRI-NotI fragment of AtRGP1 (GenBank accession no. AF013627), encoding all but the first nine amino acids from the 5′ end of the cDNA, was cloned into the EcoRI-NotI sites of pGEX-5X-2 (Pharmacia) to generate an N-terminal in-frame fusion with the 26-kD domain of GST. This fusion was overexpressed in E. coli (strain DH5α) by growing cells in Luria-Bertani medium at 37°C to an A600 of 0.7 to 0.8, then adding isopropylthio-β-d-galactosidase to a final concentration of 0.2 mm, and incubating the culture at 28°C for 4 h. The soluble GST fusion protein was purified as described previously (Bar-Peled and Raikhel, 1996). Five hundred microliters (0.4–0.5 mg) of eluted GST-AtRGP1 fusion protein was emulsified by sonication with an equal amount of TiterMax adjuvant (CytRx Corp., Norcross, GA) and injected into New Zealand White rabbits. An equal amount of GST-AtRGP1 protein was injected into the rabbits two more times to boost the immune response over a 2-month period. Blood was taken from the rabbits 1 month after the first injection and every other week thereafter and treated as described previously (Harlow and Lane, 1988).
Timed Studies of RNA and Protein Levels
A 0.5-L Arabidopsis cell-suspension culture was started by adding 50 mL of a 7-d-old culture to 450 mL of fresh suspension-culture medium. Fifteen-milliliter aliquots were taken at 24-h intervals over a 12-d span. Each fraction was centrifuged at 50g for 5 min, the supernatant was removed, and the weight of the packed cells was measured. Equal amounts of protein (50 μg/lane) were separated by SDS-PAGE (Laemmli et al., 1970), and the presence of AtRGP1 was determined by immunoblotting. Equal amounts of RNA (30 μg/lane) were separated in a 1% agarose and 2% formaldehyde gel, and the level of AtRGP1 RNA was determined by northern analysis.
Reaction with UDP-Sugars
One hundred to 250 μg of total Arabidopsis, pea, tobacco, or maize total protein from roots was labeled as described previously (Dhugga et al., 1991). First, 0.1 μCi of UDP-d-[U-14C]Glc (specific activity 263 μCi μmol−1 [ICN]) or 0.5 μCi of UDP-d-[6-3H]Gal (specific activity 10.6 mCi μmol−1 [ICN]), was added to the protein sample in a volume of 50 to 150 μL of lysis buffer containing 3 mm MgCl2. The reactions were stopped after a 10-min incubation at room temperature by adding 10 to 30 μL of SDS-PAGE loading buffer (120 mm Tris, pH 6.8, 200 mm DTT, 4% SDS, 0.02% bromphenol blue, and 20% glycerol). The protein samples were separated by SDS-PAGE and treated as described below. Since the UDP-Glc and UDP-Gal reactions gave the same results, only the UDP-Glc reactions are presented.
Reactions with purified GST-AtRGP1 protein were carried out using 2 μg of protein in the absence of Mg2+. The reactions were mixed by inversion and incubated statically at room temperature for 10 min. For displacement reactions, after a 10-min incubation with 20 pmol (0.2 μCi) of UDP-[3H]Glc (specific activity 10.9 Ci mmol−1; Sigma), UDP, Glc, UDP-Glc, UDP-Xyl, UDP-Gal, or UDP-Man was added to a final concentration of 3 mm and the reactions continued for 10 more min. To stop the reactions, SDS-PAGE loading buffer (120 mm Tris, pH 6.8, 200 mm DTT, 4% SDS, 0.02% bromphenol blue, and 20% glycerol) was added. The protein samples were separated by SDS-PAGE and treated as described below.
Immunochemical Studies
Immunoprecipitation
Immunoprecipitations were as described previously (Harlow and Lane, 1988). Anti-AtRGP1 polyclonal antibodies were cross-linked to protein A-Sepharose 6MB (Pharmacia) and mixed with total protein from roots grown in liquid culture. After the samples were washed three times with PBS and once with 10 mm potassium phosphate buffer, pH 7.0, proteins bound to the antibodies were eluted with 100 mm Gly-HCl, pH 2.5. Protoplasts labeled with [35S]Met were incubated overnight in a rotary shaker at 80 rpm, and total protein was prepared as described above, and immunoprecipitated.
SDS-PAGE and Immunoblotting
Protein concentration was determined as described previously (Bradford, 1976) using BSA as the standard. Protein molecular weight standards were purchased from Bio-Rad (low- and broad-range markers, catalog nos. 161-0304 and 161-0317, respectively). All protein samples were separated on 12.5% SDS-PAGE gels using 1× running buffer (250 mm Tris, 1 m Gly, and 0.5% SDS).
After separation, gels containing UDP-d-[U-14C]Glc- and UDP-d-[3H]Gal-labeled reactions were stained with 0.1% Coomassie blue overnight, destained with 5 volumes of 30% methanol and 10% acetic acid, washed twice with distilled water, incubated with Fluoro-Hance (Research Products International, Prospect, IL) for 15 min, and dried for 2 h in a gel drier (model 583, Bio-Rad). The dried gels were exposed to film for 7 to 10 d at −80°C prior to film development.
For immunoblotting, gels containing various protein samples (30–50 μg/lane) were washed with 5 volumes of 1× transfer buffer (63 mm Trizma base, 250 mm Gly, 0.13% SDS, and 20% methanol) and transferred in the same buffer to a 0.45-μm nitrocellulose membrane (Hybond N, Amersham) using a semidry apparatus at 2 mA cm−2 for 2 h. Filters were stained with 1× Ponceau-S (2% Ponceau-S, 30% TCA, and 30% sulfosalicylic acid) and blocked overnight in 10% (w/v) dry milk powder in PBST (1× PBS and 0.1% Tween 20). AtRGP1 polyclonal antibodies were used at a 1:1000 dilution. Anti-RD28 plasma membrane marker sera was used at 1:500 dilution and anti-binding protein ER lumen protein at 1:1000 dilution. They were a gift from Maarten Chrispeels (University of California, San Diego). Anti-AtELP (partially Golgi-localized protein, Ahmed et al., 1997) and anti-AtPEP12p (post-Golgi compartment protein, Conceição et al., 1997) polyclonal antibodies were used at a 1:500 dilution. ARA4 monoclonal antibody (a Golgi membrane-localized and -soluble protein, Ueda et al., 1996) was used at 1:500 dilution.
After primary antibody incubation for 1 to 2 h in PBST, membranes were washed three times with PBST and then reacted with a 1:2500 dilution of alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Kirkegaard and Perry Laboratories, Gaithersburg, MD) for 1 to 2 h, except the ARA4 antibody, which required alkaline phosphatase-conjugated goat anti-mouse secondary antibody. Finally, filters were washed twice with PBST, twice with PBS, and once with alkaline buffer (100 mm Tris, 100 mm NaCl, and 5 mm MgCl2, pH 9.5) prior to detection of the immune complexes using the substrates 5-bromo-4-chloro-3-indolyl phosphate (150 μg mL−1 final concentration) and nitroblue tetrazolium (300 μg mL−1 final concentration) in alkaline buffer.
Localization
Differential Centrifugation
Total protoplast protein was centrifuged at 1,000g for 20 min at 4°C. The pellet (p1) was washed with lysis buffer and centrifuged again. The supernatant (s1) was centrifuged at 5,000g for 30 min. The pellet was washed with lysis buffer and centrifuged again (now called p5). The supernatant (now s5) was treated in the same way for sequential differential centrifugations at 15,000, 25,000, 50,000, and 100,000g, with the last two centrifugations carried out for 40 min. All pellets (p1, p5, p15, p25, p50, and p100) were resuspended in lysis buffer and the protein was quantified.
Membrane Association
Total microsomes were prepared by centrifugation of total protoplast protein at 150,000g for 1 h and washing the pellet with lysis buffer. Pellets were resuspended with lysis buffer (total protein) or lysis buffer containing 0.1, 0.5, or 1.0% Triton X-100, 0.5 or 2 m urea, 0.1 m sodium carbonate, 0.1, 0.5, or 1.0 m NaCl, or 0.1 m potassium phosphate, pH 7.0, for 1 h on ice, and centrifuged again at 150,000g for 1 h. The resulting pellets were washed with lysis buffer, resuspended in loading buffer, and separated by SDS-PAGE prior to analysis by immunoblot using anti-AtRGP1 antibodies.
Suc-Density-Gradient Centrifugation
Total protoplast protein was centrifuged at 1000g for 10 min at 4°C, and 6 mL of the supernatant (S1) was loaded on top of a 16 to 55% equilibrium-density Suc gradient modified from Gibeaut and Carpita (1994). To prepare the gradient, stock solutions of 55, 40, 33.5, 26.5, and 16% (w/v) Suc buffered in 10 mm Hepes-KOH, pH 7.0, 10 mm potassium acetate, and 2 mm EDTA were prepared. Gradients were prepared by the sequential addition of 2.7 mL of 55% Suc solution, 8.8 mL of 40% Suc solution, 7.0 mL of 33.5% Suc solution, 6.0 mL of 26.5% Suc solution, and 4.5 mL of 16% Suc solution to 35-mL polyallomer tubes (catalog no. 326823, Beckman). After these solutions were carefully layered, they were centrifuged at 150,000g for 18 to 20 h at 4°C in a rotor (model SW28, Beckman). This resulted in an equilibrium-density gradient similar to one obtained using a gradient maker. Equal-volume fractions were collected from the bottom of the gradients, and the Suc concentration was estimated by measuring the refractive index of the individual fractions using a refractometer. Aliquots of the fractions (50–100 μL) were separated by SDS-PAGE, and the separation of AtRGP1 and membrane marker proteins were analyzed by immunoblotting.
RESULTS
Identification of an Arabidopsis cDNA Encoding an AtRGP1 Protein
Sequences from three purified tryptic peptides from the pea RGP protein doublet were presented by Dhugga and Ray (1994) and used to search the dBEST database. A total of 28 Arabidopsis ESTs have been obtained so far from the original and subsequent searches. Based on sequence similarity, these ESTs can be assigned to at least three different groups, denoted AtRGP1 (AF013627), AtRGP2 (AF013628), and AtRGP3 (AF034255). These cDNAs share between 93 and 99% sequence identity at the amino acid level. Because the similarity is spread through the whole sequence, only one cDNA, AtRGP1, was used for further analysis (Fig. 1A).
We used the full-length cDNA of AtRGP1 to search the dBEST databases and obtained, in addition to the 28 Arabidopsis cDNAs, 19 rice cDNAs, a full-length maize cDNA, and a full-length cowpea cDNA, all of which shared significant sequence similarity with AtRGP1. None of these sequences belonged to a gene with an assigned function at the time of the BLAST search. AtRGP1 is 84, 87, 83, and 85% identical to the pea, cowpea, maize, and rice proteins, PsRGP1, VuRGP, ZmRGP, and OsRGP, respectively. A full-length cDNA clone encoding the rice RGP has not yet been isolated, but available ESTs can be combined to make up a full-length clone (OsRGP). Searches of nonplant databases gave mixed results. Although similar sequences were found in both the yeast genome database (http://speedy.mips.biochem.mpg.de/mips/yeast/index.htmlx) and the Synechocystis sp. strain PCC6803 database (http://www.kazusa.or.jp/cyano/cyano.html), none shared significant similarity to AtRGP1 (P > 0.01), all of the sequence alignments were very short (10–20 amino acids), and none of the predicted open reading frames encoded a 41-kD protein. These findings are further supported by the immunoscreening of various species described below.
Hydropathy profile analysis (Kyte and Doolittle, 1982) predicts that AtRGP1 is a hydrophilic protein (data not shown). Visual and computer-aided (pSORT, http://psort. nibb.ac.jp/) analyses of the protein sequence predict that the protein lacks a signal sequence that might direct AtRGP1 to a membrane compartment within the cell. The same computer program predicts the existence of an N-myristoylation site in AtRGP1 (Fig. 1A). N-myristoylation has been shown to aid in the association of otherwise soluble proteins to the cytosolic surface of membranes.
The sequence of AtRGP1 was compared with the reported predicted binding domains for UDP-Glc. The determination of these UDP-Glc-binding domains is based on the observation that there is a substantial sequence conservation between the bacterial catalytic subunit and other enzymes that catalyze the polymerization of β-glycosyl residues from either UDP-Glc or UDP-GlcNAc (Saxena et al., 1995), as well as UDP-Glc-binding proteins (Delmer and Amor, 1995; Pear et al., 1996). AtRGP1 is 40% identical and 53% similar to the predicted UDP-Glc-binding domain (U1) of cotton CelA (Delmer and Amor, 1995; Pear et al., 1996; Saxena and Brown, 1997). Furthermore, AtRGP1 contained the critical Asp residue (Fig. 1B). In contrast, sequence identity between the deduced amino acid sequence of cotton CelA and AtRGP1 is very low, approximately 15%.
Singh et al. (1995) demonstrated that the Glc from UDP-Glc was attached to an Arg residue of a maize protein with high sequence similarity to AtRGP1. The tryptic peptide containing the glucosylated Arg is 93% identical to a region of AtRGP1 (Fig. 1A). Dhugga et al. (1997) noted this same similarity to PsRGP1 and postulated that Arg-158 is the location of a sugar addition in the pea protein. Similarly, we postulate that Arg-158 is the location of Glc addition in the Arabidopsis protein.
Distribution of RGP1 Transcript and Protein
The isolation of RGP proteins in both monocots and dicots suggests a general function for this protein. To confirm the presence of proteins similar to RGP, as evidenced by database searches, we used AtRGP1 antibodies in immunoblots against protein from various organisms. Immunoreactive polypeptides of 41 kD were detected in Arabidopsis, pea, tobacco, and maize (Fig. 2). Although pea RGP1 has been referred to as a polypeptide of about 40 kD (Dhugga et al., 1991, 1997), it is clear from the cloned cDNA that PsRGP1 has a predicted size of 41 kD. Since all RGPs so far identified encode proteins of approximately the same size, we will refer to them as 41-kD proteins.
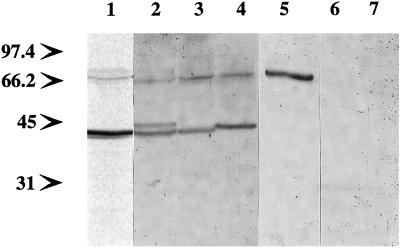
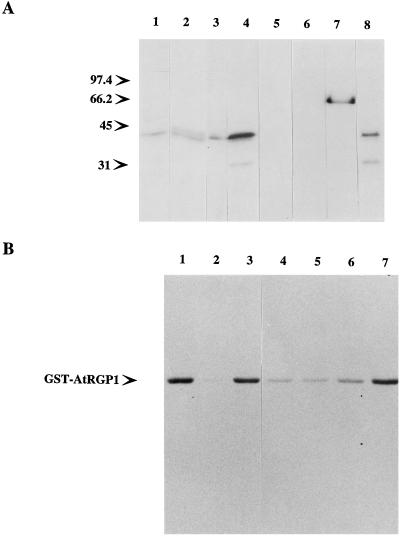
Figure 2.
Homologs of AtRGP1 from other species. Total protein (50 μg) from Arabidopsis (lane 1), pea (lane 2), tobacco (lane 3), maize (lane 4), cyanobacteria (lane 5), yeast (lane 6), and HeLa cells (lane 7) was analyzed by immunoblot using anti-AtRGP1 antibodies. All plant-derived protein extracts were obtained from root tissue. Molecular mass is indicated in kilodaltons.
A 64-kD protein was also detected by AtRGP1 antibodies in immunoblots of Arabidopsis, pea, tobacco, maize, and Synechocystis sp. strain PCC6803 (Fig. 2). The relationship of this protein to the 41-kD protein is not yet clear. Although four hypothetical proteins were found in the Synechocystis sp. strain PCC6803 database (sll0524, sll0501, sll0602, and sll1080) that shared limited sequence similarity within separate regions in AtRGP1, none was of the size detected by immunoblotting. AtRGP1 antibodies did not detect any proteins in either yeast or mammalian extracts (Fig. 2), as expected from database searches.
The AtRGP1 transcript level in different tissues of Arabidopsis was determined by northern analysis (Fig. 3A). One band of approximately 1.4 kb was detected in all tissues tested. Although AtRGP1 RNA levels were relatively uniform, the highest level was detected in suspension-cultured cells. In whole plants, roots had the highest level and stems had the lowest level of AtRGP1 RNA (Fig. 3A). Immunoblotting using anti-AtRGP1 antibodies was used to determine protein levels in the same tissues. Levels of AtRGP1 protein mirrored RNA levels, again being highest in suspension-cultured cells and roots and lowest in leaves (Fig. 3B). Consistent with these results, immunoblots of similar tissues from the various plant species tested (data not shown) showed that the highest level of RGP1 protein was found in roots (Fig. 2). Although RNA levels are similar in leaves and stems, more AtRGP1 protein can be found in stems than in leaves.
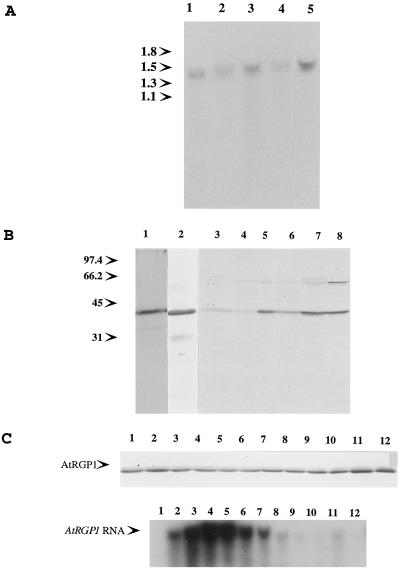
Figure 3.
AtRGP1 RNA and protein are highest in suspension-cultured cells and roots from whole plants. A, RNA levels in different tissues. Total RNA (30 μg) from A. thaliana flowers (lane 1), leaves (lane 2), roots (lane 3), stems (lane 4), and suspension-cultured cells (lane 5) was separated in a 1% agarose and 6% formaldehyde gel, transferred onto a nylon membrane, and hybridized with a random-primed, 32P-labeled, 1.0-kb EcoRI-BbvI fragment of the AtRGP1 cDNA. The single band obtained is of the expected size. Molecular mass is indicated in kilodaltons. B, Protein levels in different tissues. Immunoprecipitations using total protein from [35S]Met-labeled protoplasts (lane 1) and unlabeled protoplasts (lane 2) were performed. The former was analyzed by SDS-PAGE and autoradiography and the latter was analyzed by immunoblot. Total protein (50 μg) from A. thaliana flowers (lane 3), leaves (lane 4), roots (lane 5), stems (lane 6), root liquid culture (lane 7), and cell-suspension cultures (lane 8) was analyzed by immunoblotting using anti-AtRGP1 antibodies. Molecular mass is indicated in kilodaltons. C, AtRGP1 RNA and protein levels during the growth cycle of suspension cells. A cell-suspension culture was started by diluting a 1-week-old culture 10 times with fresh medium. Samples representing 1/40 of the original volume were collected at 1-d intervals, and protein and RNA were extracted from each sample. Top, Total AtRGP1 protein from each sample was analyzed by immunoblot; bottom, total AtRGP1 RNA was determined as described in Figure 4A.
AtRGP1 was detected only as a doublet in tissues in which it was highly expressed (Figs. 2 and 3B), but it was clearly not the same kind of doublet as that seen for pea (Fig. 2). In addition, a 64-kD protein was detected by immunoblot analysis of total protein in nearly all tissues tested (Fig. 3B), as well as in all of the other plant species tested (Fig. 2). The nature of this protein is not understood, and it does not seem to be well recognized by AtRGP1 antibodies, since immunoprecipitation studies recognized AtRGP1 almost exclusively (Fig. 3B).
Since AtRGP1 RNA and the corresponding protein were highest in suspension-cultured cells, its behavior was studied by starting cultures and then following them through a time course. As seen in Figure 3C, cells were grown for a period of 12 d and aliquots were taken at 24-h intervals to determine the amount of AtRGP1 RNA and its corresponding protein. Although there was a clear increase in RNA accumulation during the first 5 d, peaking at d 4 and almost undetectable by d 8, the corresponding protein level was constant throughout the experiment.
AtRGP1 Is Soluble and Membrane Associated
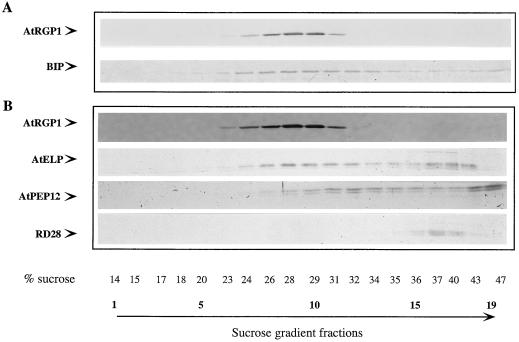
One strategy for understanding the function of AtRGP1 is to localize this protein. Hydropathy plots predict that AtRGP1 is a soluble protein (not shown) although analysis of the sequence shows that it contains a putative N-myristoylation site that may be involved in its association with membranes (Fig. 1A). Fractionation studies were carried out to determine whether AtRGP1 is soluble or membrane associated. Protoplasts derived from cell-suspension cultures were lysed and subjected to sequential differential centrifugation. The resulting pellets were analyzed by immunoblot using AtRGP1 antibodies (Fig. 4A). Several intrinsic membrane proteins (AtPEP12p, RD28, and AtELP), membrane-associated and soluble proteins (ARA4), and soluble proteins (ER lumen and BiP) were analyzed in the same fractions and used as a control. AtRGP1 was enriched in fractions composed of soluble proteins (s100) but was also detected in membrane-containing fractions (p5-p100, Fig. 4A). Our results indicate that AtRGP1 is both soluble (s100) and membrane associated.
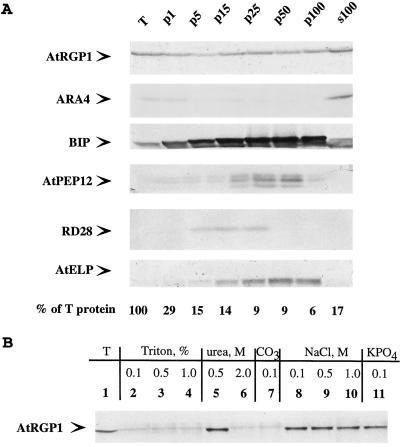
Figure 4.
AtRGP1 is soluble and membrane associated. A, Total protoplast protein from suspension-cultured cell protoplasts was centrifuged at 1,000, 5,000, 10,000, 15,000, 25,000, 50,000, and 100,000g. The resultant pellets were resuspended in lysis buffer and make up fractions p1, p5, p10, p15, p25, p50, and p100, respectively. s100 denotes the supernatant after the 100,000g centrifugation and represents total soluble proteins. Equal volumes of protein were separated by SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblot. Various soluble proteins (BiP), integral membrane proteins (AtPEP12p, RD28, and AtELP), and peripheral membrane proteins (ARA4) were compared with the membrane association of AtRGP1. The fraction of total protein (T) present in each pellet is shown. B, AtRGP1 is a peripheral membrane protein. Total microsomes were prepared by centrifuging total protein at 150,000g for 1 h and washing the pellet with lysis buffer. Pellets were resuspended with various buffers for 1 h on ice and centrifuged again at 150,000g for 1 h, and the pellets were analyzed by immunoblot using anti-AtRGP1 antibodies. Microsomes were resuspended with lysis buffer (lane 1, total protein) or lysis buffer containing 0.1, 0, or 1.0% Triton X-100 (lanes 2, 3, and 4, respectively), 0.5 or 2 m urea (lanes 5 and 6, respectively), 0.1 m sodium carbonate (lane 7), 0.1, 0.5, or 1.0 m NaCl (lanes 8, 9, and 10, respectively), or 0.1 m potassium phosphate buffer, pH 7.0, alone (lane 11).
The presence of AtRGP1 in both soluble and membrane-bound fractions is interesting and may provide hints regarding its role in the cell. Since AtRGP1 is predicted to be a soluble protein, we were specifically interested in studying the nature of its association with membranes. Equal amounts of total microsomes were prepared (p150) and pellets were resuspended under various conditions. After treated membranes were centrifuged at 150,000g, pellets were resuspended in lysis buffer to analyze by immunoblot any protein left that was associated with membranes. Total protein (Fig. 4B) and microsomes resuspended in lysis buffer containing Triton X-100, urea, sodium carbonate, NaCl, or potassium phosphate buffer alone were pelleted and resuspended in lysis buffer for immunoblot analysis (Fig. 4B). Nonionic detergents, high levels of urea (2 m), and alkaline treatment eluted AtRGP1 from the membrane (Fig. 4B), whereas low levels of urea (0.5 m), NaCl treatment, and a hypotonic wash with a neutral buffer left some AtRGP1 associated with the membrane (Fig. 4B). This suggests that AtRGP1 is a peripheral membrane protein.
As a further attempt to determine the localization of AtRGP1, a postnuclear supernatant (s1) fraction of protoplast lysates derived from cell-suspension cultures was fractionated on Suc gradients and the individual fractions were analyzed by immunoblot (Fig. 5). AtRGP1 peaked in fraction 10, corresponding to approximately 30% Suc. BiP, a soluble protein localized to the ER lumen, peaked in fraction 9, corresponding to approximately 28% Suc (Fig. 5A). Because of the extreme difficulty in separating ER from Golgi membranes, a separate gradient was performed in the presence of Mg2+. Shifts toward denser fractions in the presence of Mg2+ are characteristic of ER proteins (Lord, 1987; Bar-Peled and Raikhel, 1997), and, as expected, BiP shifted to denser fractions. The fact that AtRGP1 did not shift toward denser fractions in the presence of Mg2+, as BiP did (data not shown), suggests that AtRGP1 is not an ER protein.
Figure 5.
Membrane localization of AtRGP1. Equilibrium-density gradients were used to fractionate A. thaliana total membrane preparations from suspension-cultured cell protoplasts. One-twentieth (100 μL) of equal-volume fractions was separated by SDS-PAGE and transferred to nitrocellulose membranes. A, Fractionation of AtRGP1 was determined by immunoblot analysis and compared with that of the membrane marker BiP (ER). Because of the presence of AtRGP1 and BiP in the same fractions, similar gradients were analyzed in the presence of Mg+2 and showed that BiP shifted to a denser part of the gradient, but AtRGP1 did not (data not shown). B, AtRGP1 fractionation was also compared with that of other known membrane markers, AtELP, AtPEP12p, and R28.
The plasma membrane marker RD28 was detected at the bottom of the gradient, in fractions that lacked AtRGP1 (Fig. 5B), suggesting that AtRGP1 is not localized to the plasma membrane. AtPEP12p, a recently identified protein believed to reside in a post-Golgi, prevacuolar compartment, is present in membranes located at the bottom of the gradient. Finally, the distribution of AtRGP1 partially coincides with that of AtELP (Fig. 5B). Results from our laboratory have shown that AtELP fractionates mainly with two fractions, one of which coincides with that of Golgi-localized fucosyltransferase activity (Ahmed et al., 1997). Preliminary results of electron microscopy partially localize AtELP to the trans-Golgi (S.U. Ahmed and N.V. Raikhel, unpublished data). Our results support the Golgi localization of RGP (Dhugga et al., 1997).
Glycosylation of AtRGP1
The pea RGP1 was originally identified because it was glycosylated by UDP-d-[U-14C]Glc (Dhugga and Ray, 1994) in a manner that was reversed by using UDP-Glc, UDP-Xyl, and UDP-Gal. Similar experiments revealed a single labeled protein when total Arabidopsis soluble protein was analyzed (Fig. 6A). Reactions using total pea membrane protein yielded the expected PsRGP1 doublet (Fig. 6A). Only pea seemed to have a clear doublet, since tobacco and maize showed only a single protein labeled with UDP-Glc (Fig. 6A), and total protein from Synechocystis sp. strain PCC6803 or purified GST protein showed no labeling (Fig. 6A).
Figure 6.
AtRGP1 is reversibly autoglycosylated. A, Total protein (100–250 μg) from Arabidopsis (lane 1), pea (lane 2), tobacco (lane 3), maize (lane 4), and cyanobacteria (lane 5); 5 μg of purified GST (lane 6) and GST-AtRGP1p (lane 7) and 50 μg of the Arabidopsis Suc-gradient fraction 10 (lane 8) were incubated with 0.1 μCi of UDP-[14C]Glc before analysis by SDS-PAGE and autoradiography. Molecular mass is indicated in kilodaltons. B, Displacement of bound radiolabel from GST-AtRGP1. Two micrograms of GST-AtRGP1 was incubated with UDP-[3H]Glc for 10 min. Various substrates were then added to a final concentration of 3 mm and the incubations were continued for 10 min more. Lane 1, No substrate added; lane 2, UDP; lane 3, Glc; lane 4, UDP-Glc, lane 5, UDP-Xyl; lane 6, UDP-Gal; and lane 7, UDP-Man. After the second incubation, reactions were stopped and analyzed by SDS-PAGE and autoradiography.
In addition to the glycosylation of AtRGP1, a reaction between radiolabeled UDP-Glc and a 30-kD protein was also detected. Labeling of a 30-kD protein was detected in maize, which contains a large proportion of RGP in total protein extracts from roots, as well as fraction 10 of the Arabidopsis Suc gradient (Fig. 6A). The presence of this 30-kD protein seems to coincide with fractions containing high concentrations of RGP, since fraction 10 is where AtRGP1 is found at the highest concentration (Fig. 5). The nature of this 30-kD polypeptide is not known.
Glycosylation of AtRGP1 using UDP-d-[U-14C]Glc implies that it corresponds to the A. thaliana homolog of PsRGP1. In pea the association between PsRGP1 and the Glc from UDP-Glc was shown to be through a glycosidic bond resistant to boiling in 5% SDS (Dhugga and Ray, 1994). AtRGP1 seems to behave in a similar way, since all of the samples shown in Figure 6 were boiled in loading buffer containing 3% SDS prior to analysis. To test further the nature of this association in Arabidopsis, a purified GST fusion of AtRGP1 was prepared. The 68-kD GST fusion, GST-AtRGP1p, was incubated with UDP-d-[14C]Glc or UDP-d-[3H]Glc in the absence of metal ions and was shown to be glycosylated by UDP-Glc (Fig. 6). The reaction with UDPd-[3H]Glc was tested against UDP, Glc, UDP-Glc, UDP-Xyl, UDP-Gal, and UDP-Man. UDP, UDP-Glc, UDP-Xyl, and UDP-Gal were able to displace the label from GST-AtRGP1 (Fig. 6B), whereas Glc and UDP-Man were not (Fig. 6B). These results demonstrate that AtRGP1, as shown for PsRGP1 (Dhugga et al., 1997), is autoglycosylated independently of secondary factors.
DISCUSSION
We have isolated and characterized a cDNA clone that encodes the Arabidopsis homolog of the PsRGP1 doublet. The PsRGP1 doublet has been localized to the Golgi compartment and is shown to be reversibly glycosylated by UDP-Glc. The existence of RGP in both dicots (Dhugga et al., 1991) and monocots (Singh et al., 1995) but not in nonplant systems suggests a plant-specific function.
Database searches using AtRGP1 yielded 28 Arabidopsis, 19 rice, a full-length maize, and a full-length cowpea cDNA. Computer-assisted sequence comparisons among the Arabidopsis ESTs led to their classification into at least three distinct groups. Furthermore, portions of AtRGP have been sequenced in both chromosomes I and II of Arabidopsis, suggesting that AtRGP1 is part of a small multigene family. Our results differ from those in pea (Dhugga et al., 1997), in which it has been suggested that PsRGP1 is a single-copy gene. Elucidating the functional role of AtRGP1-related Arabidopsis proteins such as AtRGP2 will be pursued because they share a high degree of identity and yet include subtle differences, e.g. a reduction of sequence identity at the N terminus, which leads to the absence of a putative N-myristoylation site in AtRGP2 (not shown).
The existence of at least two different AtRGP cDNAs could explain the presence of a doublet in Arabidopsis, since AtRGP2 is seven amino acids longer than AtRGP1, approximately equal to the small difference in size between the two polypeptides detected by immunoblot analysis (Fig. 3B). The larger difference between the two pea polypeptides could be species specific, since no other plant examined thus far shows this kind of doublet.
Immunoblot studies using anti-AtRGP1 antibodies detected not only AtRGP1 but also a 64-kD protein, the nature of which is not understood. It could be a modified version of AtRGP1, a related protein, or a completely unrelated protein cross-reacting with antibodies raised against AtRGP1.
The use of known bacterial UDP-sugar-binding motifs to determine the corresponding motifs in plants (Pear et al., 1996) is a strong indication that these motifs are conserved in different polysaccharide-synthesizing enzymes. Sequence comparisons with previously defined UDP-Glc-binding sites (Delmer and Amor, 1995; Pear et al., 1996; Saxena and Brown, 1997) showed that AtRGP1 contains a similar motif that may be involved in its binding to UDP-sugars. This motif shares 40% sequence identity with U1, the cotton CelA UDP-Glc-binding domain.
Singh et al. (1995) isolated a sweet corn protein based on its glycosylation with UDP-Glc, which they named amylogenin. The protein was digested and the sequence from eight tryptic peptides accounted for about 40%. Dhugga et al. (1997) suggested that this protein be named ZmRGP1, since the tryptic peptide sequences were nearly identical to PsRGP1 and AtRGP1. A single amino acid, Arg-158, was found to be labeled with UDP-d-[14C]Glc, in accordance with the single glycosylation of PsRGP1 (Dhugga et al., 1991). The U1 motif (Pear et al., 1996), a UDP-Glc-binding domain predicted through its sequence conservation with the catalytic subunits of enzymes involved in the polymerization of β-glucosyl residues, as well as UDP-Glc-binding proteins in other systems, may function in UDP-Glc binding in AtRGP1 prior to the glycosylation of this protein at Arg-158.
RGP has been localized to the Golgi apparatus in pea (Dhugga et al., 1997) and possibly in maize (Epel et al., 1996). All of the membrane markers used in this study, with the exception of ARA4, have been extensively characterized in our laboratory (Ahmed et al., 1997; Bar-Peled and Raikhel, 1997; Conceição et al., 1997). We were able to show that AtRGP1 is found in membrane-containing fractions other than that of the ER or the plasma membrane, presumably the Golgi apparatus. The accumulation of ARA4 (Ueda et al., 1996) in the soluble fraction (Fig. 4) is typical of small GTP-binding proteins, which exist in both cytosolic and membrane-associated forms. Sequence analysis of AtRGP1 suggests that it is a soluble protein. Membrane-association experiments show that most of AtRGP1 is indeed soluble, but that a small fraction can be found peripherally associated with membranes. Such a distribution within the cell suggests that the reaction of AtRGP1 with UDP-sugars takes place in the cytoplasm.
Our working hypothesis is that AtRGP1 may be in some way involved in polysaccharide synthesis, possibly as a protein primer or as some form of intermediate. There is no evidence for a primer function other than an analogy to protein-primed starch and glycogen synthesis (Moreno et al., 1986). AtRGP1's reversible glycosylation, its prominent residence in the cytoplasm, where nucleotide sugars are found, and its transient association with membranes suggests that it functions as a carrier of UDP-sugars from the cytoplasm to membranes such as the Golgi apparatus.
Our understanding of the enzymes involved in cell wall biosynthesis is scant at best, and so far, little progress has been made in isolating these enzymes, which is a requirement for studying their function. With this in mind, the use of cell-suspension cultures for the study of cell wall biosynthesis may prove to be highly beneficial. Our preliminary observations show that at the transcriptional level AtRGP1 RNA can accumulate severalfold within 4 d after subculture and disappear completely after d 8. The fact that AtRGP1 protein persisted throughout this time course is not surprising, since cell-suspension systems are known to be very active. For example, processes such as the secretion of cell wall polysaccharides continue unhindered because there is less spatial limit to cell wall deposition due to cell-to-cell contact.
Further investigation is necessary to determine the function of AtRGP1. Our studies support the notion that AtRGP1 is capable of autoglycosylation using UDP-Glc and UDP-Gal, most likely from the cytoplasmic pool of sugar nucleotides, and carry these sugars to membranes, most likely the Golgi apparatus. Its function when associated with membranes is not clear but may involve the priming of polysaccharide synthesis. These observations, along with those made for the PsRGP1 doublet (Dhugga et al., 1991), suggest that AtRGP1 may play a role in cell wall biosynthesis.
ACKNOWLEDGMENTS
We thank all of the members of the Raikhel group, especially Anton Sanderfoot and Diane Bassham, for their help with technical and conceptual matters; the members of the cell wall group, including Hans Kende and Jonathan Walton, for helpful discussions; Maarten Chrispeels for the gifts of BiP and RD 28 antibodies; and Takashi Ueda for ARA-4 antibodies.
Abbreviations:
- EST
expressed sequence tag
- GST
glutathione S-transferase
- RGP
reversibly glycosylated polypeptide
Footnotes
This research was supported by a grant from the U.S. Department of Energy (no. De-FG02-91ER20021).
LITERATURE CITED
- Ahmed SU, Bar-Peled M, Raikhel NV. Cloning and subcellular localization of an Arabidopsis receptor-like protein that shares common features with protein-sorting receptors of eukaryotic cells. Plant Physiol. 1997;114:325–336. doi: 10.1104/pp.114.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albersheim P, Darvill A, Roberts K, Staehelin AL, Varner JE. Do the structures of cell wall polysaccharides define their mode of synthesis? Plant Physiol. 1997;113:1–3. doi: 10.1104/pp.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. A protocol for transient expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem. 1992;30:123–128. [Google Scholar]
- Bacic A, Harris PJ, Stone BA. Structure and function of plant cell walls. In: Preiss J, editor. The Biochemistry of Plants, Vol 14. San Diego, CA: Academic Press; 1988. pp. 297–371. [Google Scholar]
- Bar-Peled M, Raikhel NV. A method for isolation and purification of specific antibodies to a protein fused to GST. Anal Biochem. 1996;241:140–142. doi: 10.1006/abio.1996.0390. [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV. Characterization of AtSEC12 and AtSAR1 proteins likely involved in ER and Golgi traffic. Plant Physiol. 1997;114:315–324. doi: 10.1104/pp.114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of nanogram quantities of proteins utilizing the principle of dye-binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Camirand A, Maclachlan GA. Differential distribution of xyloglucan glycosyl transferases in pea Golgi dictyosomes and secretory vesicles. J Cell Sci. 1990;96:705–710. [Google Scholar]
- Campbell RE, Brett CT, Hillman JR. A xylosyltransferase involved in the synthesis of a protein-associated xyloglucan in suspension-cultured dwarf-French-bean (Phaseolus vulgaris) cells and its interaction with a glycosyltransferase. Biochem J. 1988;253:795–800. doi: 10.1042/bj2530795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of the primary cell walls of flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann MC, Griffing LR. The plant extracellular matrix: news from the cell's frontier. Plant Cell. 1996;8:1451–1463. doi: 10.1105/tpc.8.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição AS, Marty-Mazars D, Bassham DC, Sanderfoot A, Marty F, Raikhel NV. The syntaxin homologue AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell. 1997;9:571–581. [PMC free article] [PubMed] [Google Scholar]
- Delmer DP, Amor Y. Cellulose biosynthesis. Plant Cell. 1995;7:987–1000. doi: 10.1105/tpc.7.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS, Ray PM. Purification of the reversibly glycosylated polypeptides from pea. Purified polypeptides exhibit the same properties as the Golgi-bound form (abstract no. 684) Plant Physiol. 1994;105:S-126. [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM. A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning and trans Golgi localization. Proc Natl Acad Sci USA. 1997;94:7679–7684. doi: 10.1073/pnas.94.14.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS, Ulvskov P, Gallagher SR, Ray PM. Plant polypeptides reversibly glycosylated by UDP-glucose: possible components of Golgi β-glucan synthase in pea cells. J Biol Chem. 1991;266:21977–21984. [PubMed] [Google Scholar]
- Driouich A, Faye L, Staehelin LA. The plant Golgi apparatus: a factory for complex polysaccharides and glycoproteins. Trends Biochem Sci. 1993;18:210–214. doi: 10.1016/0968-0004(93)90191-o. [DOI] [PubMed] [Google Scholar]
- Epel BL, Van Lent JWM, Cohen L, Kotlizky G, Katz A, Yahalom A. A 41 kDa protein isolated form maize mesocotyl cell walls immunolocalizes to plasmodesmata. Protoplasma. 1996;191:70–78. [Google Scholar]
- Faïk A, Chileshe C, Sterling J, Maclachan G. Xyloglucan galactosyl- and fucosyltransferase activities from pea epicotyl microsomes. Plant Physiol. 1997;114:245–254. doi: 10.1104/pp.114.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut DM, Carpita NC. Biosynthesis of plant cell wall polysaccharides. FASEB J. 1994;8:904–915. doi: 10.1096/fasebj.8.12.8088456. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hayashi T. Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:139–168. [Google Scholar]
- Kleene R, Berger EG. The molecular and cell biology of glucosyltransferases. Biochim Biophys Acta. 1993;1154:283–325. doi: 10.1016/0304-4157(93)90003-7. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levy S, Staehelin LA. Synthesis, assembly and function of plant cell wall macromolecules. Curr Opin Cell Biol. 1992;4:856–862. doi: 10.1016/0955-0674(92)90111-o. [DOI] [PubMed] [Google Scholar]
- Lord JM. Isolation of endoplasmic reticulum: general principles, enzymatic markers, and endoplasmic reticulum-bound polysomes. Methods Enzymol. 1987;148:576–584. [Google Scholar]
- Moreno S, Cardini CE, Tandecarz JS. α-Glucan synthesis on a protein primer, uridine diphosphoglucose:protein transglycosylase. I. Separation from starch synthetase and phosphorylase and a study of its properties. Eur J Biochem. 1986;157:539–545. doi: 10.1111/j.1432-1033.1986.tb09700.x. [DOI] [PubMed] [Google Scholar]
- Newman T, de Brujn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel NV, Somerville S, Tomashow M and others. Genes galore. A summary of methods for accessing results from a large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant C, Houdebine L-M. An improvement of the single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Biotechniques. 1990;8:148–149. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saxena IM, Brown RM., Jr Identification of cellulose synthase(s) in higher plants: sequence analysis of processive β-glycosyltransferases with the common motif “D,D,D35Q(R,Q)XRW”. Cellulose. 1997;4:33–49. [Google Scholar]
- Saxena IM, Brown RM, Jr, Fevre M, Geremia RA, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for a mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DG, Lomako J, Lomako WM, Whelan WJ, Meyer HE, Serwe M, Metzger JW. β-Glycosylarginine: a new glucose-protein bond in a self-glucosylating protein from sweet corn. FEBS Lett. 1995;376:61–64. doi: 10.1016/0014-5793(95)01247-6. [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Moore I. The plant Golgi apparatus: structure, functional organization and trafficking mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:261–288. [Google Scholar]
- Ueda T, Anai T, Tsukaya H, Hirata A, Uchimiya H. Characterization and subcellular localization of a small GTP-binding protein (ARA-4) from Arabidopsis: conditional expression under the control of the promoter of the gene for heat-shock protein HSP81–1. Mol Gen Genet. 1996;250:533–539. doi: 10.1007/BF02174441. [DOI] [PubMed] [Google Scholar]
- Waldron KW, Brett CT. Interaction of enzymes involved in cell wall heteropolysaccharide biosynthesis. In: Brett CT, Hillman JR, editors. Biochemistry of Plant Cell Walls, SEB Seminar Series. Cambridge, UK: Cambridge University Press; 1985. pp. 79–97. [Google Scholar]
- White AR, Xin Y, Pezeshik V. Xyloglucan glucosyltransferase in Golgi membranes from Pisumsativum (pea) Biochem J. 1993;294:231–238. doi: 10.1042/bj2940231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablackis E, Huang J, Muller B, Darvill AG, Albersheim P. Characterization of the cell wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 1995;107:1129–1138. doi: 10.1104/pp.107.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]