Abstract
Background
OCT4 and Survivin are important factors for cancer cell proliferation, renewal and dedifferentiation, and correlate with resistance to radiotherapy and chemotherapy in most human cancers, but their regulatory mechanisms are not well known.
Methodology/Principal Findings
In this study, 50 patients with esophageal squamous cell carcinoma (ESCC) were retrospectively analyzed. OCT4 was expressed in 13 cases (26%), and survivin was positively expressed in 31 cases (62%), examined by immunochemistry. OCT4 was found to be an independent predictive factor for median survival time, and the patients from the subgroup with both high expression of OCT4 and Survivin had the worst prognosis investigated by log-rank test. To further explore the molecular regulatory mechanism between OCT4 and Survivin, we constructed the specific small hairpin RNA (shRNA)-expressing vectors targeting OCT4 or/and Survivin and manipulated the expression of OCT4 and Survivin. By Western blotting and RT-PCR, we found that OCT4 could up-regulate Survivin expression in the esophageal cancer cell lines Eca109 and TE1. Simultaneously knockdown of OCT4 and Survivin expression induced cell apoptosis and G2-phase decrease of cell cycle by flow cytometry, and finally exerted an enhanced anti-proliferation potency in Eca109 and TE1 cell lines by MTT assay.
Conclusions
This study shows that OCT4 and Survivin expression were correlated with poor survival in patients with ESCC. OCT4 and Survivin may be regarded as targets in ESCC biotherapy.
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of most malignant tumors with high mortality [1], [2]. Although some new molecular targets have been found and used in ESCC biotherapy, the molecular mechanisms of ESCC recurrence and metastasis are still not understood. A growing body of evidence suggested that only a small fraction of cancer initiating cells have the ability to self-renew, as well as, to drive initiation and progression of cancer, and presented strongly resistance to chemotherapy and radiotherapy [3], [4], which gives us a better understanding of molecular basis of ESCC.
Octamer-binding transcription factor 4 (OCT4) is one of the stem related transcription factors, regulating tumor proliferation and self-renewal. Poorly differentiated or undifferentiated cancer cells have been characterized by many phenotypic traits similar to undifferentiated embryonic stem cells, suggesting that OCT4 may be expressed in solid tumors as a cancer initiating cell biomarker [5]. OCT4 belongs to the family of POU-domain transcription factors, including a homeodomain which is important in embryonic development [6], [7]. It has been proved that OCT4 overexpressed in a lot of somatic cancers, such as breast cancer, prostate cancer, non-small cell lung cancer, bladder cancer, oral squamous cell carcinoma, gastric cancer, esophageal cancer [8]–[14]. OCT4 expression plays a pivotal link in tumorigenesis and maintainance of cancer cells.
Survivin is a member of the inhibitors of the apoptotic gene family, and plays an important role in tumor progression by inhibiting cell apoptosis, regulation of cell division, and induction of angiogenesis [15]. Overexpression of Survivin was proposed in various cancers including ESCC [16], [17], but rarely present in normal adult tissues. Survivin expression in circulating cancer cells in the peripheral blood of patients with ESCC was detected and provided valuable information in the prediction of cancer recurrence and poor prognosis [18], [19]. Besides, overexpression of Survivin in ESCC presented resistance to chemotherapy and shorter survival [20], and there were similar results in other cancers [21].
Previous study demonstrated that knockdown of Survivin expression in a number of human cancer cell lines, such as A549, HeLa and MCF-7 cells, resulted in a significant reduction of cell viability, and combination of Survivin-directed silencing strategy with chemotherapeutic agents constituted a valuable approach for cancer treatment with an enhanced antitumor efficacy [21], [22]. However, cancer re-growth is probably the most important feature, because the cancer initiating cells resist the conventional cancer therapies and are likely to play a major role in cancer relapse [23]. Therefore, targeting cancer initiating cells has the potential to significantly improve outcomes for cancer patients. OCT4 is a master gene that plays a key role in the self-renewal and pluripotency of stem cells. Being selectively expressed in tumor tissues, evidence suggested that OCT4 may be a promising target for development of anticancer strategies to eliminate cancer initiating cells [24].
Recently, it was reported that Survivin expression was dramatically decreased in OCT4 knockdown murine embryonic stem cells [25], suggesting that there is a relationship between OCT4 and Survivin. But the molecular regulatory mechanisms between OCT4 and Survivin are not yet clear in cancers. In the current study, we examined OCT4 and Survivin expression and analyzed the prognostic relevance of these two genes with ESCC specimens. Meanwhile, the regulatory mechanism of OCT4 and Survivin expression and their function on cell apoptosis, cell proliferation or cell cycle were investigated in ESCC cell lines.
Results
OCT4 and Survivin were Over-expressed in ESCC
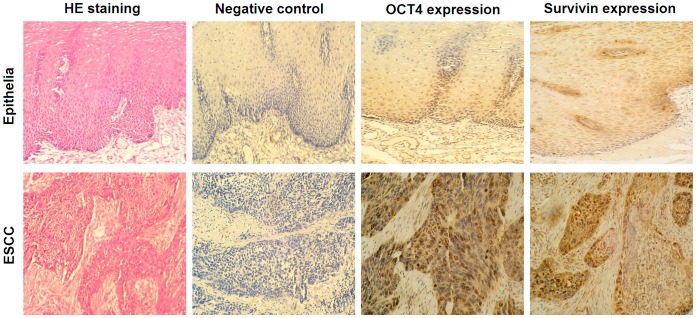
The expression of OCT4 and Survivin was detected by immunohistochemistry in the specimens of ESCC and adjacent normal esophageal tissues. OCT4 was expressed in 13 (26%) of ESCC but only 2 (4%) of normal esophageal tissues. Survivin was expressed in 31 (62%) of ESCC but only 11 (22%) of normal esophageal tissues. There were differences between ESCC and normal esophageal groups (p = 0.0051 for OCT4; p = 0.0001 for Survivin). The OCT4-positive immunoreactivity was mainly distributed in ESCC cellular nuclei and Survivin was mainly distributed in ESCC cytoplasm. The OCT4- and Survivin-positive cells were primarily located in the basal parts of the epithelia (Fig. 1). Survivin expression did not related with OCT4 expression in these ESCC samples (R = 0.276, p = 0.052; Table 1).
Figure 1. OCT4 and Survivin expression were detected by immunohistochemical staining in the resected specimens of esophageal squamous cell carcinoma (ESCC) and adjacent esophageal mucosa.
The paraffin-embedded sections were subjected to immunohistochemical examination with the primary mouse anti-human OCT4 and rabbit anti-human Survivin antibodies at working concentration of 1∶100, and the diaminobenzidine (DAB) was used to stain the positive reaction. The phosphate buffered saline (PBS), instead of the primary antibodies, was used for negative control. Percentages of positive cells were counted within 5 high-power fields, original magnification ×200.
Table 1. Relationship between OCT4 and Survivin expression in ESCC samples.
| OCT4 | Survivin | ||
| Positive | Negative | Total | |
| Positive | 11 | 2 | 13 |
| Negative | 20 | 17 | 37 |
| Total | 31 | 19 | 50 |
R = 0.276, p = 0.052.
Statistical correlation between OCT4 or Survivin expression and ESCC clinicopathological characteristics (gender, age, cell differentiation, tumor invasion depth, lymph node metastasis) was analyzed and revealed no significant differences between the OCT4- or Survivin-positive and OCT4- or Survivin-negative cases of ESCC (Table 2).
Table 2. Correlation between OCT4 or Survivin expression and ESCC clinicopathological features.
| Variables | n | OCT4Positive | OCT4 Negative | χ2 | p | Survivin Positive | Survivin Negative | χ2 | p |
| Gender | 0.002 | 0.968 | 0.002 | 0.968 | |||||
| Male | 37 | 23 | 14 | 23 | 14 | ||||
| Female | 13 | 8 | 5 | 8 | 5 | ||||
| Age | 0.071 | 0.790 | 0.071 | 0.790 | |||||
| ≤60 y | 24 | 15 | 8 | 15 | 8 | ||||
| >60 y | 26 | 16 | 10 | 16 | 10 | ||||
| Differentiation | 3.295 | 0.192 | 3.295 | 0.192 | |||||
| Grade 1 | 14 | 6 | 8 | 6 | 8 | ||||
| Grade 2 | 31 | 21 | 10 | 21 | 10 | ||||
| Grade 3 | 5 | 4 | 1 | 4 | 1 | ||||
| p-T | 1.438 | 0.230 | 1.438 | 0.230 | |||||
| T1/2 | 16 | 8 | 8 | 8 | 8 | ||||
| T3 | 34 | 23 | 11 | 23 | 11 | ||||
| p-N | 0.002 | 0.968 | 0.002 | 0.968 | |||||
| N0 | 13 | 8 | 5 | 8 | 5 | ||||
| N1/2 | 37 | 23 | 14 | 23 | 14 |
OCT4 and Survivin Correlated to Poor Prognosis of ESCC Patients
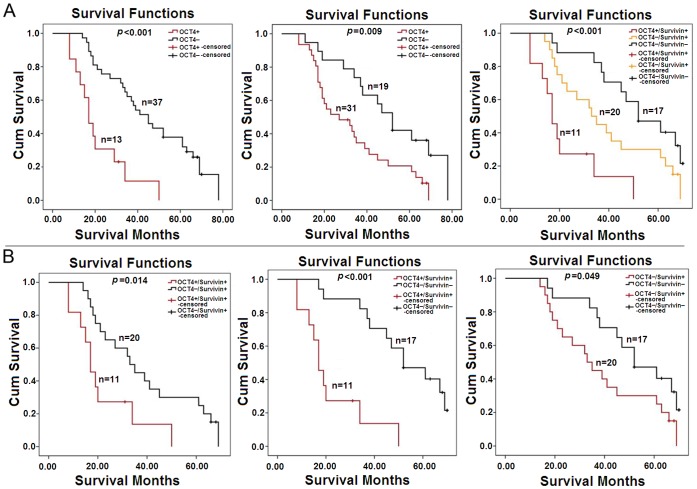
Follow-up data of 50 patients were analyzed using the Kaplan-Meier method to estimate survival curves. The median OS was 34.5 months. The median survival of ESCC patients with OCT4 positive expression was significantly less than that of patients with OCT4 negative expression (p<0.001). The similar result was showed between Survivin-positive and -negative ESCC cases (p = 0.009). Among the three subgroups (OCT4-positive/Survivin-positive, OCT4-negative/Survivin-positive, OCT4-negative/Survivin-negative), patients with OCT4-positive/Survivin-positive ESCC had a significantly poorest prognosis (p<0.001), and the longest OS was documented in OCT4-negative/Survivin-negative subgroup (Fig. 2A). Further analysis was performed between any two subgroups by log-rank test, and the results showed that OCT4 and Survivin expression were strongly associated with poor prognosis of ESCC patients (Fig. 2B).
Figure 2. Comparison of overall survival (OS) curves for 50 patients underwent the radical esophagectomy of ESCC.
(A) Fifty patients with ESCC were enrolled in the study, and the OS was defined as the time period from the operation date to the date of death or the end date of follow-up. Survival curves were calculated by the Kaplan-Meier method, and the significant difference was tested using log-rank test. (B) Every two subgroups were compared by log-rank test.
Data analysis with the univariate Cox’s proportional hazard model revealed that OCT4 and Survivin were significant prognostic factors of ESCC patients. However, the multivariate analysis showed that OCT4 was an independent prognostic value in ESCC patients, but Survivin was not (p = 0.168; Table 3).
Table 3. Univariate and Multivariate Cox’s propotional hazard models (n = 50).
| Term | Risk ratio | 95% confidence interval | p value |
| Univariate | |||
| Grade | 1.289 | 0.761–2.184 | 0.345 |
| p-T | 1.256 | 0.780–2.024 | 0.384 |
| p-N | 0.921 | 0.448–1.894 | 0.823 |
| OCT4 | 4.233 | 2.011–8.910 | 0.000 |
| Survivin | 2.352 | 1.201–4.606 | 0.013 |
| Multivariate | |||
| p- Grade | 1.313 | 0.727–2.371 | 0.367 |
| p-T | 1.225 | 0.720–2.083 | 0.454 |
| p-N | 0.988 | 0.437–2.233 | 0.977 |
| OCT4 | 3.670 | 1.598–8.431 | 0.002 |
| Survivin | 1.694 | 0.801–3.584 | 0.168 |
Inhibitory Effect of shRNA Vectors Targeting OCT4 and Survivin in ESCC Cell Lines
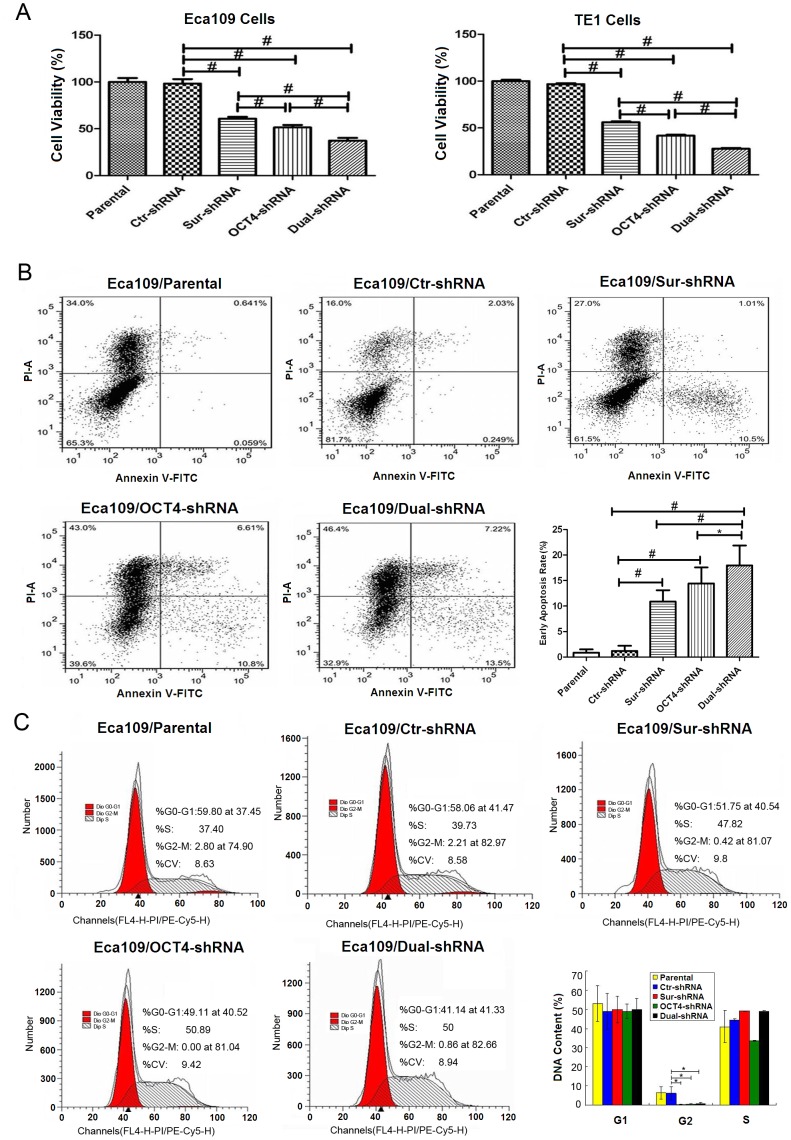
To indentify whether the specific small hairpin RNA (shRNA) targeting OCT4 (OCT4-shRNA), Survivin (Sur-shRNA), or double shRNAs (Dual-shRNA) targeting both OCT4 and Survivin, influenced esophageal cancer cell proliferation, MTT assay was performed to detect cell viability. Cell viability was obviously decreased in the Eca109 and TE1 cells transfected with OCT4-shRNA and Sur-shRNA when compared with the parental or Ctr-shRNA transfected cells. Dual-shRNA exerted an enhanced inhibitory effect on cell viability compared with the shRNA vector targeting the single factor. However, there was no difference in cell viability between OCT4-shRNA and Sur-shRNA groups (Fig. 3A).
Figure 3. Co-Suppression of OCT4 and Survivin effectively inhibited tumor cell growth.
(A) The parental and shRNA-transfected Eca109 and TE1 cells were cultured in 96-well plates at a density of 8×103 cells/well for 48 h. Cell viability was determined by methylthiazoletetrazolium (MTT) assay at a wavelength of 490 nm, # p<0.01; Dual-shRNA: vector containing double shRNAs targeting both OCT4 and Survivin. (B) After Eca109 cells were transfected with shRNA vectors, 5×106 cells/ml were harvested and stained with Annexin V-FITC and PI, and analyzed by flow cytometry. Percentages of early apoptosis were shown in histograms, data were representatives of three independent experiments, and quantitative histograms were pooled data, error bars are standard deviation (SD), *p<0.05; # p<0.01. (C) Cell cycle was measured by flow cytometry. Data of cell cycle were shown in histograms, *p<0.05.
Apoptosis of ESCC cell lines was measured by FCM with Anexin V-FITC and PI staining. After treatment with shRNA vectors for 48 h, the percentages of early cell apoptosis were increased in OCT4-shRNA (13.4±2.76%), Sur-shRNA (10.89±2.21%) and Dual-shRNA (17.79±3.89%) groups, compared with that in the parental cells (1.20±1.01%) and Ctr-shRNA (0.87±0.64%) group, especially higher in Dual-shRNA group (Fig. 3B). Compared with the parental and Ctr-shRNA groups, the percentages of G2-phase cells were significantly reduced in the OCT4-shRNA, Sur-shRNA and Dual-shRNA transfected groups, but there were no significantly differences for the G1 and S phase cells (Fig. 3C).
Survivin Expression was Associated with OCT4 in ESCC Cells
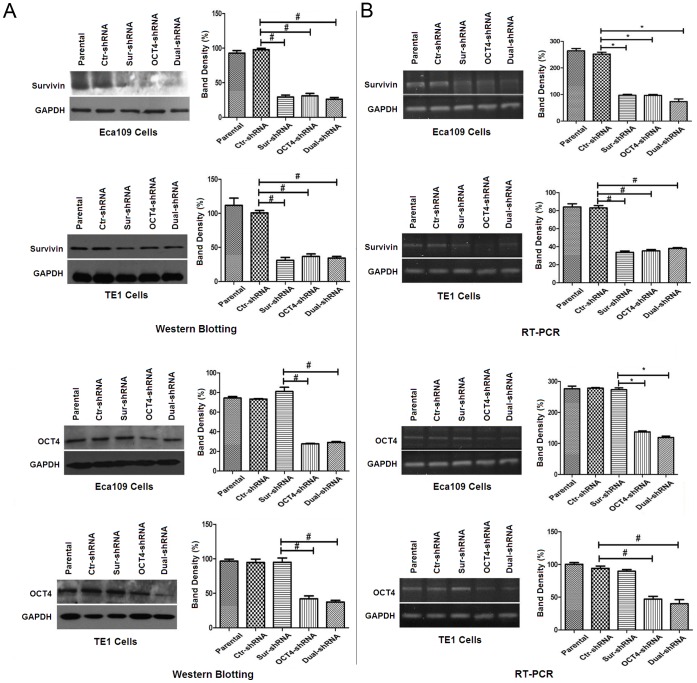
To investigate the interaction and regulation between OCT4 and Survivin, the OCT4-shRNA, Sur-shRNA and Dual-shRNA vectors were designed to manipulate the target gene expression in ESCC cell lines. By Western blot and RT-PCR analysis, OCT4 and Survivin were positively expressed in the parental Eca109 and TE1 cells. The OCT4-shRNA and Sur-shRNA vectors could inhibit the specific target gene expression, and the Dual-shRNA vector could inhibit the expression of both OCT4 and Survivin genes. The OCT4-shRNA could also down-regulate the Survivin expression in Eca109 and TE1 cells (Fig. 4A), but the Sur-shRNA did not influence the OCT4 expression (Fig. 4B).
Figure 4. Survivin expression was associated with OCT4 in Eca109 and TE1 cells.
(A) The parental and shRNA-transfected Eca109 and TE1 cells were cultured in 96-well plates at a density of 8×103 cells/well for 48 h, and expression of OCT4 and Survivin in the harvested ESCC cells was examined by Western blotting. Glyceraldehyde-3- phosphatedehydrogenase (GAPDH) was used as a loading control. The relative band intensity was analyzed by densitometry after normalized with GAPDH contents, *p<0.05; # p<0.01. (B) OCT4 and Survivin mRNA expression were tested by reverse transcription polymerase chain reaction (RT-PCR). GAPDH was used as an inner control. The relative band intensity was analyzed by densitometry after normalizing with GAPDH contents, *p<0.05; # p<0.01. All data were representatives of 3 independent experiments. Quantitative histograms were pooled data. Error bars: standard deviation (SD); Dual-shRNA: vector containing double shRNAs targeting both OCT4 and Survivin.
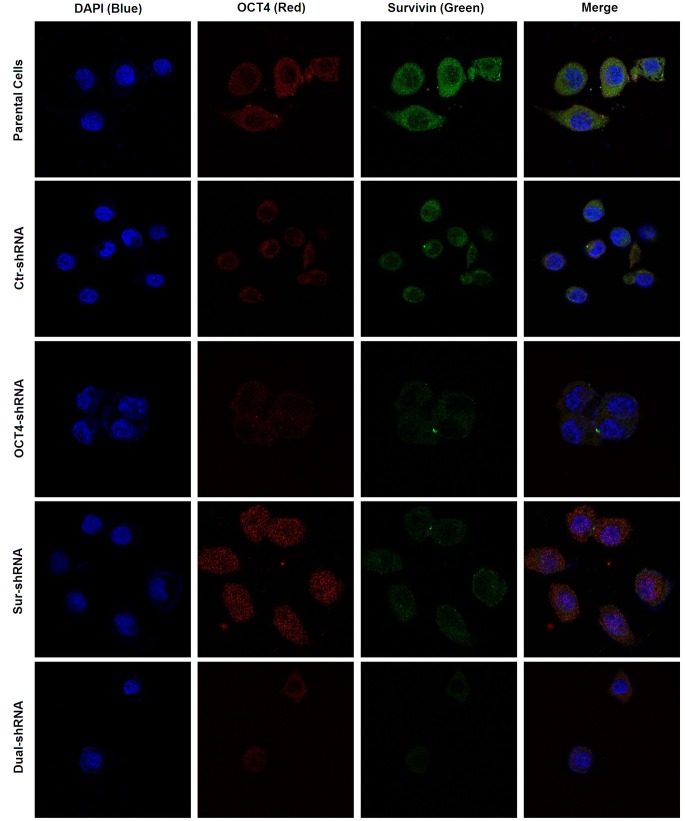
Dynamic Localization of OCT4 and Survivin Expression in ESCC Cells
We further investigated the dynamic variation of OCT4 and Survivin expression in ESCC cells by confocal assay. After 48 h transfection with OCT4-shRNA, Survivin expression was notably down-regulated in cancer cellular cytoplasm along with the decline of OCT4 expression in cellular nuclei. However, after transfection with Sur-shRNA, the OCT4 expression level was not altered along with the decrease of Survivin expression (Fig. 5).
Figure 5. Subcellular localization of OCT4 and Survivin expression in ESCC cells.
The parental and shRNA-transfected Eca109 and TE1 cells were cultured in 96-well plates at a density of 8×103 cells/well for 48 h, and the harvested cells were fixed with 4% formaldehyde, then examined OCT4 and Survivin expression by immunofluorescent confocal assay and observed under a laser-scanning confocal microscope.
Discussion
OCT4, as one of the most important transcription factors, plays a pivotal role in pluripotent stem cells [26]. OCT4, together with SOX2 and Nanog, maintains stem cell pluripotency, self-renewal and differentiation [27]. It has gradually become a promising tumor biomarker for diagnosis of germ cell tumors [28]. Recently, it is reported that OCT4 could be detected in many somatic cell cancers from esophagus, bladder, lung, and liver, and has a strong influence on patients’ prognosis. The function of OCT4 could modulate a series of signal pathways, such as the Wnt/β-catenin, TGF-β, JAK/STAT3 signal pathways [29], [30], to activate or restrain the downstream target genes. Besides, OCT4 expression in mouse embryonic stem cells is necessary for protection from apoptosis and this effect may be associated with STAT3/Survivin pathway [25].
Survivin, a recently identified member of the inhibitors of apoptosis protein (IAPs) family, regulates the essential cellular process, including suppression of apoptosis, control of cell division, and promotion of angiogenesis [31]. As one of most prominent cancer genes, Survivin is expressed in almost all tumors, but is not detected in most normal adult tissue [32]. Survivin was considered as a target gene in cancer therapy, and down-regulation of Survivin could suppress tumor growth and improve tumor cell sensitivity to radiation and chemotherapy by promoting apoptosis and inhibiting cell viability. Drugs of LY2183108 [33] and YM155 [34] targeting Survivin were put into use in clinical trials in different stages and the effect was promising. Although tumor growth speed was slowed down on the strength of silencing Survivin, tumors still possess the ability of development and expansion and deprive of patients’ life, suggesting that Survivin was not a single factor for prognosis. There remains an unknown regulatory mechanism between OCT4 and Survivin.
Over-expression of OCT4 or Survivin in ESCC has been consistently connected with disease progression, metastatic dissemination, resistance to therapy [14], [18]. Therefore, we detected both OCT4 and Survivin expression in ESCC tumor specimens, and found that OCT4 and Survivin were closely related to the surgical outcome of ESCC patients. Patients with OCT4-positive or Survivin-positive tumors presented much poorer prognosis than those with OCT4-negative or Survivin-negative tumors. Among the subgroups, patients with OCT4-positive/Survivin-positive tumors showed the shortest overall survival time. By multivariate and univariate analyses, both OCT4 and Survivin are associated with patients’ prognosis, and OCT4 is considered as an independent factor for forcasting patients’ overall survival time. From our study, we concluded that OCT4 and Survivin were jointly related to the poor prognosis of ESCC patients, but the regulatory mechanisms between OCT4 and Survivin in ESCC are not yet clear.
Inhibiting the expression of OCT4 or Survivin in ESCC cell lines with the OCT4-shRNA or Sur-shRNA vectors resulted in a reduction in G2-phase cells and an increase in cell apoptosis, and co-suppression of OCT4 and Survivin by the Dual-shRNA vector resulted in an enhanced effect. Some studies showed that the inhibition of Survivin caused cell cycle arrest in G2-M phase [35], [36], but our study found the number of G2-phase cells was decreased tremendously after suppressing Survivin expression. G1-S phase, as well as G2-M phase, was the important checkpoint in cell cycle, which controls and maintains cell process accuracy. It was reported that the over-expression of Survivin can help cancer cells to pass G2-M checkpoint [37], [38]. At the same time, Survivin possesses a capability to upregulate the expression of Cyclin D1 and ensures cancer cells to pass G1-S checkpoint fluently and obtain an ability of persistent proliferation [38], [39].
Early apoptosis was increased with the downregulation of Survivin in many reports. To further investigate the molecular mechanism of OCT4-involved cell apoptosis and cell cycle arrest in Eca109 and TE1 cells, we found that the ESCC cells expressed lower levels of OCT4 and Survivin protein after transfected with OCT4-shRNA, but Sur-shRNA only down-regulated Survivin expression and there was no change in OCT4 expression level. Therefore, we concluded that OCT4 may regulate cell apoptosis and cell division through Survivin function. OCT4 could activate multiple signaling pathways to control Survivin expression, such as the STAT3, myc and NFκB pathways [30], [40]–[42], which is critical for cancer cell survival and antiapoptosis.
In the present study, we found that the over-expression of OCT4 and Survivin are an important feature in ESCC progression, and OCT4 expression is closely correlated with Survivin expression in the regulation of cancer cell apoptosis and proliferation. However, the molecular mechanisms between OCT4 and Survivin are very complicated. We should perform work to clarify the genetic features of OCT4 and Survivin, and design more efficient antitumor therapy for ESCC.
Materials and Methods
Patients and Samples
Fifty patients with ESCC were enrolled in the study in Changhai Hospital (Shanghai, China) from Jan 1 to Dec 31, 2005. The study was approved by the Ethic Committee of Second Military Medical University. The patients consisted of 37 men and 13 women. The age ranged from 47 to 72 years old, and the median age was 62 years old. All patients provided the informed written consent. The resected lesions were diagnosed pathologically as ESCC after surgery. The follow-up for patients started from the operation date to Aug 31, 2011. The overall survival (OS) was defined as the time period from the start date of follow-up to the date of death or the end date of follow-up.
Cell Lines and Cell Culture
Human ESCC cell lines, Eca109 and TE1, were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). Cells were maintained as a monolayer in RPMI-1640 with 10% FBS (fetal bovine serum), 100 IU/ml penicillin G and 100 µg/ml streptomycin, at 37°C in a humidified 5% CO2 incubator.
Immunohistochemical Staining
The fresh tumors and adjacent normal tissues were obtained from the resected specimens during operation. The paraffin-embedded consecutive sections were subjected to immunohistochemical examination for OCT4 and Survivin expression using the primary antibodies, including mouse anti-human OCT4 (Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA) and rabbit anti-human Survivin (RD Systems Inc, Minneapolis, MN,USA), and the UltraSensitive Streptavidin Peroxidase Kit (Fuzhou Maixin Biotechnology Development Co., Fuzhou, China). The percentage of positive cells was counted within 5 high-power fields, and the evaluation of staining reaction was performed in accordance with the immunoreactive score standard proposed by Friedrichs [43].
Construction and Transfection of shRNA Vectors
The shRNA sequences for human OCT4 (OCT4-shRNA: 5′-CCCTCACTTCACTGCACTG-3′) and Survivin (Sur-shRNA: 5′-GAAAGTGCGCCGTGCCATC-3′) were synthetized and cloned, respectively, into pGenesil vector (Wuhan Genesil Biotechnology Co., Ltd., Wuhan, China), in which the encoding sequences were controlled by the U6 promoter. The Dual-shRNA vector containing double shRNAs targeting both OCT4 and Survivin and mock control shRNA vector (Ctr-shRNA, 5′-GACTTCATAAGGCGCATGC-3′) were concomitantly constructed.
ESCC cells in log phase were transfected with shRNA vectors at a concentration of 4 µg/105 cells using the Lipofectamine 2000 reagent (Invitrogen Corporation Shanghai Representative Office, Shanghai, China). The transfected cells were cultured continuously at 37°C in a CO2 incubator for another 48 h, then harvested for preparing to examine gene expression.
Western Blotting Analysis
The harvested cells were lysed in phenylmethanesulfonyl fluoride (PMSF) solution, and the isolated protein was electrophoresed on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and examined by Western blotting using the anti-Survivin and anti-OCT4 antibodies. Blots were visualled by the enhanced chemiluminescence reagent (PerkinElmer Inc., Shanghai, China).
Semiquantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR)
After 48 h of transfection, total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, California, USA) from the harvested Eca109 and TE1 cell lines, and dissolved in diethylpyrocarbonatetreated (DEPC) water. The expression of OCT4 and Survivin was examined using the Takara Reverse Transcription System Kit (Takara Biotechnology Co. Ltd., Dalian, China) by the OCT4 primers (sense: 5′-CAG TCGTCAGCGTCGTCGTTGTAAGCTGCGGCCC-3′, antisense: 5′-ACGCGTCG ACTCAGTTTGAATGCATGGGAG-3′, products 480-bp) and the Survivin primers (sense: 5′-CGGAATTCACCATGGGTGCCCCGACG-3′, antisense: 5′-GAAGATC TTCAATCCATGGCAGCCAG-3′, products 448-bp). Glyceraldehyde-3- phosphatedehydrogenase (GAPDH) was used as a loading control by the primers (sense: 5′-ACCACAGTCCATGCCATCAC-3′, antisense: 5′-TCCACCACCCTGT TGCTTGTA-3′, products 120-bp). Bands of the products electrophoresed on ethidium bromide (EB)-agarose gel were analyzed semiquantitatively by grayscale densitometry analysis.
Analysis of Cell Cycle and Apoptosis by Flow Cytometry (FCM)
After 48 h of transfection, the harvested cells washed by 0.1 M phosphate buffered saline (PBS) solution and resuspended cells with 0.1 M PBS to the concentration of 106 cells/ml. A part of cells were fixed in 1 ml pre-cooled 70% alcohol overnight at 4°C followed by incubation with propidium iodide (PI) solution at the concentration of 50 µg/ml and RNase A at the concentration of 20 µg/ml for 30 min in darkness. The cell cycle was analyzed by FCM (FACS420, BD Biosciences, San Jose, CA). Another part of cells were incubated with 5 µl Annexin V-FITC in 195 µl binding buffer in darkness for 10 min. Cells were centrifuged at 1000 g for 5 min and resuspended in 190 µl binding buffer and 10 µl PI, followed by fluorescence analysis.
Methylthiazoletetrazolium Assay
Eca109 and TE1 cells were seeded in 96-well plates at a density of 8,000 cells/well and transfected with shRNA vector as described above. After 48 h, 3-(4, 5)-dimethylthiahiazo(-z-y1)-3,5-diphenytetrazoliumromide (MTT) at 5 mg/ml was added into each well and continuously incubated for 4 h. After added 150 µl dimethyl sulfoxide (DMSO) to each well, the absorbance was measured at 490 nm.
Immunofluorescent Confocal Assay
Cancer cells were harvested and fixed with 4% formaldehyde for 15 min at room temperature, then washed in 0.1 M PBS and treated with 0.3% Triton X-100, followed by incubation with the indicated OCT4 (1∶100) and Survivin (1∶100) primary antibodies at 4°C overnight and the secondary antibodies, fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG or cyanine-3 (Cy3)-labeled goat anti-mouse IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), at room temperature for 40 min in darkness. Cells were stained by 4′,6-diamidino-2-phenylindole (DAPI) and detected under a laser-scanning confocal microscope.
Statistical Analysis
The chi-square test and Spearman’s rank correlation were used to analyze the relationship and correlation between relative gene expression and clinical pathological data. OS was assessed by the Kaplan–Meier method and the significant difference in OS was calculated by log-rank test. Univariate and multivariate analyses were performed using Cox’s proportional hazard model. The experimental data were representative of three independent experiments, and analyzed statistically by two-way analysis of variance (ANOVA) using the PASW Statistics software version 18.0 (SPSS, Chicago, Illinois, USA). Differences were considered to be significant for p<0.05.
Funding Statement
This work was supported by the National Natural Scientific Foundation of China (30801106, 81172308, 30973469). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. (2008) Cancer statistics, 2008. CA Cancer J Clin 58: 71–96. [DOI] [PubMed] [Google Scholar]
- 2. Collard JM, Otte JB, Fiasse R, Laterre PF, De Kock M, et al. (2001) Skeletonizing en bloc esophagectomy for cancer. Ann Surg 234: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao JS, Li WJ, Ge D, Zhang PJ, Li JJ, et al. (2011) Tumor initiating cells in esophageal squamous cell carcinomas express high levels of CD44. PLoS One 6: e21419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, et al. (2006) Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology 44: 240–251. [DOI] [PubMed] [Google Scholar]
- 5. Wang Q, He W, Lu C, Wang Z, Wang J, et al. (2009) Oct3/4 and Sox2 are significantly associated with an unfavorable clinical outcome in human esophageal squamous cell carcinoma. Anticancer Res 29: 1233–1241. [PubMed] [Google Scholar]
- 6. Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, et al. (2007) Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells. Stem Cells 25: 500–510. [DOI] [PubMed] [Google Scholar]
- 7. Pan GJ, Chang ZY, Scholer HR, Pei D (2002) Stem cell pluripotency and transcription factor Oct4. Cell Res 12: 321–329. [DOI] [PubMed] [Google Scholar]
- 8. Ezeh UI, Turek PJ, Reijo RA, Clark AT (2005) Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer 104: 2255–2265. [DOI] [PubMed] [Google Scholar]
- 9. Gu G, Yuan J, Wills M, Kasper S (2007) Prostate cancer cells with stem cell characteristics reconstitute the original human tumor in vivo. Cancer Res 67: 4807–4815. [DOI] [PubMed] [Google Scholar]
- 10. Chen YC, Hsu HS, Chen YW, Tsai TH, How CK, et al. (2008) Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One 3: e2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Atlasi Y, Mowla SJ, Ziaee SA, Bahrami AR (2007) OCT-4, an embryonic stem cell marker, is highly expressed in bladder cancer. Int J Cancer 120: 1598–1602. [DOI] [PubMed] [Google Scholar]
- 12. Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, et al. (2008) Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res 14: 4085–4095. [DOI] [PubMed] [Google Scholar]
- 13. Chen Z, Xu WR, Qian H, Zhu W, Bu XF, et al. (2009) Oct4, a novel marker for human gastric cancer. J Surg Oncol 99: 414–419. [DOI] [PubMed] [Google Scholar]
- 14. Zhou X, Huang GR, Hu P (2011) Over-expression of Oct4 in human esophageal squamous cell carcinoma. Mol Cells 32: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duffy MJ, O’Donovan N, Brennan DJ, Gallagher WM, Ryan BM (2007) Survivin: a promising tumor biomarker. Cancer Lett 249: 49–60. [DOI] [PubMed] [Google Scholar]
- 16. Andersen MH, Svane IM, Becker JC, Straten PT (2007) The universal character of the tumor-associated antigen survivin. Clin Cancer Res 13: 5991–5994. [DOI] [PubMed] [Google Scholar]
- 17. Mega S, Miyamoto M, Li L, Kadoya M, Takahashi R, et al. (2006) Immunohistochemical analysis of nuclear survivin expression in esophageal squamous cell carcinoma. Dis Esophagus 19: 355–359. [DOI] [PubMed] [Google Scholar]
- 18. Cao M, Yie SM, Wu SM, Chen S, Lou B, et al. (2009) Detection of survivin-expressing circulating cancer cells in the peripheral blood of patients with esophageal squamous cell carcinoma and its clinical significance. Clin Exp Metastasis 26: 751–758. [DOI] [PubMed] [Google Scholar]
- 19. Hoffmann AC, Vallbohmer D, Grimminger P, Metzger R, Prenzel KL, et al. (2010) Preoperative survivin mRNA detection in peripheral blood is an independent predictor of outcome in esophageal carcinoma. Pharmacogenomics 11: 341–347. [DOI] [PubMed] [Google Scholar]
- 20. Kato J, Kuwabara Y, Mitani M, Shinoda N, Sato A, et al. (2001) Expression of survivin in esophageal cancer: correlation with the prognosis and response to chemotherapy. Int J Cancer 95: 92–95. [DOI] [PubMed] [Google Scholar]
- 21. Trabulo S, Cardoso AM, Santos-Ferreira T, Cardoso AL, Simoes S, et al. (2011) Survivin silencing as a promising strategy to enhance the sensitivity of cancer cells to chemotherapeutic agents. Mol Pharm 8: 1120–1131. [DOI] [PubMed] [Google Scholar]
- 22. Okamoto K, Okamoto I, Hatashita E, Kuwata K, Yamaguchi H, et al. (2012) Overcoming erlotinib resistance in EGFR mutation-positive non-small cell lung cancer cells by targeting survivin. Mol Cancer Ther 11: 204–213. [DOI] [PubMed] [Google Scholar]
- 23.Prud’homme GJ (2012) Cancer Stem Cells and Novel Targets for Antitumor Strategies. Curr Pharm Des. [DOI] [PubMed]
- 24. Kim RJ, Nam JS (2011) OCT4 Expression Enhances Features of Cancer Stem Cells in a Mouse Model of Breast Cancer. Lab Anim Res 27: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo Y, Mantel C, Hromas RA, Broxmeyer HE (2008) Oct-4 is critical for survival/antiapoptosis of murine embryonic stem cells subjected to stress: effects associated with Stat3/survivin. Stem Cells 26: 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gil J, Stembalska A, Pesz KA, Sasiadek MM (2008) Cancer stem cells: the theory and perspectives in cancer therapy. J Appl Genet 49: 193–199. [DOI] [PubMed] [Google Scholar]
- 27. Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, et al. (2009) Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev 18: 1093–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng L, Sung MT, Cossu-Rocca P, Jones TD, MacLennan GT, et al. (2007) OCT4: biological functions and clinical applications as a marker of germ cell neoplasia. J Pathol 211: 1–9. [DOI] [PubMed] [Google Scholar]
- 29. Yuan F, Zhou W, Zou C, Zhang Z, Hu H, et al. (2010) Expression of Oct4 in HCC and modulation to wnt/beta-catenin and TGF-beta signal pathways. Mol Cell Biochem 343: 155–162. [DOI] [PubMed] [Google Scholar]
- 30. Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Margulis V, Lotan Y, Shariat SF (2008) Survivin: a promising biomarker for detection and prognosis of bladder cancer. World J Urol 26: 59–65. [DOI] [PubMed] [Google Scholar]
- 32. Guha M, Altieri DC (2009) Survivin as a global target of intrinsic tumor suppression networks. Cell Cycle 8: 2708–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Talbot DC, Ranson M, Davies J, Lahn M, Callies S, et al. (2010) Tumor survivin is downregulated by the antisense oligonucleotide LY2181308: a proof-of-concept, first-in-human dose study. Clin Cancer Res 16: 6150–6158. [DOI] [PubMed] [Google Scholar]
- 34. Giaccone G, Zatloukal P, Roubec J, Floor K, Musil J, et al. (2009) Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non-small-cell lung cancer. J Clin Oncol 27: 4481–4486. [DOI] [PubMed] [Google Scholar]
- 35. Zhonghong L, Lianjie L, Changqing Z, Ying H, Yu J, et al. (2008) The influence of survivin shRNA on the cell cycle and the invasion of SW480 cells of colorectal carcinoma. J Exp Clin Cancer Res 27: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu JL, Wang Y, Jiang J, Kong R, Yang YM, et al. (2010) Inhibition of survivin expression and mechanisms of reversing drug-resistance of human lung adenocarcinoma cells by siRNA. Chin Med J (Engl) 123: 2901–2907. [PubMed] [Google Scholar]
- 37. Pennati M, Binda M, Colella G, Zoppe M, Folini M, et al. (2004) Ribozyme-mediated inhibition of survivin expression increases spontaneous and drug-induced apoptosis and decreases the tumorigenic potential of human prostate cancer cells. Oncogene 23: 386–394. [DOI] [PubMed] [Google Scholar]
- 38. Pietenpol JA, Stewart ZA (2002) Cell cycle checkpoint signaling: cell cycle arrest versus apoptosis. Toxicology 181–182: 475–481. [DOI] [PubMed] [Google Scholar]
- 39. Zhao X, Guo J, Yu Y, Yi S, Yu T, et al. (2011) Overexpression of survivin and cyclin D1 in CHO cells confers apoptosis resistance and enhances growth in serum-free suspension culture. Biotechnol Lett 33: 1293–1300. [DOI] [PubMed] [Google Scholar]
- 40. Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, et al. (2007) Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A 104: 7391–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kawakami H, Tomita M, Matsuda T, Ohta T, Tanaka Y, et al. (2005) Transcriptional activation of survivin through the NF-kappaB pathway by human T-cell leukemia virus type I tax. Int J Cancer 115: 967–974. [DOI] [PubMed] [Google Scholar]
- 42. Cosgrave N, Hill AD, Young LS (2006) Growth factor-dependent regulation of survivin by c-myc in human breast cancer. J Mol Endocrinol 37: 377–390. [DOI] [PubMed] [Google Scholar]
- 43. Friedrichs K, Gluba S, Eidtmann H, Jonat W (1993) Overexpression of p53 and prognosis in breast cancer. Cancer 72: 3641–3647. [DOI] [PubMed] [Google Scholar]