Abstract
PR1 is a human leukocyte antigen (HLA)-A2 restricted peptide that has been targeted successfully in myeloid leukemia with immunotherapy. PR1 is derived from the neutrophil granule proteases proteinase 3 (P3) and neutrophil elastase (NE), which are both found in the tumor microenvironment. We recently showed that P3 and NE are taken up and cross-presented by normal and leukemia-derived antigen presenting cells, and that NE is taken up by breast cancer cells. We now extend our findings to show that P3 and NE are taken up and cross-presented by human solid tumors. We further show that PR1 cross-presentation renders human breast cancer and melanoma cells susceptible to killing by PR1-specific cytotoxic T lymphocytes (PR1-CTL) and the anti-PR1/HLA-A2 antibody 8F4. We also show PR1-CTL in peripheral blood from patients with breast cancer and melanoma. Together, our data identify cross-presentation as a novel mechanism through which cells that lack endogenous expression of an antigen become susceptible to therapies that target cross-presented antigens and suggest PR1 as a broadly expressed tumor antigen.
Introduction
Proteinase 3 (P3) and neutrophil elastase (NE) are proteases normally stored in neutrophil primary azurophil granules. They play a role in infection, leukemogenesis and autoimmune disease (e.g. Wegener’s granulomatosis) (1–4). We have identified the human leukocyte antigen (HLA)-A2 restricted nonameric peptide PR1 that is derived from P3 and NE (5, 6). PR1 has shown efficacy in the therapy of myeloid leukemia (7, 8). In addition, PR1-specific cytotoxic T-lymphocytes (CTL) were detected in peripheral blood (PB) from patients with myeloid leukemia and were correlated with positive clinical outcomes following allogeneic stem cell transplantation (allo-SCT) and interferon (IFN)-α2b therapy (7–9).
We have shown that P3 and NE are cross-presented by normal donor antigen presenting cells (APC) and leukemia, and that cross-presentation by leukemia renders cells susceptible to killing by PR1 targeting therapy (10). Cross-presentation is a mechanism by which APC take up exogenous antigen and present them on HLA class I (HLA A, B, C), thereby priming a CTL immune response. Furthermore, cross-presentation is thought to be the primary mechanism through which tumor antigens are presented to the immune system, and is believed to be restricted to subpopulations of APCs (11, 12). A recent report by Francois et al., however, showed that mesenchymal stromal cells cross-present soluble exogenous antigens (i.e. ovalbumin) in a mouse model (13). P3 and NE are both endogenously expressed in myeloid hematopoietic cells and therefore provide a source for PR1 in myeloid malignancies. Since NE was shown to be taken up by lung cancer (14) and as we have shown that breast cancer cells take up NE (15), we hypothesized that NE and P3 uptake by solid tumors may lead to PR1 cross-presentation, thereby rendering non-myeloid malignancies susceptible to killing by PR1-targeting therapy.
We first show NE and P3 uptake by a number of solid tumors. Since breast cancer was shown to contain an inflammatory component that may be the source for NE and P3 (16, 17), is susceptible to immunotherapy (18), and is the most common malignancy in women, we investigated cross-presentation of NE and P3 in breast cancer. We show that P3, like NE (15) is absent in breast cancer cell lines and primary breast cancer tumors, and is taken up by breast cancer cells. We then demonstrate that NE and P3 uptake leads to PR1 cross-presentation. PR1 cross-presentation increases susceptibility of breast cancer cells to killing by PR1 targeting therapies, including PR1-CTLs and anti-PR1/HLA-A2 (8F4) antibody. We demonstrate PR1 on the surface of primary breast tumors in association with HLA-A2 (PR1/HLA-A2) and detect PR1-CTLs in peripheral blood (PB) from breast cancer patients. We then extend our findings to melanoma, which like breast cancer also contains an inflammatory component (19) and has demonstrated susceptibility to immunotherapy (20, 21). We show NE and P3 uptake and cross-presentation by melanoma and susceptibility of melanoma to killing by PR1-CTLs following NE and P3 cross-presentation. Together, our data demonstrate the ability of solid tumors to cross-present antigen and suggest PR1 as a broadly expressed tumor antigen.
Materials and Methods
Patient tissues, cells and cell culture
Patient breast cancer frozen tissue blocks were purchased from Origene. Patient and healthy donor (HD) samples were collected after informed consent was obtained to participate in a study approved by the institutional review board at MD Anderson Cancer Center. MDA-MB-231, MCF-7, MDA-MB-453 and T47D breast cancer cell-lines, SW-620 (colorectal adenocarcinoma), OVCAR-3 (ovarian adenocarcinoma), MIA PaCa-2 (pancreatic carcinoma), Jurkat (acute T cell leukemia), T2 (B-cell/T-cell hybridoma), HL-60 (acute promyelocytic leukemia) and U-937 (histiocytic leukemia) cell lines were obtained from American Type Culture Collection. MCF-HER-18 cell line was provided by Dr. Mien-Chie Hung (MD Anderson Cancer Center). Mel 526, Mel 624, MT 2019 and MT 2333 melanoma cell lines were provided by Dr. Laszlo Radvanyi. Cell lines were authenticated by DNA finger-printing at MD Anderson Cancer Center within six months of use in experiments.
Breast cancer cells were grown in Dulbecco’s modified Eagle’s media (DMEM) with 2.5mM L-Glutamine (Hyclone) supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products), 100 U/mL Penicillin/100 μg/mL Streptomycin (Cellgro). G418 (Lonza) (0.5 mg/mL) was added to the MCF-7-HER18 cell cultures as a selective agent. RPMI-1640 with 25mM HEPES+L-Glutamine (Hyclone) was used in place of DMEM for leukemia cell line cultures. All cell lines were cultured in 5% CO2 at 37 °C. Healthy donor and patient peripheral blood mononuclear cells (PBMC) and polymorphonuclear neutrophils (PMN) were enriched using standard Histopaque 1077 and 1119 (Sigma) gradient centrifugation, respectively.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
mRNA was extracted from cell lines and laser capture microdissection (LCM) samples using RNA Stat 60 kit (TelTest). Synthesis of cDNA was performed using the Gene Amp RNA kit (Perkin Elmer). The following primers were used: P3 forward primer 5′-GACCCCACCATGGCTCAC-3′ and reverse primer 5′-ATGGGAAGGACAGACAGGAG-3′; mammaglobin (MGB)-1 forward primer 5′-AGCACTGCTACGCAGGCTCT-3 ′ and reverse primer 5 ′-ATAAGAAAGAGAAGGTGTGG-3′ ; actin forward 5′-CCAGAGCAAGAGAGCTATCC-3′ and reverse 5′-CTGTGGTGGTGAAGCTGTAG-3′; and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward 5′-TAGACGGGAAGCTCACTGGC-3′ and reverse 5′-AGGTCCACCACCCTGTTGCT-3′. Following denaturation for 5 minutes at 95 C, samples were amplified for 35 cycles using an iCycler (Bio-Rad). Samples were run on 1.5% agarose gel. Bands were imaged using GelDoc2000 (BioRad) and analyzed by Quantity One software (BioRad).
Western blotting
Whole cell lysates (WCL) were generated by suspending cell pellets in lysis buffer (10 mM/L HEPES [pH 7.9], 10 mM/L KCl, 0.1 mM/L EGTA, 0.1 mM/L EDTA, and 1 mM/LDTT) containing protease inhibitors and subsequent freeze-thaw cycles for 15 min. WCL were separated by electrophoresis on 10% SDS gels under reducing conditions, transferred onto PVDF membranes, blocked with 5% milk and stained with anti-NE (Santa Cruz), anti-P3 (NeoMarkers), anti-tubulin (Sigma) or anti-GAPDH (Sigma) antibodies. Chemiluminescence was captured on Kodak film.
Antigen cross-presentation
To determine protein uptake, cells were pulsed in reduced serum medium (0.5% FBS) containing 10 μg/mL P3, NE (both from Athens Research & Technology), EndoGrade ovalbumin (Ova) (Hyglos) or irradiated (7500 cGy) polymorphonuclear (PMN) or peripheral blood mononuclear cells (PBMC) at a ratio of 1:1 (breast cancer:irradiated-cell). Cells were then permeabilized (BD Biosciences) and stained with alexa-488 or 647 directly conjugated anti-P3 (Clone MCPR3-2; Thermo Scientific) or anti-NE (Santa Cruz) and analyzed by flow cytometry. To determine cross-presentation, cells were surface-stained with fluorescently-conjugated 8F4 as previously described (22). Alexa-488 or 647 kits (Invitrogen) were used to directly conjugate anti-P3, anti-NE and anti-PR1/HLA-A2 (8F4) antibodies. Aqua live/dead stain (Invitrogen) was used to assess viability. For all flow cytometry experiments, light scatter was used to establish the initial gating followed by aqua live dead stain. To inhibit cross-presentation, cells were co-incubated with the endoplasmic reticulum (ER) to Golgi antegrade inhibitor brefeldin A (BFA) (Sigma) or the proteasome inhibitor lactacystin (Sigma) (13, 23, 24).
Confocal imaging to show intracellular P3 localization was performed using Leica Microsystems SP2 SE confocal microscope (Leica) with 10×/25 air, 63×/1.4 oil objectives and analyzed using Leica LCS software (version 2.61). FITC-conjugated lysosome-associated membrane protein-2 (LAMP-2; eBioscience) was used to stain for lysosomes and late endosomes (25). Flow cytometry was performed using the Cytomation CyAn flow cytometer (Dako). Data were analyzed using FlowJo software (Tree Star Inc).
Immunohistochemistry
Cryopreserved breast and melanoma tumor tissues (Origene) were formalin fixed and then paraffin-embedded for immunohistochemistry. Prior to staining, tissue sections were de-paraffinized in xylene, re-hydrated and quenched for endogenous peroxidase activity. Sections were blocked with 10% normal horse serum and then incubated with primary WGM2 anti-P3 mAb clone (1:10) (Abcam) or anti-NE (Santa Cruz) for 30 minutes at room temperature. Melanoma slides were co-stained with anti-Microphthalmia-associated transcription factor (MITF) antibody (Thermoscientific). Slides were then washed and incubated with secondary anti-mouse IgG-biotin antibody (1:200) (Vector laboratories) followed by avidin-biotin peroxidase (1:100) (Vector laboratories). Chromagen 3, 3′-diaminobenzidine (Dako) was used for staining visualization. All slides were counterstained with hematoxylin. PMN staining of normal tonsil tissue was used as a positive control. Negative controls were stained as above after deletion of primary antibodies.
Peptide-specific CTL lines
PR1-specific CTLs were expanded by stimulating PBMCs from healthy HLA-A2 donors with PR1-peptide in vitro, as previously described (6, 26). Briefly, T2 cells were washed in serum-free RPMI 1640 medium and incubated with PR1 peptide at 20 μg/mL for 90 minutes at 37°C. Peptide-loaded T2 cells were irradiated with 7500 cGy, washed, and cultured with freshly isolated PBMCs at a 1:1 ratio in RPMI 1640 medium supplemented with 10% human AB serum. Cultures were re-stimulated with peptide-pulsed T2 cells on days 7, 14, and 21, and the following day 20 IU/mL of recombinant human interleukin-2 (rhIL-2; Biosource International) was added.
Cell-mediated cytotoxicity assay
A standard cytotoxicity assay was used to determine specific lysis as described previously (5, 27). Briefly, 1000 target cells in 10 μl (1.0 × 105 cells/ml) were stained with calcein-AM (Invitrogen) for 90 minutes at 37 °C, washed 3 times with RPMI-1640 and then co-incubated with 10 μl of peptide-specific CTLs at varying effector to target (E:T) ratios. After a 4-hour incubation period at 37 C in 5% CO2, 5 μl of Trypan blue was added to each well and fluorescence was measured using an automated CytoFluor II plate reader (PerSeptive Biosystems). Percent specific cytotoxicity was calculated as follows: (1−(Fluorescence Target+Effector − Fluorescencemedia)/(FluorescenceTarget alone − Fluorescence media)) × 100.
Complement-mediated cytotoxicity assay
To determine whether cross-presentation increases breast cancer susceptibility to 8F4, we performed complement mediated cytotoxicity assay, as previously described (22, 28). MDA-MB-231 cells were cultured in NE (10 μg/ml) or P3 (10 μg/ml) containing media for 24 hours. Cells were incubated with calcein AM (Invitrogen), washed 3 times and resuspended in serum-free RPMI. One million cells were mixed with increasing doses of 8F4 antibody (0.624, 1.25, 2.5, 5, and 10 μg/mL) or isotype antibody (negative control) at a final concentration of 10 μg/ml and incubated for 10 minutes at 37 °C. Standard rabbit complement (C′) (5 μL) (Cedarlane Labs) was then added and cells were incubated for 60 minutes at 37 °C. Supernatant from BB7.2 hybridoma (source for anti-HLA-A2) and digitonin (Promega) were used as positive controls. Fluorescence was measured and specific killing was calculated as described above.
Laser-capture microdissection (LCM) and RNA extraction from breast tumor tissue
Laser capture microdissection (LCM) was preformed to isolate breast cancer cells from breast tumor biopsy tissue with an Arcturus PixCell laser capture microscope with an IR diode laser (Life Technologies, Applied Biosystems). Tissue was sectioned (5 μm thickness), placed on non-charged glass slides and fixed in 75% ethanol and DEPC water. Hematoxylin was used to stain nuclei after tissue hydration. Samples were stored in xylene after graded alcohol dehydration until ready for LCM. The areas used for microdissection were identified using hematoxylin and eosin staining. Tissue was pulsed with a laser beam with power adjusted between 30 – 70 mW to maintain a 10 μm diameter. Approximately 5,000 breast cancer cells were captured in Arcturus Capsure HS LCM caps (Life Technologies, Applied Biosystems). Total RNA was extracted and purified using the Arcturus PicoPure RNA Isolation Kit (Life Technologies, Applied Biosystems). RNA integrity and quantity were determined with a Nano Drop ND-1000 Spectrophotometer (Thermo Scientific). Arcturus RiboAmp RNA Amplification Kit was used to amplify RNA using two rounds of T7-based amplification. This yielded 2.5 μg of amplified RNA (aRNA). cDNA was synthesized from 1 μg of aRNA using the Roche Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science), per manufacturer instructions.
Staining for PR1-CTLs in breast cancer patients
PBMC from patients were stained with the following fluorescent antibodies: CD8 APC-H7(BD), CD3 PE Cy7 (BD), CD4 pacific orange (Invitrogen), PE-conjugated PR1/HLA-A2-dextramer (Immudex) and the following pacific blue conjugated lineage antibodies: CD14 (BD), CD16 (BD) and CD19 (Biolegend). Aqua live dead stain (Invitrogen) was used to exclude dead cells. Samples were fixed with 4% PFA. Data were acquired on Canto flow cytometer (BD) and analyzed using FlowJo software (Tree Star Inc). The frequency of PR1-CTLs was determined as the percentage of live cells that were lineage, CD4−, CD3+, CD8+ and PR1-dextramer+.
Confocal imaging of patient tissues
Cryopreserved tissue sections were fixed with cold acetone. Breast cancer tissues were stained with the breast cancer marker Alexa-488 conjugated mouse anti-Cytokeratin-7 (CK7) antibody (Abcam) and Alexa-647 conjugated 8F4 antibody (22). To confirm that the PR1/HLA-A2 expression is by breast cancer cells and not by the infiltrating leukocytes, consecutive breast cancer tissue sections were also stained with Alexa-647 conjugated mouse anti-CD45 antibody (Invitrogen) as a leukocyte marker. Human tonsil tissue sections (Origene) were used as positive staining control for CD45. For melanoma, tissue sections were fixed with cold acetone, permeabilized with 0.5% triton x-100 (Sigma) for 15 minutes, and blocked with 5% normal goat serum (Jackson Immunoresearch). Sections were then incubated with the melanoma marker mouse anti-MITF (Thermoscientific) for 1 hour, washed with PBS and then incubated with Alexa-488 conjugated goat-anti mouse IgG (Jackson Immunoresearch). Slides were then washed, blocked with 5% normal mouse serum (Jackson Immunoresearch) and incubated with Alexa-647 conjugated 8F4 antibody. ProLong Gold antifade reagent with dapi (Invitrogen) was added. Confocal imaging was performed using Leica Microsystems SP2 SE confocal microscope (Illinois, USA) with 10×/25 air, 63×/1.4 oil objectives. Leica LCS software (version 2.61) was used for image analysis.
Results
Solid tumors take up NE and P3
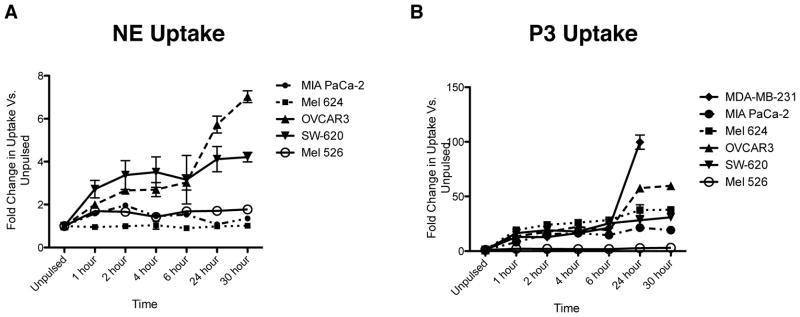
To determine whether uptake of NE and P3 is a ubiquitous phenomenon, we co-cultured multiple solid tumor cell lines with 10 μg/mL of NE or P3 and then used flow cytometry to assess for intracellular uptake. We show that not all tumor types take up NE and P3, and furthermore the degree of uptake varies among different tumor types (Fig. 1A and B). In addition, NE uptake appears to plateau over time and is much lower than P3 uptake, indicating different uptake mechanisms and suggesting a receptor-mediated process that may be involved in NE uptake.
Figure 1.
Solid tumor cell lines take up NE and P3. Cell lines representing solid tumors were incubated with (A) NE (10 μg/mL) or (B) P3 (10 μg/mL) and then permeabilized and stained with anti-NE or anti-P3 antibodies. Data represent mean ± SEM fold increase in NE or P3 uptake vs. unpulsed cells from triplicate wells from 2 independent experiments. MDA-MB-231, breast carcinoma; MIA PaCa-2, pancreatic carcinoma; Mel 624 and Mel 526, melanoma; OVCAR3, ovarian adenocarcinoma; SW-620, colon adenocarcinoma.
P3 is absent in breast cancer
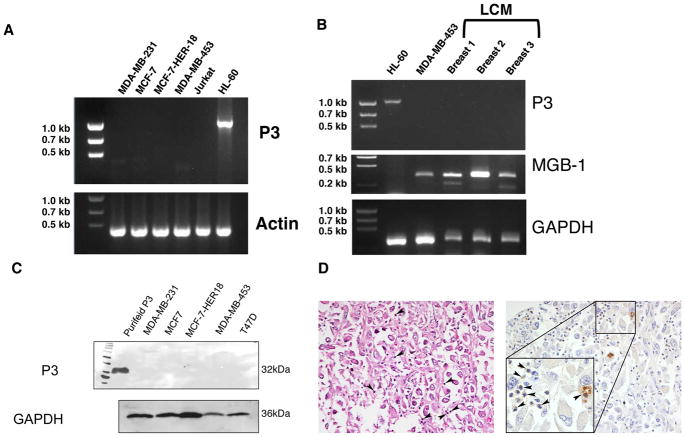
Since we have previously shown that NE is absent in breast cancer and is taken up by breast cancer cells (15) and to differentiate P3 uptake from endogenous expression, we analyzed breast cancer cell lines and primary tumor tissues for P3 expression at the mRNA and protein levels. PCR shows that the breast cancer cell lines MDA-MB-231, MCF-7, MCF-7-HER18 (HER18), and MDA-MB-453 all lack P3 mRNA (Fig. 2A). Similarly, breast cancer cells extracted from 3 different breast tumors (Fig. 2B and Table I) also lack P3 mRNA. Immunoblots of whole cell lysates (WCL) from cell lines confirmed the absence of P3 protein in breast cancer cells (Fig. 2C). Immunohistochemistry staining of primary breast cancer detected P3 in breast cancer tissue, but the P3 was limited to the inflammatory component within the breast tumor, and not in the breast tumor cells (Fig. 2D). These data are consistent with previous reports showing P3 in breast cancer (29), although our data suggest that the source of P3 is inflammatory cells within the tumor, and not the breast cancer cells.
Figure 2.
Breast cancer does not endogenously express P3. mRNA was extracted from (A) breast cancer cell lines and (B) primary breast cancer tissue. RT-PCR was performed using P3 primers, which shows lack of P3 mRNA expression in breast cancer cell lines and primary breast cancer. Jurkat and HL-60 leukemia cell lines were used as negative and positive controls, respectively. Primary breast cancer cells from patient tissues, samples Breast 1–3, were obtained by laser capture microdissection (LCM) performed on tumor obtained from patients at the time of surgical resection. Mammaglobin (MGB)-1 was used to confirm analysis of breast cancer cells. β-actin and GAPDH were used as loading controls. C, Immunoblots demonstrate lack of P3 protein in whole cell lysates from 5 different breast cancer cell lines. Gels were loaded with 20 μg of protein. Purified P3 (5 μg) was used as positive control and GAPDH was used as a loading control. D, Immunohistochemistry showing absence of P3 in patient breast cancer tissue (Breast 3). Left panel is an H&E section (200X) showing poorly differentiated carcinoma with admixed neutrophils. Right panel shows positive staining of P3 in the admixed neutrophils but not in the breast cancer cells. The inset (400X) shows a rare tumor cell engulfing a neutrophil. Both images are taken from the same patient and are representative of 5 tissues. Arrowheads indicate neutrophils.
Table I.
Pathologic characteristics of breast and melanoma tumor tissues used for laser capture microdissection and confocal microscopy.
| Patient | Histology | ER/PR/HER2 Status | TNM Staging | HLA-A2 Status |
|---|---|---|---|---|
| Breast #1 | IDC | ER−/PR−/HER2− | T1cNXMX | Positive |
| Breast #2 | IDC | ER−/PR−/HER2+ | T2N1bMX | N/D |
| Breast #3 | IDC/ILC | ER+/HER2+ | T2N1aMX | Negative |
| Breast #4 | IDC | ER−/HER2− | T3N0MX | Positive |
| Melanoma #1 | Nodular | N/A | T3N2M1c | Positive |
| Melanoma #2 | N/D | N/A | T3N2M1b | Negative |
Abbreviations: IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; ER, estrogen receptor; PR, progesterone receptor; HER2, HER2/neu; N/D, not determined.
P3 is taken up by breast cancer cells
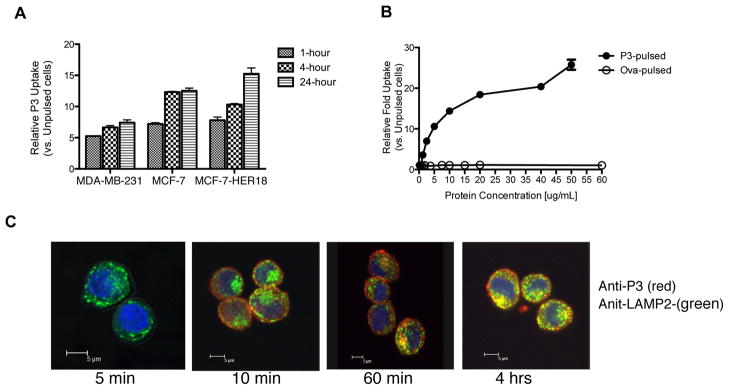
Since we showed that P3 is not expressed endogenously by breast cancer cells, we hypothesized that P3 may be taken up by breast cancer cells, as we have previously shown for NE (15). The HLA-A2 positive cell lines MDA-MB-231, MCF-7 and HER18 were co-cultured with 10 μg of P3 at 1, 4 and 24 hours and then analyzed using flow cytometry for intracellular uptake of P3 (Fig. 3A). We detected a time-dependent increase in P3 uptake in all three cell lines. We also demonstrated a dose dependent uptake of P3 that does not appear to plateau, suggesting a non-receptor mediated process for P3 uptake (Fig. 3B). To further characterize P3 uptake as it relates to antigen cross-presentation, which occurs in distinct cellular compartments (30), we performed laser confocal microscopy and showed that following uptake, P3 localizes within lysosomes, as shown by P3 co-staining with lysosomal-associated membrane protein 2 (LAMP-2) (Fig. 3C). Uptake into lysosomal compartments occurred at early time points (1–4 hours) and may be the initial step in antigen degradation as it is being processed for cross-presentation on HLA class-I molecules (31).
Figure 3.
P3 is taken up by breast cancer cell lines and localizes to lysosomal compartments. A, MDA-MB-231, MCF-7, and MCF-7-HER18 cell lines were incubated with soluble P3 (10 μg/mL) for 1, 4, and 24 hours and then intracellularly stained with anti-P3 antibody. Median fluorescence intensity (MFI) was measured for triplicate experimental groups and was normalized to the MFI of unpulsed cells. Fold increase in MFI vs. unpulsed cells is plotted on the Y-axis. Data are means ± SEM and represent 2 independent experiments. B, MDA-MB-231 cells were incubated with increasing doses of soluble P3 or ovalbumin (ova) and analyzed by flow cytometry for intracellular uptake of P3 or ova using anti-P3 or anti-ova antibodies, respectively. Data are means ± SEM from duplicate experiments. C, MDA-MB-231 cells were cultured with soluble P3 (10 μg/mL) and then stained intracellularly for P3 (red) and LAMP-2 (green). Confocal microscopy images demonstrate localization of P3 in lysosomal compartments 4 hours following uptake as shown by overlay images (yellow). Nuclei appear blue using DAPI.
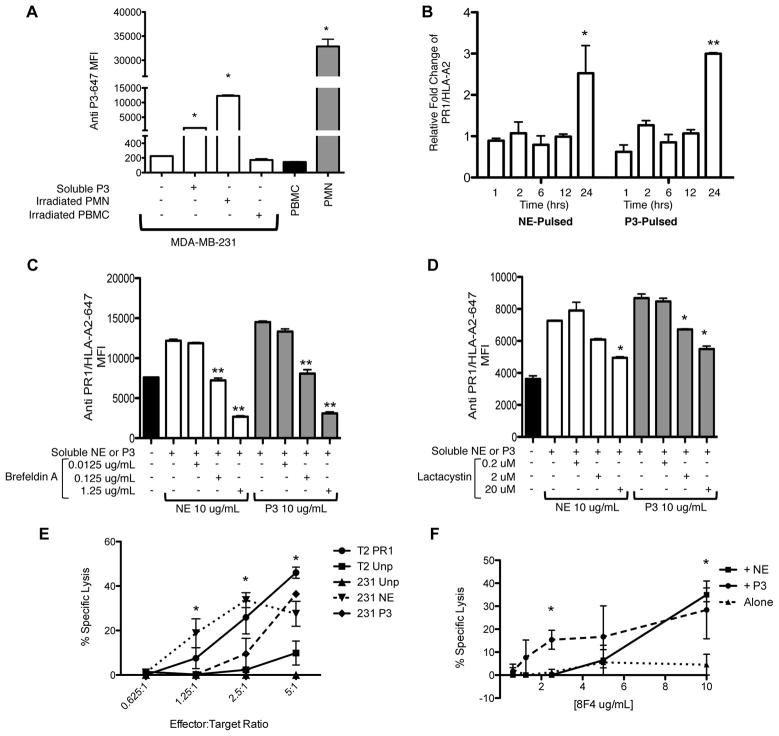
Because different cellular pathways are involved in uptake and processing of soluble and cell-associated proteins, which can determine whether or not they are cross-presented (32), and since neutrophils were reported in tumor tissues including breast cancer (16, 17), we evaluated if there was difference in the uptake of soluble and cell-associated P3 by breast cancer cells. To examine this, MDA-MB-231 cells were co-cultured for 4 hours with soluble P3 (10 μg/mL) or with irradiated polymorphonuclear (PMN) or peripheral blood mononuclear (PBMC) cells at a 1:1 ratio (Fig. 4A and Supplemental Fig. 1). Data demonstrated that breast cancer cells can take up both soluble P3 as well as cell-associated P3. In fact, uptake from cell-associated P3 appears to be more efficient compared with uptake of soluble protein (MFI= 12,292 vs. 1,356; P<0.05), which may be due to the association of P3 with other proteins that could facilitate uptake.
Figure 4.
Uptake of P3 and cross-presentation of P3 and NE increases breast cancer susceptibility to killing by PR1-CTLs and anti-PR1/HLA-A2. A, MDA-MB-231 breast cancer cells were incubated with soluble P3, irradiated PMNs or PBMC for 4 hours. Cells were permeabilized, stained with anti-P3 antibody and analyzed by flow cytometry. For cell-associated uptake, light scatter seen on flow cytometry provided a clear distinction between PBMC, PMN and MDA-MB-231 cells. PBMC and PMN cells alone were used as negative and positive controls, respectively. ANOVA followed by Tukey test was performed using Prism 5.0 software (*P<0.05). Data are means ± SEM from duplicate experiments. B, MDA-MB-231 breast cancer cells were cultured with soluble P3 or NE (10 μg/mL) at increasing time-points and then analyzed for expression of PR1/HLA-A2. Mean ± SEM fold increase of the median fluorescence intensity (MFI) of PR1/HLA-A2 vs. unpulsed cells is shown from duplicate experiments. ANOVA followed by Tukey test was performed using Prism 5.0 software (*P=0.01); (**P<0.0001). C and D, MDA-MB-231 cells were cultured for 24 hours in media containing NE or P3 (10 μg/mL) and the antigen presentation inhibitors brefeldin A or lactacystin. Cells were then analyzed for expression of PR1/HLA-A2. Mean ± SEM of the median fluorescence intensity (MFI) of PR1/HLA-A2 is shown from duplicate wells of a representative experiment. ANOVA followed by Tukey test were performed using Prism 5.0 software (*P<0.01); (**P<0.0001). E, MDA-MB-231 cells were cultured overnight in media containing P3 or NE (10 μg/mL), loaded with calcein-AM, and then co-cultured with PR1-CTLs for 4 hours. Cytotoxicity was determined by measuring released calcein-AM. NE- or P3-pulsed cells show higher killing vs. unpulsed MDA-MB-231 cells. PR1-pulsed and unpulsed T2 cells were used as positive and negative controls, respectively. Data are means ± SEM from duplicate wells from a representative experiment. F, MDA-MB-231 cells were cultured with NE (10 μg/mL) or P3 (10 μg/mL) for 24 hours. Cells were then incubated with anti-PR1/HLA-A2 (8F4) antibody for 60 minutes and then complement was added. Complement-dependent cytotoxicity was measured using calcein-AM release and shows specific killing of NE- or P3-pulsed MDA-MB-231 cells by 8F4 antibody. Cytotoxicity data are means ± SEM from duplicate wells from a representative experiment. Unpaired t test was performed using Prism 5.0 software (*P<0.05). Abbreviations; PMN, polymorphonuclear cells; PBMC, peripheral blood mononuclear cells.
P3 and NE are cross-presented by breast cancer cells
Since we have shown that NE is also taken up by breast cancer (15) and since PR1 is derived from both of the neutrophil azurophil granule proteases NE and P3, we investigated whether NE and P3 are cross-presented by breast cancer cells following uptake. The HLA-A2+ MDA-MB-231 cells were co-cultured with soluble P3 or NE at increasing time points and subsequently analyzed for PR1/HLA-A2 expression using the mouse anti-PR1/HLA-A2 antibody 8F4 (22). Our data show that breast cancer cells can cross-present PR1 from both NE and P3. Significant PR1 cross-presentation was primarily seen at 24 hours (Fig. 4B) with a 2.5- and 3.0-fold increase in PR1/HLA-A2 on breast cancer cell surface following culture with NE and P3, respectively, compared with unpulsed cells. There was no significant increase in HLA-A2 expression on the cell surface (data not shown).
Furthermore, to investigate the intracellular mechanisms that are involved in NE and P3 cross-presentation, we studied whether the proteasome and the ER/Golgi are involved in NE and P3 cross-presentation as was previously shown for other antigens (13, 23, 24). Our data show that the ER/Golgi and proteasome are both involved in NE and P3 cross-presentation, since incubation of cells with BFA, which inhibits ER to Golgi antegrade transport, and with lactacystin, a proteasome inhibitor, both decreased PR1/HLA-A2 expression by MDA-MB-231 breast cancer cells after co-culturing with NE or P3 (Fig. 4C and D). This is similar to our previous results demonstrating proteasome and ER/Golgi involvement in NE and P3 cross-presentation by APCs (10).
PR1 cross-presentation renders breast cancer susceptible to PR1-targeting therapies
Since PR1 has been effectively targeted in leukemia using a PR1 peptide vaccine (8), PR1-CTLs (33, 34), and anti-PR1/HLA-A2 antibody (8F4) (22), we investigated whether PR1/HLA-A2 expression on breast cancer cells following cross-presentation would render these cells susceptible to killing by PR1-CTLs and 8F4 antibody. The HLA-A2+ MDA-MB-231 cells were cultured in media containing 10 μg/mL of NE or P3 for 24 hours and then incubated with healthy donor expanded PR1-CTLs for 4 hours in a standard calcein-AM cytotoxicity assay (5, 35) (Fig. 4E). Our data demonstrate that cross-presentation of NE and P3 increased the susceptibility of MDA-MB-231 cells to killing by PR1-CTLs after NE- or P3-pulsing, in comparison with unpulsed MDA-MB-231 cells. Similarly, using 8F4 antibody in a complement dependent cytotoxicity assay (Fig. 4F) (22), we observed a dose-dependent killing of MDA-MB-231 cells following NE or P3 cross-presentation in comparison with unpulsed cells. The greatest killing was noted at the highest dose of 8F4 antibody (10 μg/mL).
PR1/HLA-A2 and PR1-CTL are detected in patients with breast cancer
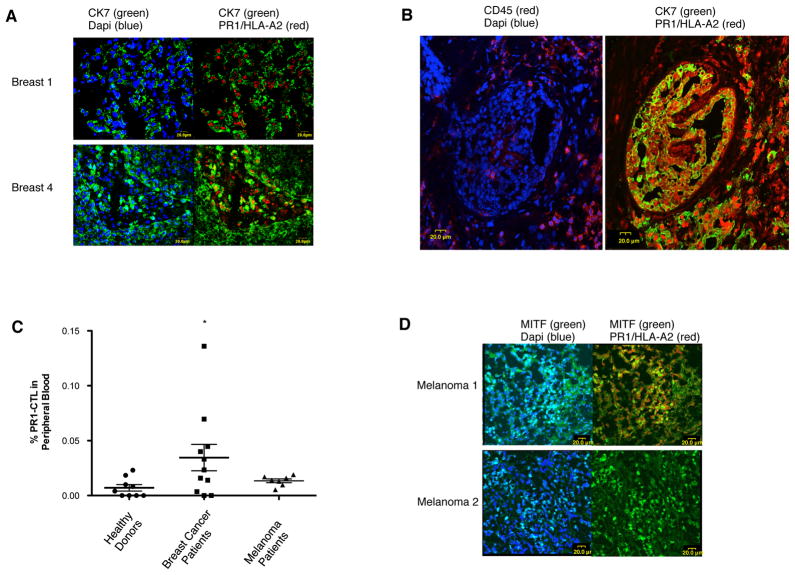
Since we showed that cultured breast cancer cell lines and tumor tissues lack endogenous NE and P3, and because we observed in vitro evidence of NE and P3 cross-presentation by breast cancer cells and subsequent susceptibility to PR1-targeting therapies, we investigated whether PR1 could be detected in primary beast cancer patient tissues and whether PR1-CTLs could be detected in peripheral blood from patients with breast cancer. Laser confocal microscopy of two HLA-A2 positive breast cancer tissues demonstrated 8F4 in both tumor tissues (Fig. 5A). 8F4 staining was absent in HLA-A2 negative tissue (data not shown). Moreover, to verify that the expression of PR1/HLA-A2 is by breast cancer cells and not by infiltrating leukocytes, we stained consecutive breast cancer tissue sections with the leukocyte marker CD45. We show the absence of CD45 staining in the areas of the breast cancer tissue that co-stained with 8F4 and CK7, further confirming that the PR1/HLA-A2 expression was by breast cancer cells, not by adjacent inflammatory cells (Fig. 5B).
Figure 5.
PR1/HLA-A2 and PR1-CTL are detected in breast cancer and melanoma patients. A, Resected HLA-A2+ patient breast cancer tissues (Breast 1 and Breast 4) were stained with anti-PR1/HLA-A2 (8F4)-647 (red) and anti-cytokeratin-7(CK-7)-FITC (green) and then imaged using confocal laser microscopy. PR1/HLA-A2 appears to be expressed by breast cancer cells as shown by the co-staining of 8F4 with CK-7. Dapi-blue was used to stain cell nuclei. B, Consecutive sections from resected HLA-A2+ breast cancer tissue were stained with anti-CD45-647 (red) (left panel) or anti-CK7-FITC (green) and 8F4-647 (red) (right panel) and then imaged using confocal laser microscopy. PR1/HLA-A2 is expressed by breast cancer cells (8F4+/CK7+) in areas that have minimal leukocytes (CD45−), thereby confirming PR1/HLA-A2 expression by breast cancer cells. Dapi-blue was used to stain cell nuclei. C, Box and whisker plot shows PR1-CTLs in peripheral blood from HLA-A2+ patients with breast cancer (n=11), melanoma (n=7) and healthy (n=9) HLA-A2+ donors. Mann-Whitney U test was performed using Prism 5.0 software (*P<0.05). D, Resected HLA-A2+ (Melanoma 1) and HLA-A2−(Melanoma 2) patient tissues were stained with 8F4-647 (red) and anti-microphthalmia-associated transcription factor (MITF)-FITC (green) and then imaged using confocal laser microscopy. PR1/HLA-A2 appears to be expressed in the HLA-A2+ melanoma sample (Melanoma 1) as shown by the co-staining of 8F4 with MITF. Dapi-blue was used to stain for cell nuclei.
To determine whether PR1-CTL could be detected in breast cancer patients, we used PR1/HLA-A2 dextramer staining of 11 peripheral blood samples from early stage breast cancer patients (Fig. 5C). The median frequency of PR1-CTLs in these HLA-A2+ patients was 0.05% of CD8+ T cells (Range, 0.02%–0.2%), slightly higher than the frequency of PR1-CTLs in healthy donors (1/15,000 to 1/350,000 CD8+ cells) (26). The gating strategy used to determine the frequency of PR1-CTL is shown in Supplemental Fig. 2. Taken together, these in vivo data suggest that the serine proteases NE and P3 present in the tumor microenvironment can be taken up and cross-presented by breast cancer cells, which may contribute to an adaptive immune response against the NE and P3-derived epitope PR1.
PR1/HLA-A2 and PR1-CTL in melanoma patients
Since melanoma tissues were also shown to have inflammatory cells that may be a source for NE and P3 (19), and because melanoma is known to be susceptible to immunotherapy (20, 21), we next investigated whether cross-presentation of NE and P3 could also be detected in melanoma. To determine whether PR1-CTL are also detected in melanoma, we stained PBMC from melanoma patients with PR1/HLA-A2 dextramer and detected PR1-CTLs in all 7 patients at a median frequency of 0.014% of CD8+ T cells (Range, 0.0053%–0.019%) (Fig. 5C), similar to what was seen in blood form normal donors. We also detected PR1/HLA-A2 expression in one HLA-A2+ (Melanoma 1) but not HLA-A2− (Melanoma 2) melanoma tissue (Fig. 5D). The gating strategy used to determine the frequency of PR1-CTL is shown in Supplemental Fig. 2.
Cross-presentation of NE and P3 by melanoma increases susceptibility to PR1-CTL
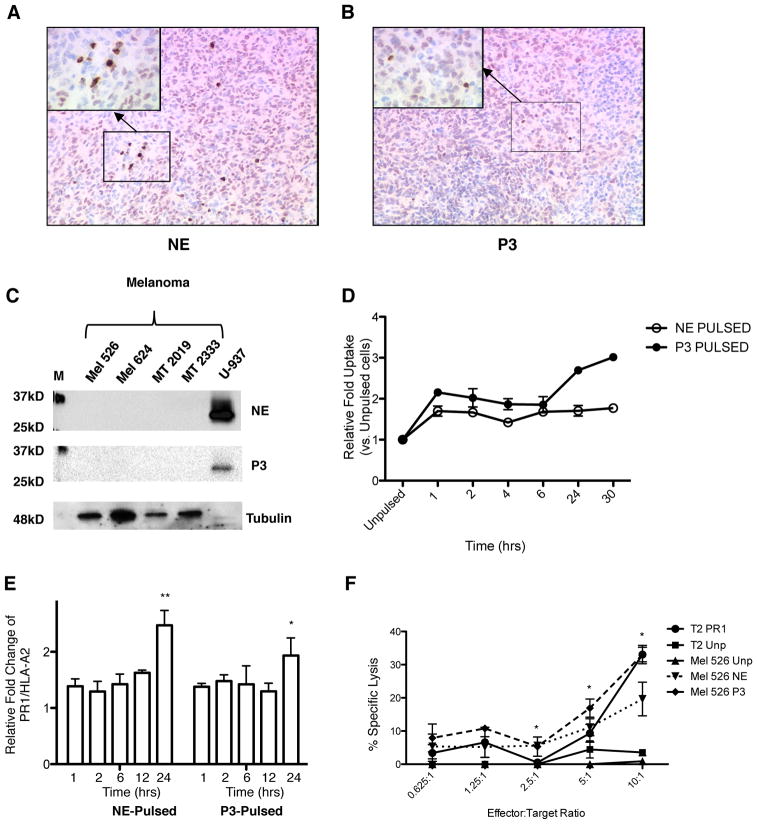
To determine whether melanoma expresses NE and P3, we stained melanoma tissue obtained from patients for NE and P3 and showed the absence of NE and P3 (Fig. 6A and B). We also analyzed NE and P3 expression in 4 melanoma cell lines, MEL526, MEL624, MT2019 and MT2333. Western blot analysis shows absence of NE and P3 in melanoma cell lines (Fig. 6C). Similar to breast cancer, we demonstrate uptake and cross-presentation of NE and P3 by the HLA-A2+ Mel 526 cell line (Fig. 6D and E). Since 8F4 antibody binds to the HLA-A2 molecule(22, 36), which composes a significant portion of the conformational PR1/HLA-A2 epitope, Mel 526 cells do show staining with 8F4 prior to co-culture with NE or P3 (Supplemental Fig. 3). However, staining with 8F4 increases after co-culture with NE or P3, without an increase in HLA-A2 surface staining (Supplemental Fig. 3), indicating an increase in PR1/HLA-A2 expression on the cell surface. Furthermore, cross-presentation of NE and P3 increased the susceptibility of the HLA-A2+ Mel 526 cell line to killing by PR1-CTL, with the highest killing noted at the highest effector: target (E:T) ratio (Fig. 6F).
Figure 6.
Cross-presentation of P3 and NE by melanoma cells increases susceptibility to PR1-CTL. A, Double staining of NE (brown) and MITF(pink), or B, P3 (brown)and MITF(pink) in a primary melanoma patient samples shows lack of NE and P3 in melanoma. The images were taken at 100× magnification. Inset, 400× magnification, shows scattered NE or P3 positive cells, which are likely inflammatory cells. C, Western blot showing absence of NE and P3 in melanoma cell lines. U-937 leukemia cell line was used as a positive control for NE and P3. Tubulin was used as loading control. M=molecular weight marker. D and E, 526 HLA-A2+ melanoma cell line was cultured with soluble NE (10 μg/mL) or P3 (10 μg/mL) at increasing time points and then analyzed for (D) uptake of NE and P3 and (E) cross-presentation (i.e. PR1/HLA-A2 expression). Fold increase of the median fluorescence intensity (MFI) of NE or P3 (D) or PR1/HLA-A2 (E) vs. unpulsed cells is shown on the Y-axis. ANOVA followed by Tukey test was performed using Prism 5.0 software (**P=0.0001); (*P<0.05). Data represent mean ± SEM from duplicate experiments. F, Calcein-AM cytotoxicity assay shows killing of NE (10 μg/mL) and P3 (10 μg/mL) 24-hour pulsed 526 HLA-A2+ melanoma cell line by PR1-CTLs vs. unpulsed (Unp) Mel 526. Unpulsed (T2 Unp) and PR1-pulsed (T2 PR1) T2 cells were used as negative and positive controls, respectively. Data are means ± SEM from duplicate wells from a representative experiment. Unpaired t test was performed using Prism 5.0 software (*P<0.05).
Discussion
P3 and NE are serine proteases that are normally expressed in hematopoietic cells and are abundant in leukemia and the microenvironment of non-hematopoietic tumors (16, 17, 19, 37). In this study, we show that breast cancer cells and melanoma, which lack endogenous P3 and NE expression, take up soluble and PMN-associated P3 and NE. After uptake, PR1, a P3- and NE-derived peptide, is cross-presented on HLA-A2+ breast cancer and melanoma cells and these cells become susceptible to lysis by PR1-CTL and anti-PR1/HLA-A2 monoclonal antibody (8F4). Our study provides evidence of a novel mechanism whereby hematopoietic antigens can be taken up and cross-presented on major histocompatibility (MHC) class I by non-hematopoietic tumors, and it suggests that in addition to leukemia, exogenous P3 and NE may also be tumor antigens in non-hematopoietic tumors. We show PR1/HLA-A2 in primary breast cancer and melanoma tissues as well as PR1-CTL in peripheral blood from patients, providing further evidence that studies evaluating PR1-targeting therapies in breast cancer and melanoma are warranted. Our data also highlight the role of cross-presentation in expanding the number of tumor types that could be targeted by existing immunotherapeutic modalities.
Antigen cross-presentation is a critical step in the elicitation of immune responses and it is an important function of hematopoietic cells, including dendritic cells, B cells macrophages and neutrophils, which are professional APCs critical for generating effective CTL responses (12, 38–40). This process is necessary for immunity to most tumors. Antigen cross-presentation can also result from the direct transfer of antigen between cells, as was shown in melanoma cells which transferred preprocessed antigenic peptides to APC through gap junctions induced by Salmonella infection (41). Furthermore cross-presentation is also required for the maintenance of peripheral tolerance, and the balance of immunity versus tolerance is thought to depend on the activation state of the APC (42). Tolerance is also induced by the cross-presentation of tissue antigens by non-hematopoietic cells, which occurs in the thymus and is facilitated by medullary thymic epithelial cells (19). However, little is understood about cross-presentation of hematopoietic antigens by non-hematopoietic tumors.
We showed previously that DCs take up soluble P3 and NE and cross-present PR1 on HLA-A2 by 4 hours (10). Di Pucchio et al. also showed cross-presentation by plasmacytoid DCs as early as 4 hours following antigen pulsing (43). In our study, while PR1 expression could be observed as early as 1 hour after antigen pulsing of breast cancer and melanoma cells, maximal expression was not observed until 24 hours after uptake of P3 or NE. Since breast cancer and melanoma cells are not naturally APCs, they may not be optimally equipped for rapid antigen cross-presentation. Our results expand on two previous reports showing cross-presentation by non-APCs, specifically mesodermally-derived mesenchymal stromal (13) and endothelial cells (44). Consistent with our findings, both of these studies showed cross-presentation at later time points (>12 hours), in contrast to the earlier cross-presentation by DCs (10, 43). Therefore, the temporal pattern of PR1 expression might be important for regulating immunity.
Because co-stimulation following antigen presentation is a requirement for immune priming and since co-stimulatory molecules including CD83, CD86 and HLA-DR are limited to distinct APC populations (45), it is likely that P3 and NE cross-presentation by solid tumors would facilitate cross-tolerance in vivo. This conclusion is indirectly supported by the observation that NE expression in breast cancer is a negative prognostic factor (29). Together with our data, this suggests that targeting PR1 or other epitopes within P3 and NE in active immunization strategies for breast cancer might not be effective. However, passive immunotherapy strategies with PR1-CTL or with anti-PR1/HLA-A2 monoclonal antibody might have antitumor activity. It is therefore important to understand whether normal tissues can also cross-present P3 and NE and express PR1 and whether this mechanism plays a role in maintaining tolerance to these tissue antigens, which is being currently investigated in our laboratory.
While P3 and NE are not expressed in breast cancer or melanoma cells, they are expressed in tumor-associated neutrophils, which are present in breast cancer, melanoma and many other non-hematopoietic tumors (16, 17, 19, 37). Here we show NE and P3 uptake by non-hematopoietic tumors, to include breast cancer (15) and melanoma cells. These data are consistent with a study by Houghton et al. that showed NE uptake and localization to endosomal compartments in a Lewis lung carcinoma tumor model, which resulted in increased proliferation via increased activity through the PI3K pathway (14). Our data demonstrate the localization of P3 and NE (15) to lysosomal and endosomal compartments, respectively, which are both known to play a role in antigen cross-presentation (31, 46, 47), thus providing further support to NE and P3 cross-presentation by non-APCs.
In conclusion, our results show that NE and P3 are cross-presented by breast cancer and melanoma cells thereby rendering them susceptible to PR1-targeting therapies, that PR1/HLA-A2 is expressed in primary patient breast cancer and melanoma tissues and that PR1-CTL are present in peripheral blood from patients with these malignancies. Furthermore, NE and P3 uptake can be seen in a variety of non-hematopoietic tumors. Since inflammatory cells, to include monocytes and PMNs, are found in numerous tissues and can provide a source for NE and P3, our findings suggest the broad applicability for PR1 immunotherapies in non-myeloid malignancies and identify cross-presentation as a novel mechanism that renders tumors susceptible to therapies that target cross-presented antigens.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by NIH grant 4R00CA133244-03200 and 5P50CA116199-05.
Gheath Alatrash received partial salary support from the MD Anderson Barbara Rattay Advanced Scholars Program.
References
- 1.Lane AA, Ley TJ. Neutrophil elastase is important for PML-retinoic acid receptor alpha activities in early myeloid cells. Mol Cell Biol. 2005;25:23–33. doi: 10.1128/MCB.25.1.23-33.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brouwer E, Stegeman CA, Huitema MG, Limburg PC, Kallenberg CG. T cell reactivity to proteinase 3 and myeloperoxidase in patients with Wegener’s granulomatosis (WG) Clin Exp Immunol. 1994;98:448–453. doi: 10.1111/j.1365-2249.1994.tb05511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apenberg S, Andrassy K, Worner I, Hansch GM, Roland J, Morcos M, Ritz E. Antibodies to neutrophil elastase: a study in patients with vasculitis. Am J Kidney Dis. 1996;28:178–185. doi: 10.1016/s0272-6386(96)90299-1. [DOI] [PubMed] [Google Scholar]
- 4.Pham CT. Neutrophil serine proteases: specific regulators of inflammation. Nat Rev Immunol. 2006;6:541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 5.Molldrem J, Dermime S, Parker K, Jiang YZ, Mavroudis D, Hensel N, Fukushima P, Barrett AJ. Targeted T-cell therapy for human leukemia: cytotoxic T lymphocytes specific for a peptide derived from proteinase 3 preferentially lyse human myeloid leukemia cells. Blood. 1996;88:2450–2457. [PubMed] [Google Scholar]
- 6.Molldrem JJ, Lee PP, Wang C, Felio K, Kantarjian HM, Champlin RE, Davis MM. Evidence that specific T lymphocytes may participate in the elimination of chronic myelogenous leukemia. Nat Med. 2000;6:1018–1023. doi: 10.1038/79526. [DOI] [PubMed] [Google Scholar]
- 7.Rezvani K, Grube M, Brenchley JM, Sconocchia G, Fujiwara H, Price DA, Gostick E, Yamada K, Melenhorst J, Childs R, Hensel N, Douek DC, Barrett AJ. Functional leukemia-associated antigen-specific memory CD8+ T cells exist in healthy individuals and in patients with chronic myelogenous leukemia before and after stem cell transplantation. Blood. 2003;102:2892–2900. doi: 10.1182/blood-2003-01-0150. [DOI] [PubMed] [Google Scholar]
- 8.Rezvani K, Yong AS, Mielke S, Savani BN, Musse L, Superata J, Jafarpour B, Boss C, Barrett AJ. Leukemia-associated antigen-specific T-cell responses following combined PR1 and WT1 peptide vaccination in patients with myeloid malignancies. Blood. 2008;111:236–242. doi: 10.1182/blood-2007-08-108241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burchert A, Wolfl S, Schmidt M, Brendel C, Denecke B, Cai D, Odyvanova L, Lahaye T, Muller MC, Berg T, Gschaidmeier H, Wittig B, Hehlmann R, Hochhaus A, Neubauer A. Interferon-alpha, but not the ABL-kinase inhibitor imatinib (STI571), induces expression of myeloblastin and a specific T-cell response in chronic myeloid leukemia. Blood. 2003;101:259–264. doi: 10.1182/blood-2002-02-0659. [DOI] [PubMed] [Google Scholar]
- 10.Alatrash G, Ono Y, Sergeeva A, Sukhumalchandra P, Zhang M, St John LS, Yang TH, Ruisaard K, Armistead PM, Mittendorf EA, He H, Qiao N, Rodriguez-Cruz T, Liang S, Clise-Dwyer K, Wieder ED, Lizee G, Lu S, Molldrem JJ. The role of antigen cross-presentation from leukemia blasts on immunity to the leukemia-associated antigen PR1. J Immunother. 2012;35:309–320. doi: 10.1097/CJI.0b013e31824b3b14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 12.Robson NC, Donachie AM, Mowat AM. Simultaneous presentation and cross-presentation of immune-stimulating complex-associated cognate antigen by antigen-specific B cells. Eur J Immunol. 2008;38:1238–1246. doi: 10.1002/eji.200737758. [DOI] [PubMed] [Google Scholar]
- 13.Francois M, Romieu-Mourez R, Stock-Martineau S, Boivin MN, Bramson JL, Galipeau J. Mesenchymal stromal cells cross-present soluble exogenous antigens as part of their antigen-presenting cell properties. Blood. 2009;114:2632–2638. doi: 10.1182/blood-2009-02-207795. [DOI] [PubMed] [Google Scholar]
- 14.Houghton AM, Rzymkiewicz DM, Ji H, Gregory AD, Egea EE, Metz HE, Stolz DB, Land SR, Marconcini LA, Kliment CR, Jenkins KM, Beaulieu KA, Mouded M, Frank SJ, Wong KK, Shapiro SD. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat Med. 16:219–223. doi: 10.1038/nm.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mittendorf EA, Alatrash G, Qiao N, Wu Y, Sukhumalchandra P, St John LS, Philips AV, Xiao H, Zhang M, Ruisaard K, Clise-Dwyer K, Lu S, Molldrem JJ. Breast cancer cell uptake of the inflammatory mediator neutrophil elastase triggers an anticancer adaptive immune response. Cancer Res. 2012;72:3153–3162. doi: 10.1158/0008-5472.CAN-11-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Queen MM, Ryan RE, Holzer RG, Keller-Peck CR, Jorcyk CL. Breast cancer cells stimulate neutrophils to produce oncostatin M: potential implications for tumor progression. Cancer Res. 2005;65:8896–8904. doi: 10.1158/0008-5472.CAN-05-1734. [DOI] [PubMed] [Google Scholar]
- 17.Jensen HK, Donskov F, Marcussen N, Nordsmark M, Lundbeck F, von der Maase H. Presence of intratumoral neutrophils is an independent prognostic factor in localized renal cell carcinoma. Journal of Clinical Oncology. 2009;27:4709–4717. doi: 10.1200/JCO.2008.18.9498. [DOI] [PubMed] [Google Scholar]
- 18.Peoples GE, Holmes JP, Hueman MT, Mittendorf EA, Amin A, Khoo S, Dehqanzada ZA, Gurney JM, Woll MM, Ryan GB, Storrer CE, Craig D, Ioannides CG, Ponniah S. Combined clinical trial results of a HER2/neu (E75) vaccine for the prevention of recurrence in high-risk breast cancer patients: U.S. Military Cancer Institute Clinical Trials Group Study I-01 and I-02. Clin Cancer Res. 2008;14:797–803. doi: 10.1158/1078-0432.CCR-07-1448. [DOI] [PubMed] [Google Scholar]
- 19.Jensen TO, Schmidt H, Moller HJ, Donskov F, Hoyer M, Sjoegren P, Christensen IJ, Steiniche T. Intratumoral neutrophils and plasmacytoid dendritic cells indicate poor prognosis and are associated with pSTAT3 expression in AJCC stage I/II melanoma. Cancer. 2011 doi: 10.1002/cncr.26511. [DOI] [PubMed] [Google Scholar]
- 20.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, Robinson MR, Raffeld M, Duray P, Seipp CA, Rogers-Freezer L, Morton KE, Mavroukakis SA, White DE, Rosenberg SA. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartzentruber DJ, Lawson DH, Richards JM, Conry RM, Miller DM, Treisman J, Gailani F, Riley L, Conlon K, Pockaj B, Kendra KL, White RL, Gonzalez R, Kuzel TM, Curti B, Leming PD, Whitman ED, Balkissoon J, Reintgen DS, Kaufman H, Marincola FM, Merino MJ, Rosenberg SA, Choyke P, Vena D, Hwu P. gp100 peptide vaccine and interleukin-2 in patients with advanced melanoma. N Engl J Med. 2011;364:2119–2127. doi: 10.1056/NEJMoa1012863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sergeeva A, Alatrash G, He H, Ruisaard K, Lu S, Wygant J, McIntyre BW, Ma Q, Li D, St John L, Clise-Dwyer K, Molldrem JJ. An anti-PR1/HLA-A2 T-cell receptor-like antibody mediates complement-dependent cytotoxicity against acute myeloid leukemia progenitor cells. Blood. 2011;117:4262–4272. doi: 10.1182/blood-2010-07-299248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovacsovics-Bankowski M, Rock KL. A phagosome-to-cytosol pathway for exogenous antigens presented on MHC class I molecules. Science. 1995;267:243–246. doi: 10.1126/science.7809629. [DOI] [PubMed] [Google Scholar]
- 24.Mukai T, Maeda Y, Tamura T, Matsuoka M, Tsukamoto Y, Makino M. Induction of cross-priming of naive CD8+ T lymphocytes by recombinant bacillus Calmette-Guerin that secretes heat shock protein 70-major membrane protein-II fusion protein. J Immunol. 2009;183:6561–6568. doi: 10.4049/jimmunol.0803857. [DOI] [PubMed] [Google Scholar]
- 25.Kuronita T, Eskelinen EL, Fujita H, Saftig P, Himeno M, Tanaka Y. A role for the lysosomal membrane protein LGP85 in the biogenesis and maintenance of endosomal and lysosomal morphology. Journal of cell science. 2002;115:4117–4131. doi: 10.1242/jcs.00075. [DOI] [PubMed] [Google Scholar]
- 26.Molldrem JJ, Lee PP, Wang C, Champlin RE, Davis MM. A PR1-human leukocyte antigen-A2 tetramer can be used to isolate low-frequency cytotoxic T lymphocytes from healthy donors that selectively lyse chronic myelogenous leukemia. Cancer Res. 1999;59:2675–2681. [PubMed] [Google Scholar]
- 27.Molldrem JJ, Clave E, Jiang YZ, Mavroudis D, Raptis A, Hensel N, Agarwala V, Barrett AJ. Cytotoxic T lymphocytes specific for a nonpolymorphic proteinase 3 peptide preferentially inhibit chronic myeloid leukemia colony-forming units. Blood. 1997;90:2529–2534. [PubMed] [Google Scholar]
- 28.Prang N, Preithner S, Brischwein K, Goster P, Woppel A, Muller J, Steiger C, Peters M, Baeuerle PA, da Silva AJ. Cellular and complement-dependent cytotoxicity of Ep-CAM-specific monoclonal antibody MT201 against breast cancer cell lines. Br J Cancer. 2005;92:342–349. doi: 10.1038/sj.bjc.6602310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desmedt C, Ouriaghli FE, Durbecq V, Soree A, Colozza MA, Azambuja E, Paesmans M, Larsimont D, Buyse M, Harris A, Piccart M, Martiat P, Sotiriou C. Impact of cyclins E, neutrophil elastase and proteinase 3 expression levels on clinical outcome in primary breast cancer patients. Int J Cancer. 2006;119:2539–2545. doi: 10.1002/ijc.22149. [DOI] [PubMed] [Google Scholar]
- 30.Cresswell P, Ackerman AL, Giodini A, Peaper DR, Wearsch PA. Mechanisms of MHC class I-restricted antigen processing and cross- presentation. Immunol Rev. 2005;207:145–157. doi: 10.1111/j.0105-2896.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- 31.Basha G, Lizee G, Reinicke AT, Seipp RP, Omilusik KD, Jefferies WA. MHC class I endosomal and lysosomal trafficking coincides with exogenous antigen loading in dendritic cells. PLoS One. 2008;3:e3247. doi: 10.1371/journal.pone.0003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgdorf S, Lukacs-Kornek V, Kurts C. The mannose receptor mediates uptake of soluble but not of cell-associated antigen for cross-presentation. J Immunol. 2006;176:6770–6776. doi: 10.4049/jimmunol.176.11.6770. [DOI] [PubMed] [Google Scholar]
- 33.Rezvani K, Price DA, Brenchley JM, Kilical Y, Gostick E, Sconocchia G, Hansmann K, Kurlander R, Douek DC, Barrett AJ. Transfer of PR1-specific T-cell clones from donor to recipient by stem cell transplantation and association with GvL activity. Cytotherapy. 2007;9:245–251. doi: 10.1080/14653240701218524. [DOI] [PubMed] [Google Scholar]
- 34.Ma Q, Wang C, Jones D, Quintanilla KE, Li D, Wang Y, Wieder ED, Clise-Dwyer K, Alatrash G, Mj Y, Munsell MF, Lu S, Qazilbash MH, Molldrem JJ. Adoptive transfer of PR1 cytotoxic T lymphocytes associated with reduced leukemia burden in a mouse acute myeloid leukemia xenograft model. Cytotherapy. 2010;12:1056–1062. doi: 10.3109/14653249.2010.506506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang YZ, Mavroudis D, Dermime S, Hensel N, Couriel D, Molldrem J, Barrett AJ. Alloreactive CD4+ T lymphocytes can exert cytotoxicity to chronic myeloid leukaemia cells processing and presenting exogenous antigen. Br J Haematol. 1996;93:606–612. doi: 10.1046/j.1365-2141.1996.d01-1676.x. [DOI] [PubMed] [Google Scholar]
- 36.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 37.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 38.Delamarre L, Holcombe H, Mellman I. Presentation of exogenous antigens on major histocompatibility complex (MHC) class I and MHC class II molecules is differentially regulated during dendritic cell maturation. J Exp Med. 2003;198:111–122. doi: 10.1084/jem.20021542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovacsovics-Bankowski M, Clark K, Benacerraf B, Rock KL. Efficient major histocompatibility complex class I presentation of exogenous antigen upon phagocytosis by macrophages. Proc Natl Acad Sci U S A. 1993;90:4942–4946. doi: 10.1073/pnas.90.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beauvillain C, Delneste Y, Scotet M, Peres A, Gascan H, Guermonprez P, Barnaba V, Jeannin P. Neutrophils efficiently cross-prime naive T cells in vivo. Blood. 2007;110:2965–2973. doi: 10.1182/blood-2006-12-063826. [DOI] [PubMed] [Google Scholar]
- 41.Saccheri F, Pozzi C, Avogadri F, Barozzi S, Faretta M, Fusi P, Rescigno M. Bacteria-induced gap junctions in tumors favor antigen cross- presentation and antitumor immunity. Science translational medicine. 2010;2:44ra57. doi: 10.1126/scitranslmed.3000739. [DOI] [PubMed] [Google Scholar]
- 42.Boussiotis VA, Gribben JG, Freeman GJ, Nadler LM. Blockade of the CD28 co-stimulatory pathway: a means to induce tolerance. Curr Opin Immunol. 1994;6:797–807. doi: 10.1016/0952-7915(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 43.Di Pucchio T, Chatterjee B, Smed-Sorensen A, Clayton S, Palazzo A, Montes M, Xue Y, Mellman I, Banchereau J, Connolly JE. Direct proteasome-independent cross-presentation of viral antigen by plasmacytoid dendritic cells on major histocompatibility complex class I. Nat Immunol. 2008;9:551–557. doi: 10.1038/ni.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagai R, Valujskikh A, Canaday DH, Bailey E, Lalli PN, Harding CV, Heeger PS. Mouse endothelial cells cross-present lymphocyte-derived antigen on class I MHC via a TAP1- and proteasome-dependent pathway. J Immunol. 2005;174:7711–7715. doi: 10.4049/jimmunol.174.12.7711. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 46.Burgdorf S, Scholz C, Kautz A, Tampe R, Kurts C. Spatial and mechanistic separation of cross-presentation and endogenous antigen presentation. Nat Immunol. 2008;9:558–566. doi: 10.1038/ni.1601. [DOI] [PubMed] [Google Scholar]
- 47.Chatterjee B, Smed-Sorensen A, Cohn L, Chalouni C, Vandlen R, Lee BC, Widger J, Keler T, Delamarre L, Mellman I. Internalization and endosomal degradation of receptor-bound antigens regulate the efficiency of cross presentation by human dendritic cells. Blood. 2012;120:2011–2020. doi: 10.1182/blood-2012-01-402370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.