Abstract
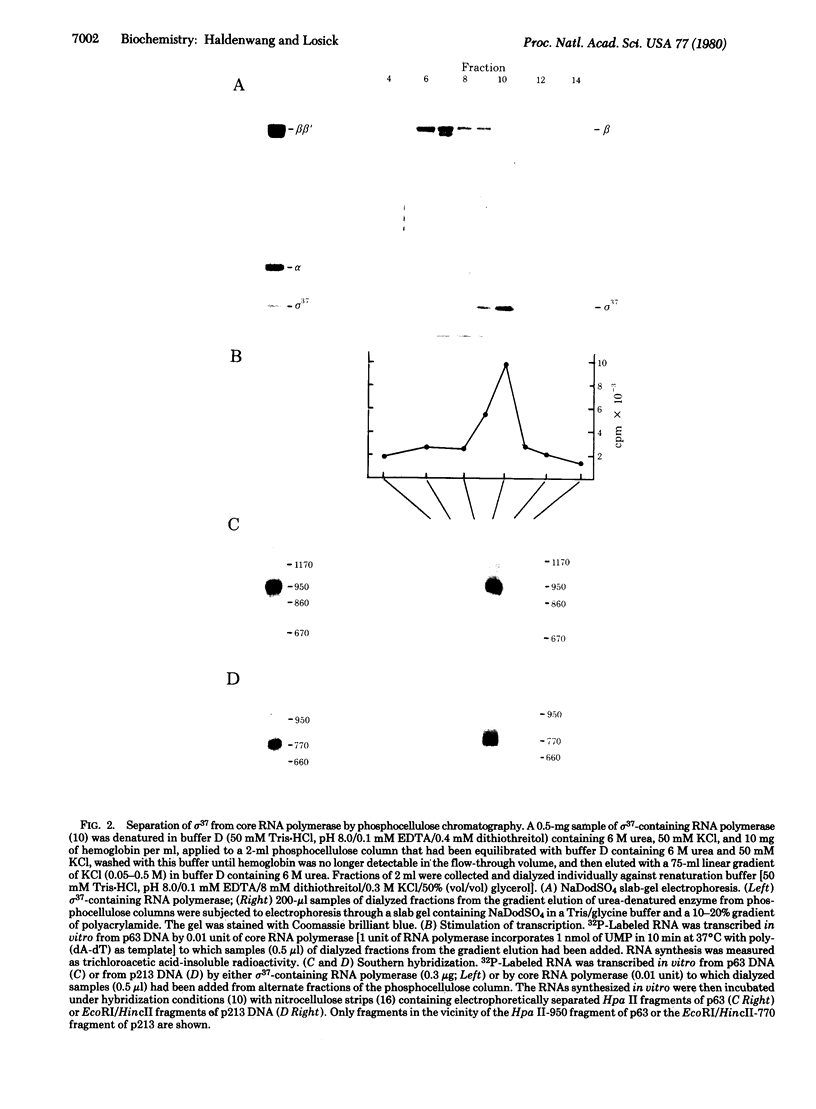
A modified form of Bacillus subtilis RNA polymerase (RNA nucleotidyltransferase) has been isolated that exhibits distinctive transcriptional specificity. This modified enzyme transcribes two cloned genes from the purA-cysA region of the B. subtilis chromosome whose expression in vivo is associated with the process of sporulation. Neither of these genes is transcribed by the usual form of B. subtilis RNA polymerase holoenzyme containing a sigma factor of 55,000 daltons (sigma 55). The modified RNA polymerase lacks sigma 55 but contains a newly identified subunit of 37,000 daltons termed sigma 37. A reconstitution experiment in which sigma 37 was added to core RNA polymerase strongly suggests that sigma 37 is responsible for the transcriptional specificity of the modified RNA polymerase. Sigma 37 apparently acts at the level of promoter recognition; this transcriptional determinant enabled core RNA polymerase to form stable binary and ternary ("initiation") complexes with endonuclease restriction fragments containing promoters for the cloned B. subtilis genes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avila J., Hermoso J. M., Vinuela E., Salas M. Purification and properties of DNA-dependent RNA polymerase from Bacillus subtilis vegetative cells. Eur J Biochem. 1971 Aug 25;21(4):526–535. doi: 10.1111/j.1432-1033.1971.tb01498.x. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Travers A. A., Dunn J. J., Bautz E. K. Factor stimulating transcription by RNA polymerase. Nature. 1969 Jan 4;221(5175):43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- Duffy J. J., Geiduschek E. P. Purification of a positive regulatory subunit from phage SP01-modified RNA polymerase. Nature. 1977 Nov 3;270(5632):28–32. doi: 10.1038/270028a0. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Identification of phage SP01 proteins coded by regulatory genes 33 and 34. Nature. 1976 Aug 26;262(5571):748–753. doi: 10.1038/262748a0. [DOI] [PubMed] [Google Scholar]
- Fox T. D., Losick R., Pero J. Regulatory gene 28 of bacteriophage SPO1 codes for a phage-induced subunit of RNA polymerase. J Mol Biol. 1976 Mar 5;101(3):427–433. doi: 10.1016/0022-2836(76)90157-1. [DOI] [PubMed] [Google Scholar]
- Fukuda R., Iwakura Y., Ishihama A. Heterogeneity of RNA polymerase in Escherichia coli. I. A new holoenzyme containing a new sigma factor. J Mol Biol. 1974 Mar;83(3):353–367. doi: 10.1016/0022-2836(74)90284-8. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Banner C. D., Ollington J. F., Losick R., Hoch J. A., O'Connor M. B., Sonenshein A. L. Mapping a cloned gene under sporulation control by inserttion of a drug resistance marker into the Bacillus subtilis chromosome. J Bacteriol. 1980 Apr;142(1):90–98. doi: 10.1128/jb.142.1.90-98.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenwang W. G., Losick R. A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature. 1979 Nov 15;282(5736):256–260. doi: 10.1038/282256a0. [DOI] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972 Sep 28;70(2):157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. II. The kinetics of the binding reaction. J Mol Biol. 1972 Sep 28;70(2):187–195. doi: 10.1016/0022-2836(72)90532-3. [DOI] [PubMed] [Google Scholar]
- Ishihama A., Saitoh T. Subunits of RNA polymerase in function and structure. IX. Regulation of RNA polymerase activity by stringent starvation protein (SSP). J Mol Biol. 1979 Apr 25;129(4):517–530. doi: 10.1016/0022-2836(79)90466-2. [DOI] [PubMed] [Google Scholar]
- Jaehning J. A., Wiggs J. L., Chamberlin M. J. Altered promoter selection by a novel form of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5470–5474. doi: 10.1073/pnas.76.11.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn T., Greenleaf A. L., Losick R. RNA polymerase from sporulating Bacillus subtilis. Purification and properties of a modified form of the enzyme containing two sporulation polypeptides. J Biol Chem. 1975 Dec 25;250(24):9256–9261. [PubMed] [Google Scholar]
- Moran C. P., Jr, Losick R., Sonenshein A. L. Identification of a sporulation locus in cloned Bacillus subtilis deoxyribonucleic acid. J Bacteriol. 1980 Apr;142(1):331–334. doi: 10.1128/jb.142.1.331-334.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Williamson V., Burtis K., Doi R. H. Purification and properties of two RNA polymerases from sporulating cells of Bacillus subtilis. Eur J Biochem. 1978 Jul 17;88(1):155–164. doi: 10.1111/j.1432-1033.1978.tb12433.x. [DOI] [PubMed] [Google Scholar]
- Pribnow D. Bacteriophage T7 early promoters: nucleotide sequences of two RNA polymerase binding sites. J Mol Biol. 1975 Dec 15;99(3):419–443. doi: 10.1016/s0022-2836(75)80136-7. [DOI] [PubMed] [Google Scholar]
- Richardson J. P. Enzymic synthesis of RNA from T7 DNA. J Mol Biol. 1966 Oct 28;21(1):115–127. doi: 10.1016/0022-2836(66)90083-0. [DOI] [PubMed] [Google Scholar]
- Segall J., Losick R. Cloned Bacillus subtilis DNA containing a gene that is activated early during sporulation. Cell. 1977 Aug;11(4):751–761. doi: 10.1016/0092-8674(77)90289-6. [DOI] [PubMed] [Google Scholar]
- Shorenstein R. G., Losick R. Purification and properties of the sigma subunit of ribonucleic acid polymerase from vegetative Bacillus subtilis. J Biol Chem. 1973 Sep 10;248(17):6163–6169. [PubMed] [Google Scholar]
- Simpson R. B. The molecular topography of RNA polymerase-promoter interaction. Cell. 1979 Oct;18(2):277–285. doi: 10.1016/0092-8674(79)90047-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Talkington C., Pero J. Distinctive nucleotide sequences of promoters recognized by RNA polymerase containing a phage-coded "sigma-like" protein. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5465–5469. doi: 10.1073/pnas.76.11.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington C., Pero J. Promoter recognition by phage SP01-modified RNA polymerase. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1185–1189. doi: 10.1073/pnas.75.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijan R., Pero J. Bacteriophage SP01 regulatory proteins directing late gene transcription in vitro. Nature. 1976 Aug 26;262(5571):753–757. doi: 10.1038/262753a0. [DOI] [PubMed] [Google Scholar]
- Tjian R., Losick R., Pero J., Hinnebush A. Purification and comparative properties of the delta and sigma subunits of RNA polymerase from Bacillus subtilis. Eur J Biochem. 1977 Mar 15;74(1):149–154. doi: 10.1111/j.1432-1033.1977.tb11376.x. [DOI] [PubMed] [Google Scholar]
- Tjian R., Stinchcomb D., Losick R. Antibody directed against Bacillus subtilis rho factor purified by sodium dodecyl sulfate slab gel electrophoresis. Effect on transcription by RNA polymerase in crude extracts of vegetative and sporulating cells. J Biol Chem. 1975 Nov 25;250(22):8824–8828. [PubMed] [Google Scholar]
- Young M. Use of temperature-sensitive mutants to study gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1976 May;126(2):928–936. doi: 10.1128/jb.126.2.928-936.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]