Abstract
Incubation of quercetin with Streptomyces rimosus subsp. rimosus ATCC 10970 yielded an unusual glycosylated derivative. The structure of the product was determined to be quercetin-7-O-β-4″-deoxy-hex-4″-enopyranosiduronic acid based on the spectral data. Quercetin was completely converted into the glycoside in 72 h.
Keywords: Biotransformation, Glycosylation, Quercetin, Quercetin-7-O-β-4″-deoxy-hex-4″-enopyranosiduronic acid, Streptomyces rimosus subsp. rimosus
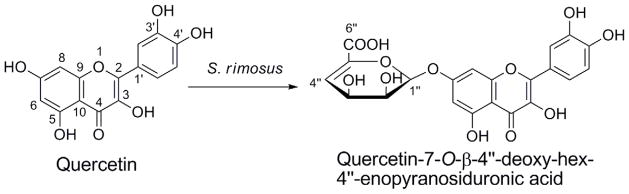
Quercetin (Fig. 1A) is a natural antioxidant flavonoid widely distributed in many plants, such as green tea, fruits and leaf vegetables. It has displayed a variety of biological activities, including anticancer (1), anti-inflammatory (2), antiviral (3), and antihypertensive (4) properties. Due to its interesting biological activities, structural modification of quercetin has been conducted using different organisms including plant and microbial cells to prepare new derivatives (5, 6). Streptomyces rimosus subsp. rimosus (hereafter abbreviated as S. rimosus) ATCC 10970 is a strain of Streptomycetaceae isolated from the soil, and is known to produce the well-known antibiotic oxytetracycline and a polyene antifungal antibiotic rimocidin (7). Both compounds belong to an important family of natural products called polyketides. Oxytetracycline is synthesized through a type II polyketide biosynthetic pathway and we have recently investigated the involved biosynthetic enzymes (8, 9). Rimocidin was found to be assembled by a type I polyketide synthase (PKS) and further modified by tailoring enzymes such as a glycosyltransferase (10). We have previously reported that quercetin can be converted into quercetin-4′-O-methyl-7-O-β-D-glucopyranoside and quercetin-3-O-β-D-glucopyranoside by the filamentous fungi Beauveria bassiana ATCC 7159 and Cunninghamella elegans ATCC 9245, respectively (11, 12). Since S. rimosus contains abundant natural product biosynthetic enzymes, we proposed that some of its endogenous enzymes may be able to accept quercetin as a substrate to generate new hybrid compounds that contain structure characteristics from both plants and bacteria. Herein, we report the bioconversion of quercetin by S. rimosus ATCC 10970 into an unusual quercetin glycoside, namely, quercetin-7-O-β-4″-deoxy-hex-4″-enopyranosiduronic acid (Figs. 1A and 1B).
FIG. 1.
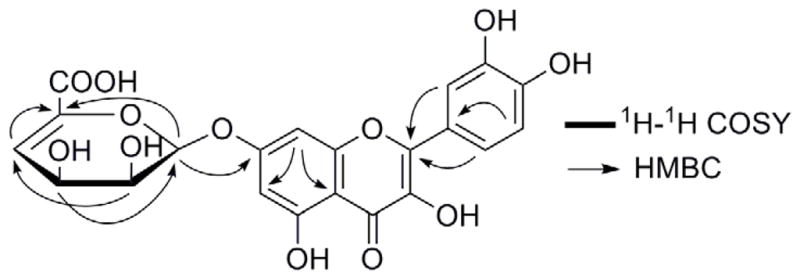
Bioconversion of quercetin into a novel glycosylated derivative by S. rimosus ATCC 10970. (A) Microbial glycosylation of quercetin. (2) 1H-1H COSY and key HMBC correlations of quercetin-7-O-β-4″-deoxy-hex-4″-enopyranosiduronic acid.
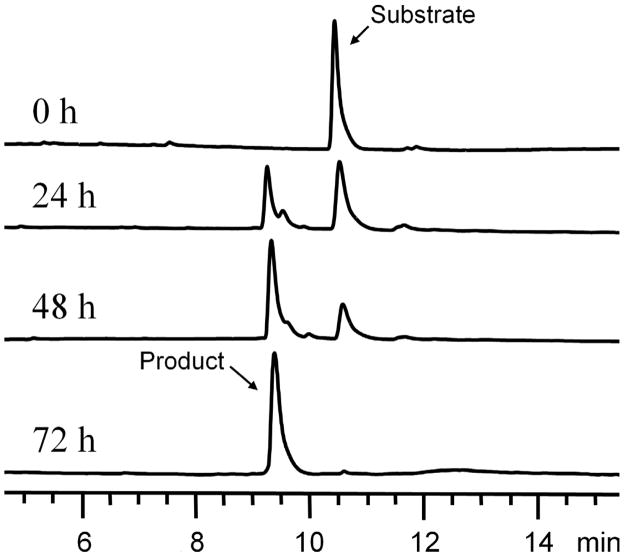
To test the ability of S. rimosus ATCC 10970 to biotransform quercetin, this bacterium was grown in 50 mL of yeast extract –malt extract (YM) medium and incubated on a rotary shaker at 250 rpm and 28°C for 3 days. 2 mg of quercetin was dissolved in 200 μL of dimethyl sulfoxide (DMSO) and was then added into the culture, which was maintained under the same conditions for an additional 4 days. The culture was harvested by centrifugation, and the supernatant was extracted twice with 50 mL of ethyl acetate. After evaporation of solvent, the residue was dissolved in 300 μL of methanol. The sample was analyzed by HPLC on an Agilent Eclipse Plus C18 column (5 μm, 4.6 × 150 mm) at 365 nm, eluted with a linear gradient of 5–95% acetonitrile-water (containing 0.1% trifluoroacetic acid) over 20 min at a flow rate of 1 mL/min. The HPLC trace revealed that a more polar metabolite was synthesized while the substrate disappeared (Fig. 2). The UV absorption pattern of the product was similar to that of quercetin, suggesting that it is a derivative of the substrate.
FIG. 2.
Time course analysis of the bioconversion process of quercetin by S. rimosus ATCC 10970.
To isolate the biotransformation product for structure elucidation, S. rimosus ATCC 10970 was cultivated in four 2-L Erlenmeyer flasks, each containing 500 mL of YM medium, to biotransform 200 mg of quercetin. After 4 days, the supernatant was extracted with ethyl acetate to yield 0.92 g residue. The extract was then subjected to Diaion HP-20 resin column chromatography, successively eluted with 20%, 40%, 60%, and 80% aqueous methanol (v/v) to give 4 fractions. The 60% aqueous methanol fraction was further separated by reversed-phase preparative HPLC (SOPELCO C18 column, 5 μm, 10 ×250 mm), eluted with 40% aqueous acetonitrile containing 0.1% trifluoroacetic acid (v/v) to yield the product (110 mg).
NMR spectra were acquired on a JEOL instrument (300 MHz) and high resolution mass spectra were recorded on an Agilent 6210 LC-MS instrument. The biotransformation product was obtained as a yellow powder, −6.50° (c 0.1, MeOH). Its molecular formula was determined to be C21H16O12 on the basis of the [M+H]+ peak at m/z 461.0711 (calcd. 461.0715) in the HRESIMS. In the 1H NMR spectrum (DMSO-d6) of the product (Table 1), there are four phenolic hydroxyl signals (δ 12.52, 9.67, 9.56, and 9.32) and a series of sugar signals including one anomeric proton (δ 5.81), indicating that it is a glycosylation product. This was further confirmed by the 21 carbon signals in the 13C NMR spectrum, among which 15 carbons are from the aglycon, and 6 carbons from the sugar moiety. A complete comparison of the 1H NMR spectrum of the product with that of the substrate revealed that the product lacks the signal of 7-OH, suggesting that the sugar moiety was introduced at C-7, which was further confirmed by the chemical shift of C-7 (δ 162.8 in the product vs. δ 164.6 in quercetin) and the HMBC correlation of the anomeric proton H-1″ to C-7 (Fig. 1B). After a careful check of the NMR data of the sugar unit of the product, and a comparison of its NMR data with those sugar units of alginate oligosaccharides reported previously (13, 14), the sugar part of the product was speculated as an unusual glucuronic acid derivative, containing a double bond between C-4″ and C-5′. The signals of four protons and six carbons of the sugar moiety were assigned based on the 1H-1H COSY, HMQC and HMBC corrections. The 1H-1H COSY correlations revealed a spin system consisting of CH-1″-CH-2″-CH-3″-CH-4″ in the sugar moiety, and the HMBC corrections were observed between H-1″ and C-5″, H-2″ and C-4″, H-3″ and C-1″, as well as H-4″ and C-5″ (Fig. 1B). The ROSEY correlations of H-1″ to H-2″ and H-3″ indicated that these protons are on the same side. Since the coupling constant of H-1″ is 5.81 Hz, the glycosidic bond should be in the β configuration. Hence, the structure of the product was established as quercetin-7-O-β-4″-deoxy-hex-4″-enopyranosiduronic acid.
TABLE 1.
1H (300 MHz) and 13C (75 Hz) NMR data of quercetin-7-O-β-4″-deoxy-hex-4″-enopyranosiduronic acid in DMSO-d6
| Position | δC | δH (J in Hz) |
|---|---|---|
| 2 | 147.6 | |
| 3 | 136.2 | |
| 4 | 176.0 | |
| 5 | 161.7 | |
| 6 | 99.0 | 6.50 (1H, d, 2.1) |
| 7 | 162.8 | |
| 8 | 94.4 | 6.88 (1H, d, 2.1) |
| 9 | 155.6 | |
| 10 | 105.0 | |
| 1′ | 121.8 | |
| 2′ | 115.2 | 7.72 (1H, d, 2.1) |
| 3′ | 145.1 | |
| 4′ | 148.0 | |
| 5′ | 115.6 | 6.89 (1H, d, 8.2) |
| 6′ | 120.1 | 7.69 (1H, dd, 8.2, 2.1) |
| 1″ | 97.9 | 5.81 (1H, d, 6.2) |
| 2″ | 70.1 | 3.72 (1H, m) |
| 3″ | 66.6 | 4.13 1H, (m) |
| 4″ | 113.7 | 5.97 (1H, d, 3.0) |
| 5″ | 140.0 | |
| 6″ | 160.4 | |
| 3-OH | 9.56 (1H, s) | |
| 5-OH | 12.52 (1H, s) | |
| 3′-OH | 9.67 (1H, s) | |
| 4′-OH | 9.32 (1H, s) | |
| 2″-OH | 5.76 (1H, d, 4.8) | |
| 3″-OH | 5.28 (1H, d, 5.8) |
In order to further understand the bioconversion process, a time course analysis was conducted. S. rimosus ATCC 10970 was grown in a 250-mL Erlenmeyer flask containing 50 mL of YM medium. A total of 2 mg of quercetin was added into the broth after 3 days. A fraction (1 mL) of the fermentation broth was taken every day after the substrate addition. As shown in Fig. 2, quercetin was efficiently metabolized by the strain. After 24 h, the ratio of the product to the substrate reached 1:1. After 48 h, the ratio became approximately 3:1. After 72 h, quercetin was completely converted into quercetin-7-O-β-4″-deoxy-hex-4″-enopyranosiduronic acid by S. rimosus ATCC 10970.
Interactions of flavonoids with S. rimosus have been previously studied. The addition of rutin to the culture of the bacterium resulted in a 28% higher yield in antibiotic production than the controls. S. rimosus was found to decompose quercetin within hours into hydroxybenzoic acid and dihydroxyphenylacetic acid (15). However, in our work, we did not observe obvious degradation of quercetin, but instead found a direct enzymatic conversion of the substrate into a novel glycoside. Recently, many studies have been reported on alginate-degrading enzymes that catalyze the degradation of alginate polysaccharides, which were isolated from many sources including brown algae, mollusks, and marine bacteria. It is known that many lyases catalyze the degradation of alginate polysaccharide by a β-elimination mechanism, in which a double bond is formed between the C-4 and C-5 at the nonreducing terminal (16). However, the novel sugar moiety of the biotransformation product has never been found in natural products except polysaccharides, and thus may represent a useful structural/functional component in bioactive agents. There is no report of 4″-deoxy-hex-4″-enopyranosiduronic acid in the host, which may be biosynthesized from its precursor, glucuronic acid by a β-elimination mechanism. In addition, the relative stereochemistry of the C-2 and C-3 of the sugar moiety is different from that of glucuronic acid, which might be catalyzed by a hydroxyl isomerase or other endogenous enzymes.
Microbial whole-cell biotransformation has attracted more attention in recent years for the production of pharmaceutical derivatives. One of its advantages is to eliminate the need to isolate, purify and stabilize enzymes (17). New molecules can be prepared with high stereo- and regio-selectivity under mild conditions (18). Moreover, this technique can be used for the creation of molecular diversity. Flavonoids are polyphenolic metabolites of plants and have been proposed to exert a wide range of beneficial effects in human health, preventing chronic and degenerative diseases such as cardiovascular, cerebrovascular, Parkinson’s and Alzheimer’s diseases (19). Some evidences suggest that flavonoids exercise cellular effects, many of which are linked to their interactions with specific proteins pathways (20). Although S. rimosus ATCC 10970 is a well-known producer of the pharmaceutically important antibiotic oxytetracycline, in this work, we have identified it as a useful and efficient biocatalytic tool to convert quercetin into a novel glycoside. This whole-cell biocatalyst may be further used to modify other similar flavonoids to yield new biologically active molecules.
Acknowledgments
This work was supported by a National Institutes of Health grant (AI065357).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paliwal S, Sundaram J, Mitragotri S. Induction of cancer-specific cytotoxicity towards human prostate and skin cells using quercetin and ultrasound. Brit J Cancer. 2005;92:499–502. doi: 10.1038/sj.bjc.6602364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rotelli AE, Aguilar CF, Pelzer LE. Structural basis of the anti-inflammatory activity of quercetin: inhibition of the 5-hydroxytryptamine type 2 receptor. Eur Biophys J. 2009;38:865–871. doi: 10.1007/s00249-009-0453-x. [DOI] [PubMed] [Google Scholar]
- 3.Kaul TN, Middleton E, Ogra PL. Antiviral effect of flavonoids on human viruses. J Med Virol. 1985;15:71–79. doi: 10.1002/jmv.1890150110. [DOI] [PubMed] [Google Scholar]
- 4.Larson AJ, Symons JD, Jalili T. Therapeutic potential of quercetin to decrease blood pressure: review of efficacy and mechanisms. Adv Nutr. 2012;3:39–46. doi: 10.3945/an.111.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimoda K, Otsuka T, Morimoto Y, Hamada H, Hamada H. Glycosylation and malonylation of quercetin, epicatechin, and catechin by cultured plant cells, Chem. Lett. 2007;36:1292–1293. [Google Scholar]
- 6.Eula Maria MBC, Fabiana CP, Wolf CL, Valeria O. Selection of filamentous fungi of the Beauveria genus able to metabolize quercetin like mammalian cells. Braz J Microbiol. 2008;39:405–408. doi: 10.1590/S1517-838220080002000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davisson JW, Tanner FW, Jr, Finlay AC, Solomons IA. Rimocidin, a new antibiotic. Antibiot Chemother. 1951;1:289–290. [PubMed] [Google Scholar]
- 8.Zhang W, Wilke BI, Zhan J, Watanabe K, Boddy CN, Tang Y. A new mechanism for benzopyrone formation in aromatic polyketide biosynthesis. J Am Chem Soc. 2007;129:9304–9305. doi: 10.1021/ja0736919. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Watanabe K, Cai X, Jung ME, Tang Y, Zhan J. Identifying the minimal enzymes required for anhydrotetracycline biosynthesis. J Am Chem Soc. 2008;130:6068–6069. doi: 10.1021/ja800951e. [DOI] [PubMed] [Google Scholar]
- 10.Seco EM, Perez-Zuniga FJ, Rolon MS, Malpartida F. Starter unit choice determines the production of two tetraene macrolides, rimocidin and CE-108, in Streptomyces diastaticus var. 108. Chem Biol. 2004;11:357–366. doi: 10.1016/j.chembiol.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Zhan J, Gunatilaka AAL. Selective 4′-O-methylglycosylation of the pentahydroxy-flavonoid quercetin by Beauveria bassiana ATCC 7159. Biocatal Biotransfor. 2006;24:396–399. [Google Scholar]
- 12.Zi J, Valiente J, Zeng J, Zhan J. Metabolism of quercetin by Cunninghamella elegans ATCC 9245. J Biosci Bioeng. 2011;112:360–362. doi: 10.1016/j.jbiosc.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Chaki T, Kakimi H, Shibata A, Baba T. Detection of alginate oligosaccharides from mollusks. Biosci Biotechnol Biochem. 2006;70:2793–2796. doi: 10.1271/bbb.60313. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Yu G, Guan H, Zhao X, Da Y, Jiang X. Preparation and structural elucidation of alginate oligosaccharides degraded by alginate lyase from Vibro sp 510. Carbohydr Res. 2004;339:1475–1481. doi: 10.1016/j.carres.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Tokodi I. Flavone utilization by Streptomyces rimosus. Biol Kozlemenyek. 1960;8:151–158. [Google Scholar]
- 16.Wong TY, Preston LA, Schiller NL. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu Rev Microbiol. 2000;54:289–340. doi: 10.1146/annurev.micro.54.1.289. [DOI] [PubMed] [Google Scholar]
- 17.Mulinacci N, la Marca G, Innocenti M, Vincieri FF, Crespi-Perellino N, Minghetti A. Cell cultures of Ajuga reptans L. to bioconvert emodin and aloe-emodin: an HPLC/ESI/MS investigation. Enzyme Microb Technol. 2005;36:399–408. [Google Scholar]
- 18.Keppler AF, Porto ALM, Schoenlein-Crusius IH, Comasseto JV, Andrade LH. Enzymatic evaluation of different Aspergillus strains by biotransformation of cyclic ketones. Enzyme Microb Technol. 2005;36:967–975. [Google Scholar]
- 19.Havsteen BH. The biochemistry and medical significance of the flavonoids. Pharmacol Ther. 2002;96:67–202. doi: 10.1016/s0163-7258(02)00298-x. [DOI] [PubMed] [Google Scholar]
- 20.Walle T. Absorption and metabolism of flavonoid. Free Radic Biol Med. 2004;36:829–837. doi: 10.1016/j.freeradbiomed.2004.01.002. [DOI] [PubMed] [Google Scholar]