Abstract
In response to pathogen insult, CD8 T cells undergo expansion and a dynamic differentiation process into functionally different subpopulations. In this report, we show that during the effector response to influenza virus infection lung CD8 T cell subsets expressing killer cell lectin-like receptor G1 (KLRG1)high or KLRG1low had similar effector functions and immediate recall efficacy. The KLRG1 expression profile of lung CD8 T cells was not permanent after adoptive transfer and recall. Airway CD8 T cells exhibited a unique phenotype expressing low levels of KLRG1 together with high levels of markers of cellular activation. We investigated the functional characteristics of these cells by analyzing their capacity to survive and to respond to a secondary challenge outside of the airway environment. KLRG1high CD8 T cells isolated from the lung during the peak of the effector T cell response could survive for over a month in the absence of cognate viral antigens after systemic adoptive transfer, and these “rested” CD8 T cells proliferated and participated in a recall response to influenza virus infection. These data highlight the unique phenotype and plasticity of effector CD8 T cell responses in the lung.
Introduction
The airway is a major portal for pathogen entry in the body, and CD8 T cells present in the bronchoalveolar space have an important role in the control of respiratory infections. The initiation of a CD8 T cell response after respiratory virus infection requires the migration of lung dendritic cells carrying viral antigens to the draining lymph node, and the subsequent priming of naïve antigen-specific CD8 T cells. After activation and clonal expansion, these CD8 T cells differentiate into effector cells that migrate to the mucosal site of infection to mediate pathogen clearance. This process is mostly mediated through the secretion of antiviral cytokines and the lysis of infected airway epithelial cells (1, 2). The effector CD8 T cell response is heterogeneous and consists of two major subpopulations: KLRG1high CD127low are terminally differentiated SLECs (3–6), and KLRG1low CD127high have been referred as memory precursor effector cells (MPECs) (6). It has been suggested that TEM are derived largely from the KLRG1highCD127low effector subpopulation, whereas KLRG1lowCD127high cells give rise to central-memory Tcells (TCM) (6, 7). The plasticity or secondary replicative function of CD8 T cells with an effector phenotype has been described during acute (8) and persistent viral infections (9–11). Memory CD8 T cells isolated from the airways can participate in recall responses (12) but whether this occurs during the effector phase of the response remains unclear.
KLRG1 is used as a surrogate marker of terminally differentiated SLECs (6, 13–15). KLRG1 expression on CD8 T cells correlates with replicative senescence and impaired proliferative potential (4, 16), suggesting that KLRG1 expression may reflect a common program of terminal cell differentiation. Upon ITIM-phosphorylation, KLRG1 recruits SHIP-1 and SHIP-2 and inhibits suboptimal TCR signaling (17), implying that KLRG1 signaling may also dampen cytokine production and killing (4, 16). The ligands of KLRG1 have recently been described and are the ubiquitous E-, N-, and R-cadherins (17–19). KLRG1 expression may define unique subpopulations of effector and memory CD8 T cells (20–22), but the relative contribution of KLRG1 subsets to immunity in non-lymphoid tissues is still a matter of debate. In this report, we show that during the effector response to influenza virus infection lung CD8 T cell subsets expressing KLRG1high or KLRG1low have similar effector functions, including IFNγ production, degranulation, and recall efficacy. KLRG1high CD8 T cells are capable of surviving long-term in the absence of cognate antigen, and can also proliferate during recall. Our findings shed light on the anatomical differences and plasticity of CD8 T cell responses.
Materials and Methods
Mice and viral infection
C57BL/6J, B6.SJL-Ptprca Pep3/BoyJ (Pep/boy, CD45.1) and B6.PL-Thy1a/CyJ (B6.PL, CD90.1) mice were obtained from The Jackson Laboratory, or were bred at the Research Institute at Nationwide Children’s Hospital. Mice were housed in BL2 containment under pathogen-free conditions. Mice were anesthetized with 2,2,2,-tribromoethanol and intranasally inoculated with 3000 50% egg infectious doses (EID50) influenza A virus strain X31 (H3N2) in 30 μl of HBSS. The Institutional Animal Care and Use Committee approved all of the animal studies described in this work.
Flow cytometry analysis
Single-cell suspensions were stained with Fc-block (CD16/32), and then washed and stained with a combination of the influenza virus-specific tetramer NP366-374/Db and antibodies against CD8, CD44, Ly-6C (Biolegend), CD90.2, CD62L, KLRG1, CD69, CD103, CD27 (eBioscience), CD43-activation associated glycoform (BD Biosciences) and CXCR3 (R&D Systems). For intracellular cytokine staining, a total of 2×106 cells/sample were incubated in tissue culture medium in the presence of IL-2 (10 U/ml) and brefeldin A (10 mg/ml) for 5 h at 37°C. As a positive control, cells were stimulated with PMA/ionomycin. After Fc blocking, the cells were stained with antibodies against CD8 and CD107ab, fixed, permeabilized, and stained with antibodies against IFNγ or isotype control antibodies. Flow cytometry data were acquired on a BD LSR II, and analyzed using FlowJo software. Gates were set using negative controls and isotype controls.
FACS-sorting and adoptive cell transfers
Lung cells from 10 donor mice/group (CD45.1/CD90.2 and CD45.2/CD90.2) at day 10 post-infection were FACS-purified into KLRG1high or KLRG1low CD44+ CD8 T cells. Sorted subsets were combined in equal numbers for each donor, and a total of 1–2.5×105 cells were intravenously transferred into naïve recipient mice (CD45.2/CD90.1). The recipient mice were rested during 24 h (immediate recall) or during 30 d (memory recall), and then intranasally challenged with influenza virus. The indicated tissues were analyzed 7 d later.
Reverse transcriptase quantitative PCR (RT-qPCR)
Total mRNA was purified using TRIzol Reagent (Invitrogen), and 1 μg of each sample was reverse transcribed using high capacity cDNA reverse transcription kit (Applied Biosystems). Primer sequences were used as follows: Bmi-1, 5′ATGGCCGCTTGGCTCGCATT-3′ and 5′AAAGATCCCGGAAAGAGCGGC-3′; KLRG1, 5′AGCCTCGTCCCGTAGACAAAA-3′ and 5′ TGGCAACAATCTCCACTTTGC-3′; Blimp-1, 5′GTCGCGGAGACGCAAGAGCA-3′ and 5′CTTGGGGGCAGCCAAGGTCG-3′; T-bet, 5′CCTCGCCTGGTGAAATGTGC-3′ and 5′GAGGGACTTCATGGCTTTCTGC-3′; Eomes 5′GGTGGGGTTGAGTCCGTTTATGTT-3′ and 5′GGCCAGGGTTCTCCGCTCTA-3′; GAPDH 5′AGCCTCGTCCCGTAGACAAAA3′ and 5′TGGCAACAATCTCCAAAACTTTGC 3′. PCR amplifications were performed on an ABI Prism 7500 Real Time PCR system using 96-well microtiter plates.
Results
Reduced KLRG1 expression on airway CD8 T cells
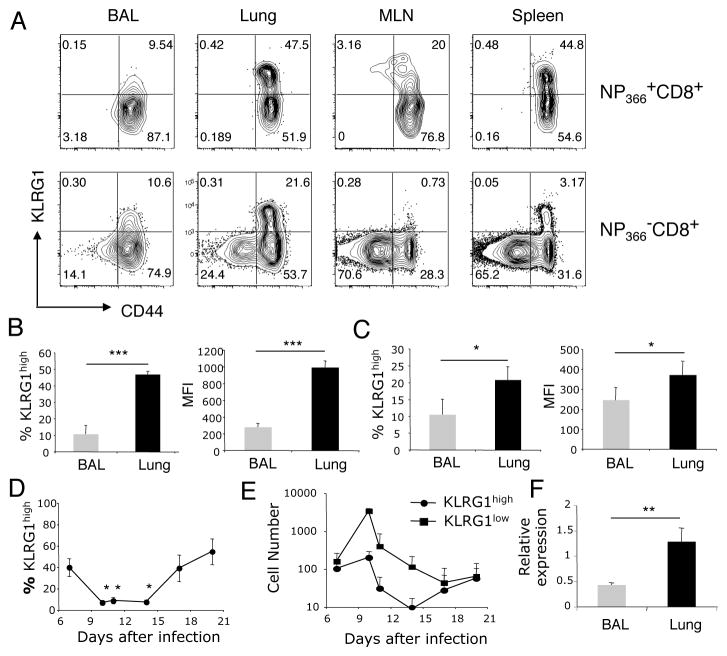
We examined influenza virus-specific CD8 T cells for cell surface KLRG1 expression on day 10 post- infection, which is at the peak of influenza virus-specific CD8 T cell expansion. While two populations of CD8 T cells (KLRG1high and KLRG1low) could be discriminated in both the NP366-374/Db tetramer-positive and tetramer-negative CD8 T cells in lung parenchyma, MLN and spleen, a conspicuous KLRG1high population was missing in the BAL (Figure A). Less than 10% of the influenza virus NP366-374/Db-specific CD8 T cells located in the airways expressed KLRG1 compared to 40–50% in the lung parenchyma on day 10 after infection (Figure 1B). The intensity of KLRG1 expression in airway NP366-374/Db-specific CD8 T cells, measured as MFI, was 5-fold lower than in their lung parenchyma counterparts (Figure 1B). Airway NP366-374/Db-negative CD8 T cells also expressed less KLRG1 than their lung parenchyma counterparts (Figure 1C). A time course analysis of KLRG1 expression on influenza virus NP366-374/Db-specific CD8 T cells in the bronchoalveolar space showed a significant decrease in the frequency of virus-specific KLRG1high CD8 T cells during the peak of T cell expansion (days 10–14, Figure 1D). Next, we analyzed absolute T cell numbers at different times after infection (Figure 1E). This analysis shows an increase in the number of airway KLRG1low NP366-374/Db-specific CD8 T cells, and that KLRG1high NP366-374/Db-specific CD8 T cells are ten times less abundant that their KLRG1low counterparts in the airways during the peak of the T cell response to influenza virus infection. Interestingly, at early (day 7) and late (day 17–20) times after infection the number of virus-specific KLRG1low and KLRG1high CD8 T cells present in the airways was similar. To determine if the reduced KLRG1 expression observed in airway CD8 T cells when compared to their lung parenchyma counterparts was due to protein cleavage or to transcriptional control we determined the expression level of KLRG1 mRNA on FACS-sorted CD8 T cells from BAL and lung parenchyma by RT-qPCR. The data show significantly reduced KLRG1 mRNA expression levels of in CD8 T cells isolated from the airways when compared with those from the lung parenchyma (Figure 1F). Altogether, these data demonstrate that airway CD8 T cells have low expression of KLRG1 during the effector phase of the T cell response to influenza virus infection, and this process is at least partially controlled at the transcriptional level.
Figure 1. Reduced KLRG1 expression in airway CD8 T cells during influenza virus infection.
(Panel A) Representative contour plots show KLRG1 cell surface expression in NP366-374/Db-specific (top) and on tetramer-negative (bottom) CD8 T cells in BAL, lung, MLN and spleen on day 10 after influenza virus infection. (Panel B) Bar diagrams show the frequency (left panel) and median fluorescence intensity (MFI, right panel) of KLRG1 expression in NP366-374/Db-specific CD8 T cells on day 10 after influenza virus infection. (Panel C) Bar diagrams show the frequency (left panel) and MFI (right panel) of KLRG1 expression in tetramer-negative CD8 T cells on day 10 after influenza virus infection. (Panel D) Dynamics of KLRG1 expression in airway NP366-374/Db-specific CD8 T cells following influenza virus infection. (Panel E) Representation of KLRG1high and KLRG1low NP366-374/Db-specific CD8 T cell numbers in the airways following influenza virus infection. (Panel F) Relative expression of KLRG1 mRNA in FACS-sorted CD8 T cells on day 10 after influenza virus infection. Data are representative from 2–3 independent experiments from 3 or more mice at each time point. Error bars indicate SD. * p ≤ 0.05, ** p ≤ 0.005, *** p ≤ 0.0005.
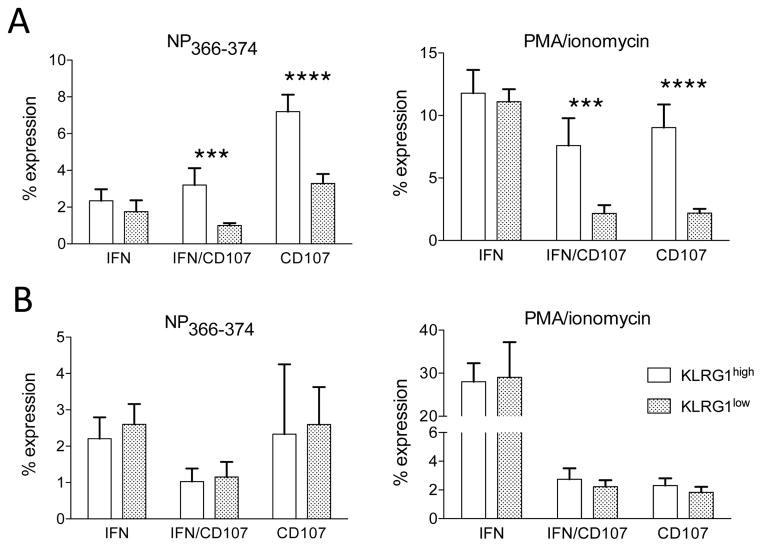
Given the association of KLRG1 expression with effector T cell differentiation, we wondered if KLRG1 expression in lung CD8 T cells could be associated with changes in effector attributes or with the generation of memory cells. First, we analyzed cytokine production and degranulation in KLRG1high and KLRG1low CD8 T cells isolated from the spleen and lung. The data show that splenic KLRG1high cells have a significantly higher frequency of cells capable of mobilizing CD107ab and of polyfunctional cells (IFNγCD107-double positive) than spleen KLRG1low cells. These findings are pertinent to both NP366-374-specific CD8 T cell splenocytes and to bulk CD8 T cell responses after PMA/ionomycin stimulation (Figure 2A), and are in concordance with published studies indicating that KLRG1 expression is associated with effector T cell differentiation (6, 13–15). Surprisingly, we did not observed statistically significant differences in the capacity of lung KLRG1high and KLRG1low CD8 T cell subsets to produce IFNγ or to degranulate after stimulation with NP366-374 or with PMA/ionomycin (Figure 2B). These results indicate that lung KLRG1high and KLRG1low CD8 T cells have similar effector functions. Altogether, our findings demonstrate that differential KLRG1high and KLRG1low expression is associated with distinct effector potential in splenic CD8 T cells but not in lung CD8 T cells.
Figure 2. Lung CD8 T cell subpopulations display similar effector function.
(Panel A) Spleen and lung cells we isolated from influenza virus infected mice on day 10 after infection. After stimulation with influenza virus-specific NP366-374 peptide or with PMA/ionomycin, we measured intracellular IFNγ production and cell surface CD107a/b mobilization on previously gated KLRG1high and KLRG1low CD8 T cells. Bar diagrams show the frequency of IFNγ+CD107ab−, polyfunctional IFNγ+CD107ab+, and IFNγ−CD107ab+ cells. (Panel A) Spleen CD8 T cells. (Panel B) Lung CD8 T cells. Data are representative from 2 independent experiments with 3 mice/group. Error bars indicate SD. *** p ≤ 0.0005, **** p ≤ 0.00005.
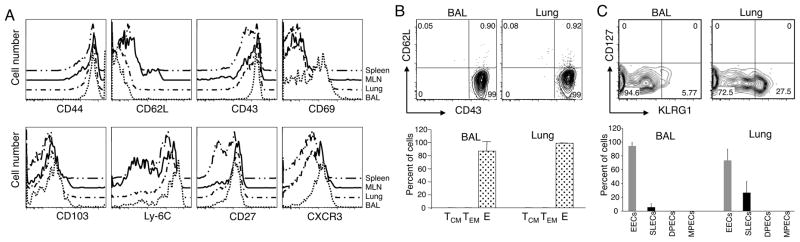
Second, we analyzed the expression of several activation markers on influenza virus-specific CD8 T cells. In accordance with previous studies, airway NP366-374/Db-specific CD8 T cells expressed high levels of CD44, CD43, CD69, CD103, Ly-6C, CD27 and CXCR3, and lacked CD62L on day 10 after infection (Figure 3A). The expression of high levels of CD69 also differentiated influenza virus-specific CD8 T cells in the BAL from their counterparts in the lung parenchyma, draining lymph node and spleen. Importantly, we could not detect down-regulation of any of the markers of effector T cell function analyzed on BAL CD8 T cells, which are predominantly KLRG1low. Third, we analyzed the presence of phenotypic memory CD8 T cells in the airways on day 10 after influenza virus infection. Our results show that central-memory T cells (TCM, CD43lowCD62Lhigh) and TEM (CD43lowCD62Llow) did not constitute a sizable subpopulation of NP366-374/Db-specific CD8 T cells in the airways on day 10 after infection (Figure 3B), at least as defined by CD43/CD62L expression. The data showed that 99% of the influenza virus-specific CD8 T cells were effectors (CD43highCD62Llow). A complementary analysis using different memory markers showed that CD127highKLRG1high (double-positive effector cell, DPEC) and CD127highKLRG1low (MPEC) NP366-374/Db-specific CD8 T cells could not be detected in the airways and lung parenchyma, and that early effector cells (EEC, CD127lowKLRG1low) and SLECs (CD127lowKLRG1high) constituted the totality of influenza virus specific CD8 T cells on day 10 after infection (Figure 3C). Altogether, our analyses support the conclusion that KLRG1low CD8 T cells in the lung and airways have a highly activated phenotype during the effector response to influenza virus, and differential KLRG1 expression in the respiratory tract does not correlate with a loss of phenotypic and functional effector attributes nor with the acquisition of cell surface markers representative of a memory phenotype.
Figure 3. Low KLRG1 expression on influenza virus-specific airway CD8 T cells is not associated with loss of activation markers or with the acquisition of a memory phenotype.
(Panel A) Histograms show the expression profile of different activation markers on NP366-374/Db-specific CD8 T cells isolated from spleen, MLN, lung parenchyma, and airways. (Panel B) Percentage of effector (E, CD43+CD62L−), TEM (CD43−CD62L−), and TCM (CD43−CD62L+) NP366-374/Db-specific CD8 T cells in BAL and lung parenchyma. (Panel C) Percentage of EECs (CD127lowKLRG1low), SLECs (CD127lowKLRG1high), DPECs (CD127highKLRG1high), and MPECs (CD127highKLRG1low) NP366-374/Db-specific CD8 T cells in BAL and lung parenchyma. Data are representative from 2 independent experiments with 3 mice/group performed on day 10 after influenza virus infection. Error bars indicate SD.
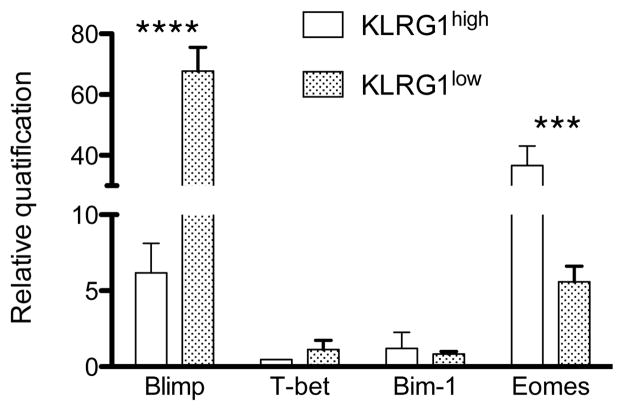
Finally, in order to examine if differential KLRG1 expression in the lung could be correlated with distinct pathways of cellular differentiation, we analyzed the expression of several transcription factors implicated in determining T cell fate in FACS-purified lung KLRG1high and KLRG1low CD8 T cells. Blimp-1 is a transcriptional repressor that promotes the development of SLECs and regulates clonal exhaustion, being absent in long-lived TCM (23–25). Expression of T-bet is necessary and sufficient to promote the development of SLECs (6, 26). The polycomb finger ring oncogene bmi-1 is necessary for efficient self-renewing and it is not expressed in KLRG1high CD8 T cells (5). Eomes has a role in promoting memory persistence (26, 27). Our analysis show that both KLRG1high and KLRG1low CD8 T cells expressed blimp-1 and eomes, and neither subset expressed substantial levels of T-bet or bmi-1 regardless of their KLRG1 profile (Figure 4). In addition, blimp-1 was more abundant in KLRG1low cells and KLRG1high cells expressed significant higher levels of eomes than their KLRG1low counterparts. In summary, both KLRG1 subsets expressed eomes and blimp-1 although with quantitative differences, and no other distinctions in effector function or activation/memory phenotype were found between KLRG1high and KLRG1low CD8 T cells in the respiratory tract that could reveal the existence of differences in their biological function.
Figure 4. Expression of transcription factors in lung KLRG1high and KLRG1low CD8 T cell subsets.
Lung cells were sorted as CD8 T cells into KLRG1high and KLRG1low subsets on day 10 after influenza virus infection. Blimp-1, T-bet, Bmi-1, and Eomes mRNA levels were determined by RT-qPCR. Data are representative from 3 independent experiments using 3 mice/group. Error bars indicate SD. *** p ≤ 0.0005, **** p ≤ 0.00005.
Comparative analysis of KLRG1 subsets contribution to recall and memory
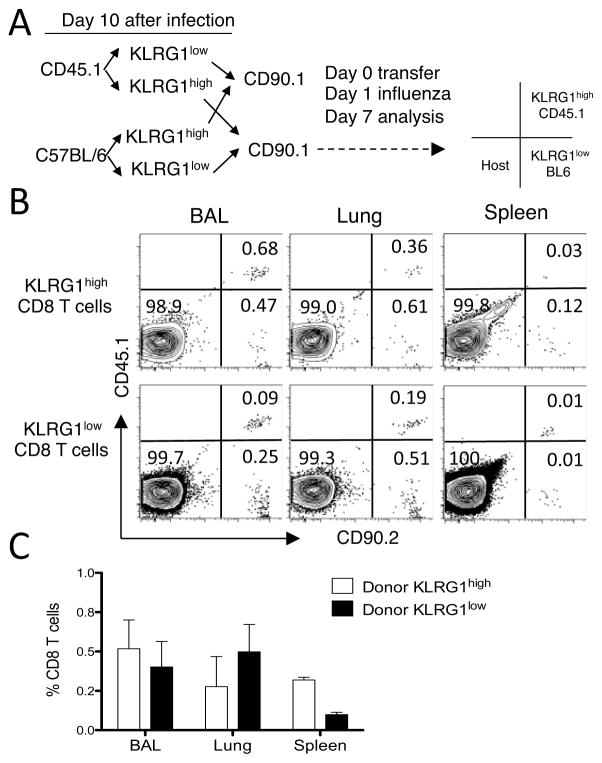
T cell memory is a functional definition, and CD8 T cell responses to secondary pulmonary infections are characterized by increased accumulation of activated CD8 T cells within the respiratory tract (28). To compare the contribution of KLRG1high and KLRG1low CD8 T cells to this accumulation in the lung parenchyma and the airways, we used a dual adoptive transfer approach. We FACS sorted KLRG1low and KLRG1high CD8 T cells from the lungs of Pep/boy mice (CD45.1/CD90.2) and BL6 mice (CD45.2/CD90.2) on day 10 after influenza virus infection. Sorted subsets from each donor were mixed in equal numbers, and transferred into naive Thy1.1 recipient mice (CD45.2/CD90.1). The strength of this approach is that the response of two different populations of KLRG1high and KLRG1low cells is compared in the same animal under identical conditions, and the two donor and host populations can be discriminated on the basis of CD90 and CD45 expression. The limitation of this approach is that only low numbers of cells can be adoptively transferred (~2×105) after sorting of lung subpopulations, which makes it problematic to elucidate the overall influence of the donor cells in the host response. First, we compared the immediate recall efficiency of donor KLRG1high and KLRG1low CD8 T cells. To this extent, the naïve CD90.1 recipient mice were intranasally challenged with influenza virus 1 d after adoptive transfer, and lung parenchyma, airways, and spleen analyzed 7 days later at the peak of the T cell recall response. An example of the experimental strategy and of the data generated is presented in Figure 5A–B. The data show the presence of KLRG1low and KLRG1high donor cells on day 7 for both sets of donor mouse strains (B6 and Pep/boy) in the same Thy1.1 recipient mouse. This finding demonstrates that the KLRG1 phenotype of the donor lung cells has changed into KLRG1low or KLRG1high during recall regardless of their previous phenotype at the time of adoptive transfer. In addition, the frequency of CD8 T cells of KLRG1low donor origin was zero to threefold higher than that of KLRG1high in the airways, lung parenchyma, and spleen (Figure 5C). Altogether, these results suggest that KLRG1 expression is not permanent, and that both lung KLRG1low and KLRG1high CD8 T cells participate in an immediate recall response with similar efficacy, which correlates with the lack of differences observed between both subpopulations regarding effector function and activation/memory phenotype.
Figure 5. Immediate recall efficacy of lung KLRG1high and KLRG1low CD8 T cells.
Donor mice (CD45.1/CD90.2 and CD45.2/CD90.2) were infected with influenza virus and lung cells isolated 10 d later. CD8 CD8 T cells were sorted into KLRG1high and KLRG1low subsets by FACS. Sorted subsets were combined in equal numbers for each donor, and a total of 1–2.5 × 105 cells were intravenously transferred into naïve recipient mice (CD45.2/CD90.1). 1 d after transfer, the recipient mice were then intranasally challenged with influenza virus and the indicated tissues were analyzed 7 d later. (Panel A) Experimental transfer strategy, which includes a template to interpret the CD45/CD90 phenotype of the representative mouse shown in panel B. (Panel B) Representative plots of the flow cytometric profiles of donor and host cells on d 7 gated as KLRG1high (top row) and KLRG1low (bottom row) CD8 T cells. (Panel C) Bar diagram shows the relative response of each KLRG1 subset at the time of transfer in each tissue measured as the percentage donor cells of the total CD8 T cell population on d 7. Data are representative of two independent sorting experiments including a total of 8 adoptively transferred mice. Error bars indicate SEM.
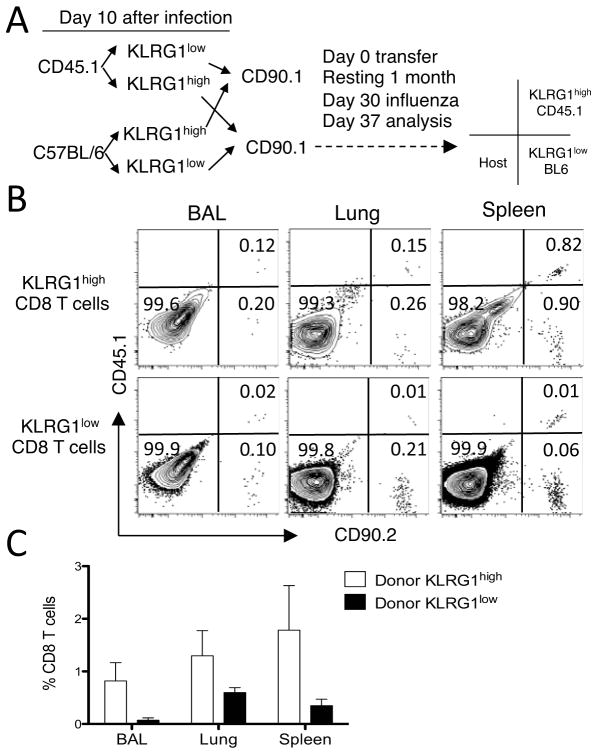
Using a similar approach, we next compared the memory and recall efficiency of donor KLRG1high and KLRG1low CD8 T cells. To this extent, after adoptive transfer of equal numbers of lung KLRG1low and KLRG1high CD8 T cells from day 10 influenza virus-infected mice into naïve CD90.1 mice the recipient mice were rested for 30 days in the absence of cognate viral antigens. Next, the recipient mice were intranasally challenged with influenza virus, and lung parenchyma, airways, and spleen analyzed 7 days later (Figure 6A). The data show, first, that both KLRG1low and KLRG1high donor cells can be detected at the time of analysis (Figure 6B), indicating that both subpopulations can survive up to 30 days in the absence of cognate influenza virus antigens. Second, donor cells can be KLRG1low or KLRG1high regardless of their original phenotype before transfer, which supports the concept that KLRG1 expression is not enduring. And third, CD8 T cells of KLRG1high donor origin were three to tenfold more efficient than KLRG1low cells at accumulating in the airways, lung parenchyma, and spleen (Figure 6C), although these differences were not statistically significant. Altogether, our results indicate that lung CD8 T cell subsets expressing KLRG1high or KLRG1low have similar recall efficacy, and that the KLRG1 expression profile of lung CD8 T cells is not permanent.
Figure 6. Memory recall efficacy of lung KLRG1high and KLRG1low CD8 T cells.
Donor mice (CD45.1/CD90.2 and CD45.2/CD90.2) were infected with influenza virus and lung cells isolated 10 d later. CD8 T cells were sorted into KLRG1high and KLRG1low subsets by FACS. Sorted subsets were combined in equal numbers for each donor, and a total of 1–2.5 × 105 cells were intravenously transferred into naïve recipient mice (CD45.2/CD90.1). The recipient mice were rested during 30 d, and then intranasally challenged with influenza virus. The indicated tissues were analyzed 7 d later. (Panel A) Experimental transfer strategy, which includes a template to interpret the CD45/CD90 phenotype of the representative mouse shown in panel B. (Panel B) Representative plots of the flow cytometric profiles of donor and host cells on d 37 after transfer gated as KLRG1high (top row) and KLRG1low (bottom row) CD8 T cells. (Panel C) Bar diagram shows the relative response of each KLRG1 subset at the time of transfer in each tissue measured as the percentage donor cells of the total CD8 T cell population on d 37. Data are representative of two independent sorting experiments including a total of 6 adoptively transferred mice. Error bars indicate SEM.
Discussion
Our results demonstrate that KLRG1high and KLRG1low CD8 T cell subsets isolated from the lung have similar effector functions, while splenic KLRG1high CD8 T cells are better effectors than their KLRG1low counterparts. We also observed that the majority of airway CD8 T cells during primary infection lack substantial KLRG1 expression on their cell surface. Several observations indicate that pulmonary KLRG1low CD8 T cells maintain their effector attributes. First, they secreted IFNγ and degranulated upon restimulation, expressed high levels of activation markers (CD69, CD43), and lacked memory markers (CD127, CD62L). Second, they expressed high levels of blimp-1. Third, this population failed to efficiently proliferate after transfer and recall. Yet, our studies revealed the capacity of KLRG1low CD8 T cells isolated at the peak of the effector response to survive long-term in the absence of cognate antigen and to subsequently proliferate during antigenic recall. These results highlight the plasticity of CD8 T cell responses revealing that cells recruited to the lung at the peak of the effector response are not terminally differentiated and retain the capacity to survive and participate in recall responses under adequate conditions.
The role of KLRG1 on CD8 T cell function has not been fully elucidated. In spite of its frequent use as a surrogate marker of terminal differentiation of T cells (6, 13–15), lack of KLRG1 has no significant impact on the generation of CD8 T cell responses and anti-viral control (6, 29). Our findings support the concept that KLRG1 subpopulations have similar effector potential in the lung, while in the spleen KLRG1high CD8 T cells are better effectors than KLRG1low cells. In addition, our dual adoptive transfer experiments demonstrate that KLRG1 expression of lung cells can change after adoptive transfer regardless of the original KLRG1high and KLRG1low phenotype at the time of transfer. Lung effector CD8 T cells have a limited proliferative capacity but can survive up to 30 days in the absence of cognate viral antigen independently of their KLRG1 expression profile at the time of adoptive transfer. Overall, these findings suggest that the KLRG1 phenotype is not permanent and does not commit lung CD8 T cells to an irreversible differentiation program.
Influenza virus-specific CD8 T cells present in the airways have a unique KLRG1lowCD62LlowCD127lowCD69highCD43high phenotype, and that due to the low expression of KLRG1 they do not fit current definitions of SLEC, DPEC, EECs, MPEC, TEM or central-memory cell (TCM) subsets defined by using some the above markers (6, 21, 22). It is possible that airway CD8 T cells down-regulate KLRG1 expression during acute influenza virus infection to avoid KLRG1 interaction with epithelial cadherins that could result in diminished effector function. In this sense, it has been shown that the airway environment alters the expression of other surface markers on CD8 T cells, such as CD11a and Ly-6C, although a mechanism for this process has not yet been proposed (30). It is also possible that lung KLRG1low CD8 T cells are selectively recruited into the airway spaces during the effector phase of the response. Regardless of the mechanisms, our data demonstrate airway CD8 T cells express low levels of KLRG1 during the effector phase of the antiviral response.
The significance of reduced KLRG1 expression on airway CD8 T cells during the effector phase of the response is poorly understood. Our data show CD8 T cells isolated during the acute phase of influenza virus infection can survive for over a month in the absence of cognate viral antigens after adoptive transfer into the circulation. This finding supports the concept that lung CD8 T cells recruited during the effector phase of the response have the potential to endure in the absence of cognate antigen. Furthermore, our data support the idea that these “rested” CD8 T cells can participate in a proliferative recall. A limitation of our adoptive transfer experiments is that the site of proliferation of these influenza virus-specific donor CD8 T cells cannot be discerned. Nevertheless, this secondary replicative function of CD8 T cells is in concordance with other studies that show that effector CD8 T cells may follow a continuum linear differentiation pathway into memory cells with replicative function (6, 8, 31). In summary, our findings indicate that CD8 T cells recruited to the airways at the peak of the effector response are not simply end-stage cells, and have relevant implications for the development of strategies to promote cellular immunity in the lung.
Acknowledgments
This work was supported in part by NIH grant AI082962 and by The Research Institute.
We thank the Flow Cytometry Core Laboratory at the Research Institute for help with flow cytometry and sorting.
Abbreviations
- BAL
bronchoalveolar lavage
- MLN
mediastinal lymph node
- KLRG1
killer cell lectin-like receptor G1
- SLEC
short lived effector cell
- MPEC
memory precursor effector cell
- DPEC
double-positive effector cells
- EEC
early effector cells
- TEM
effector-memory T cell
- TCM
central-memory T cells
- RT-qPCR
reverse transcriptase quantitative PCR
References
- 1.Hou S, Hyland L, Ryan KW, Portner A, Doherty PC. Virus-specific CD8+ T-cell memory determined by clonal burst size. Nature. 1994;369:652–654. doi: 10.1038/369652a0. [DOI] [PubMed] [Google Scholar]
- 2.Flynn KJ, Belz GT, Altman JD, Ahmed R, Woodland DL, Doherty PC. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 3.Marcolino I, Przybylski GK, Koschella M, Schmidt CA, Voehringer D, Schlesier M, Pircher H. Frequent expression of the natural killer cell receptor KLRG1 in human cord blood T cells: correlation with replicative history. Eur J Immunol. 2004;34:2672–2680. doi: 10.1002/eji.200425282. [DOI] [PubMed] [Google Scholar]
- 4.Voehringer D, Koschella M, Pircher H. Lack of proliferative capacity of human effector and memory T cells expressing killer cell lectinlike receptor G1 (KLRG1) Blood. 2002;100:3698–3702. doi: 10.1182/blood-2002-02-0657. [DOI] [PubMed] [Google Scholar]
- 5.Heffner M, Fearon DT. Loss of T cell receptor-induced Bmi-1 in the KLRG1(+) senescent CD8(+) T lymphocyte. Proc Natl Acad Sci USA. 2007;104:13414–13419. doi: 10.1073/pnas.0706040104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtulus S, Tripathi P, Moreno-Fernandez ME, Sholl A, Katz JD, Grimes HL, Hildeman DA. Bcl-2 allows effector and memory CD8+ T cells to tolerate higher expression of Bim. J Immunol. 2011;186:5729–5737. doi: 10.4049/jimmunol.1100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science. 2009;323:505–509. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cush SS, Flano E. Protective antigen-independent CD8 T cell memory is maintained during {gamma}-herpesvirus persistence. J Immunol. 2009;182:3995–4004. doi: 10.4049/jimmunol.0803625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bannard O, Kraman M, Fearon DT. Cutting edge: Virus-specific CD8+ T cell clones and the maintenance of replicative function during a persistent viral infection. J Immunol. 2010;185:7141–7145. doi: 10.4049/jimmunol.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cush SS, Flano E. KLRG1(+)NKG2A(+) CD8 T cells mediate protection and participate in memory responses during gamma-herpesvirus infection. J Immunol. 2011;186:4051–4058. doi: 10.4049/jimmunol.1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ely KH, Roberts AD, Woodland DL. Cutting edge: effector memory CD8+ T cells in the lung airways retain the potential to mediate recall responses. J Immunol. 2003;171:3338–3342. doi: 10.4049/jimmunol.171.7.3338. [DOI] [PubMed] [Google Scholar]
- 13.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodland DL. Memories of Santa Fe. Symposium on immunologic memory. EMBO Rep. 2007;8:823–828. doi: 10.1038/sj.embor.7401033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voehringer D, Blaser C, Brawand P, Raulet DH, Hanke T, Pircher H. Viral infections induce abundant numbers of senescent CD8 T cells. J Immunol. 2001;167:4838–4843. doi: 10.4049/jimmunol.167.9.4838. [DOI] [PubMed] [Google Scholar]
- 17.Tessmer MS, Fugere C, Stevenaert F, Naidenko OV, Chong HJ, Leclercq G, Brossay L. KLRG1 binds cadherins and preferentially associates with SHIP-1. Int Immunol. 2007;19:391–400. doi: 10.1093/intimm/dxm004. [DOI] [PubMed] [Google Scholar]
- 18.Ito M, Maruyama T, Saito N, Koganei S, Yamamoto K, Matsumoto N. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J Exp Med. 2006;203:289–295. doi: 10.1084/jem.20051986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundemann C, Bauer M, Schweier O, von Oppen N, Lassing U, Saudan P, Becker KF, Karp K, Hanke T, Bachmann MF, Pircher H. Cutting edge: identification of E-cadherin as a ligand for the murine killer cell lectin-like receptor G1. J Immunol. 2006;176:1311–1315. doi: 10.4049/jimmunol.176.3.1311. [DOI] [PubMed] [Google Scholar]
- 20.Beyersdorf NB, Ding X, Karp K, Hanke T. Expression of inhibitory “killer cell lectin-like receptor G1” identifies unique subpopulations of effector and memory CD8 T cells. Eur J Immunol. 2001;31:3443–3452. doi: 10.1002/1521-4141(200112)31:12<3443::aid-immu3443>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 21.Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, Woodland DL. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med. 2007;204:1625–1636. doi: 10.1084/jem.20070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obar JJ, Jellison ER, Sheridan BS, Blair DA, Pham QM, Zickovich JM, Lefrancois L. Pathogen-induced inflammatory environment controls effector and memory CD8(+) T cell differentiation. J Immunol. 2011;187:4967–4978. doi: 10.4049/jimmunol.1102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J Exp Med. 2008;205:1959–1965. doi: 10.1084/jem.20080526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 25.Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 27.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 28.Hikono H, Kohlmeier JE, Ely KH, Scott I, Roberts AD, Blackman MA, Woodland DL. T-cell memory and recall responses to respiratory virus infections. Immunol Rev. 2006;211:119–132. doi: 10.1111/j.0105-2896.2006.00385.x. [DOI] [PubMed] [Google Scholar]
- 29.Grundemann C, Schwartzkopff S, Koschella M, Schweier O, Peters C, Voehringer D, Pircher H. The NK receptor KLRG1 is dispensable for virus-induced NK and CD8+ T-cell differentiation and function in vivo. Eur J Immunol. 2010;40:1303–1314. doi: 10.1002/eji.200939771. [DOI] [PubMed] [Google Scholar]
- 30.Ely KH, Cookenham T, Roberts AD, Woodland DL. Memory T cell populations in the lung airways are maintained by continual recruitment. J Immunol. 2006;176:537–543. doi: 10.4049/jimmunol.176.1.537. [DOI] [PubMed] [Google Scholar]
- 31.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, Von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;3:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]