Summary
Eukaryotic genomes are colonized by transposons whose uncontrolled activity causes genomic instability. The piRNA pathway silences transposons in animal gonads, yet how this is achieved molecularly remains controversial. Here, we show that the HMG protein Maelstrom is essential for Piwi-mediated silencing in Drosophila. Genome-wide assays revealed highly correlated changes in RNA polymerase II recruitment, nascent RNA output, and steady-state RNA levels of transposons upon loss of Piwi or Maelstrom. Our data demonstrate piRNA-mediated trans-silencing of hundreds of transposon copies at the transcriptional level. We show that Piwi is required to establish heterochromatic H3K9me3 marks on transposons and their genomic surroundings. In contrast, loss of Maelstrom affects transposon H3K9me3 patterns only mildly yet leads to increased heterochromatin spreading, suggesting that Maelstrom acts downstream of or in parallel to H3K9me3. Our work illustrates the widespread influence of transposons and the piRNA pathway on chromatin patterns and gene expression.
Graphical Abstract
Highlights
► Piwi-RISC guides transcriptional silencing of transposons in trans ► Piwi-mediated silencing triggers H3K9me3 heterochromatin formation ► Maelstrom is required for transcriptional silencing, but not for H3K9 trimethylation ► Transposon silencing by the piRNA pathway broadly affects gene expression
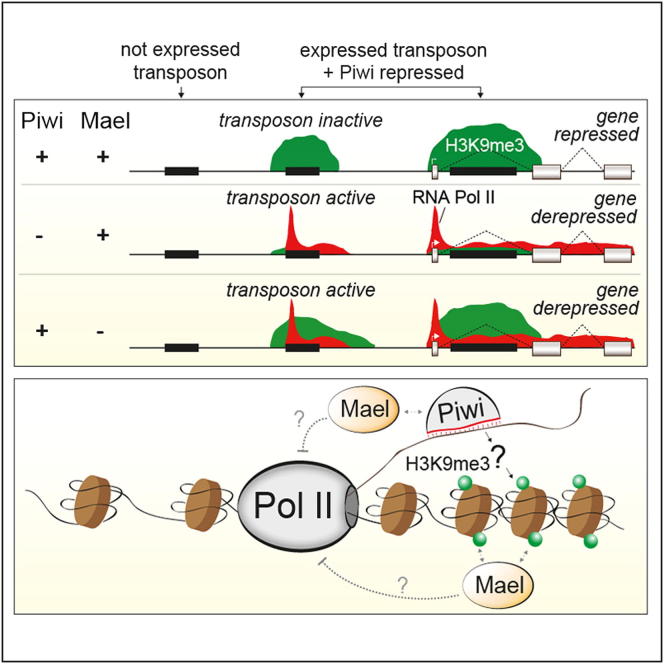
The piRNA-interacting protein Piwi and the HMG protein Maelstrom collaborate genome-wide to silence transcription of transposons in part through trimethylation of H3K9, which depends on Piwi. Silencing broadly affects the expression of genes neighboring the transposons, and Maelstrom restricts spreading of the repressive histone mark.
Introduction
A major selection force during evolution is the maintainance of genomic integrity over generations. Transposable elements (TEs) are threatening genomic stability due to their mobile character and their creating repetitive sequence islands that can initiate ectopic recombination (Kazazian, 2004). Small-RNA-based silencing pathways are universally employed by eukaryotes to silence TEs (Slotkin and Martienssen, 2007). In animals, this is of particular importance in germ cells. The PIWI-interacting RNA (piRNA) pathway serves as the main line of defense in the animal gonad, and defects in it result in TE derepression, genomic instability, and sterility (Malone and Hannon, 2009; Siomi et al., 2011).
At the core of the pathway is the piRNA-induced silencing complex (pi-RISC) that consists of a single-stranded piRNA bound by a PIWI family protein. piRNAs are typically processed from TE RNAs and so-called piRNA cluster transcripts that are enriched in TE sequences. Thus, by virtue of their sequence, piRNAs guide the specific silencing of TEs. (Senti and Brennecke, 2010; Siomi et al., 2011).
Conceptually, two major silencing modes are distinguished, namely transcriptional silencing (TGS) and posttranscriptional silencing (PTGS). Most animals express multiple PIWI proteins, and these might employ different silencing modes. The Drosophila genome encodes the PIWI proteins Piwi, Aubergine (Aub), and Argonaute 3 (AGO3). Aub and AGO3 piRISCs are cytoplasmic, possess slicer activity, and are the major players in a piRNA amplification loop that requires reciprocal cleavage of TE RNAs and piRNA cluster transcripts (Senti and Brennecke, 2010; Siomi et al., 2011). As TE sense RNAs are consumed during this amplification loop, Aub/AGO3-mediated silencing represents a PTGS process.
The third family member Piwi, however, is enriched in the nucleus, and its silencing mode is much less understood. Genetically, Piwi-mediated TE silencing depends on its nuclear localization, but not on its slicer activity (Klenov et al., 2011; Saito et al., 2010). These observations indicate that Piwi might induce TGS via triggering repressive chromatin modifications. Indeed, changes in chromatin marks and nascent RNA levels have been observed for some TEs in piRNA pathway mutants (Klenov et al., 2011; Shpiz et al., 2011; Wang and Elgin, 2011). On the other hand, a specific chromatin association of PIWI proteins in germ cells has not been demonstrated. Some studies even challenge a role for Piwi in chromatin regulation. For example, no significant changes in heterochromatin protein 1 (HP1) occupancy on TEs were observed upon Piwi knockdown (KD) (Moshkovich and Lei, 2010), and a genetic study in flies concluded that Piwi triggers PTGS rather than TGS (Dufourt et al., 2011). A systematic understanding of the silencing mode employed by nuclear PIWI proteins is therefore a major open question in the field.
All three Drosophila PIWI family proteins are coexpressed in germline cells. Due to their interdependence in terms of piRNA biogenesis and TE silencing, the precise genetic and mechanistic dissection of Piwi's nuclear role is challenging. Somatic support cells of the ovary, however, express a simplified piRNA pathway based exclusively on nuclear Piwi. Importantly, a stable cell line derived from ovarian somatic cells (these cultured cells are called OSSs or OSCs) has been established (Niki et al., 2006; Saito et al., 2009). These cells harbor a piRNA pathway that, in every aspect analyzed, mirrors the pathway acting in ovarian somatic cells. In OSCs, piRNAs antisense to TEs are derived from piRNA clusters such as flamenco. piRNA biogenesis depends on several cytoplasmic factors, and defects in it result in loss of Piwi, presumably due to destabilization of unloaded Piwi (Siomi et al., 2011). Upon loss of Piwi-RISC, several TEs, which are normally silenced by the piRNA pathway, are derepressed.
We took advantage of this linear piRNA pathway and dissected the underlying silencing process in detail. Our data demonstrate that Piwi-RISC mediates TE silencing at the transcriptional level and that this is accompanied by local heterochromatin formation. Remarkably, most euchromatic H3K9me3 islands are due to piRNA-mediated silencing of TE insertions, and spreading of this heterochromatic mark into flanking genomic regions has striking effects on the expression of nearby genes.
Results
Maelstrom Is Required for Piwi-Mediated Silencing, but Not for piRNA Biogenesis
While the process of piRNA biogenesis within the somatic pathway is being increasingly dissected at the molecular level and multiple involved factors are known, not a single protein has been linked to Piwi-mediated silencing in the nucleus.
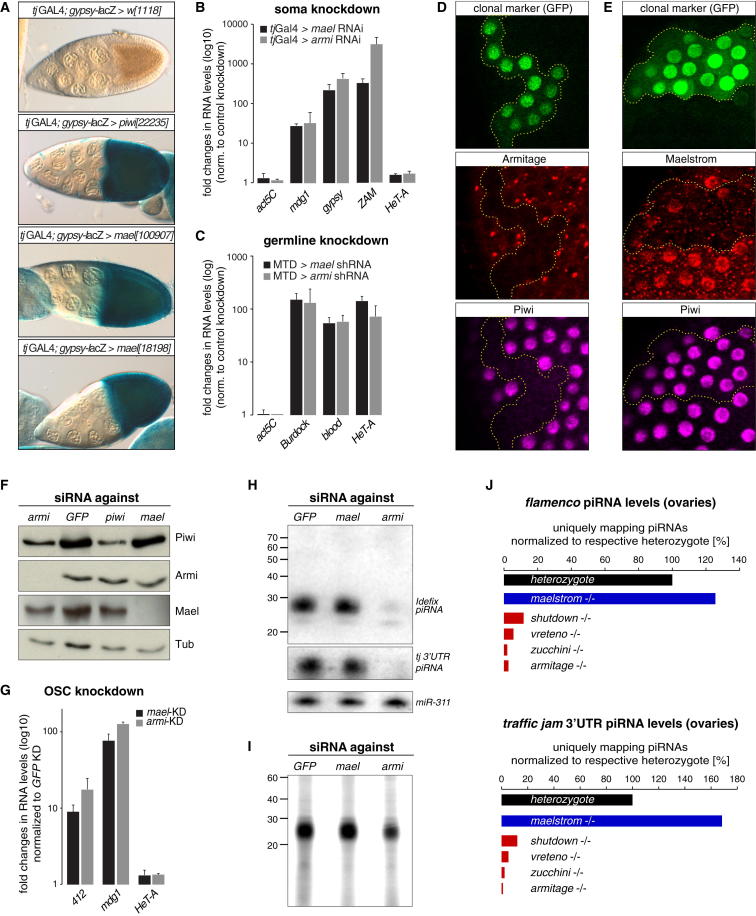
To identify such factors, we utilized an assay system based on transgenic RNAi and a lacZ reporter that monitors silencing of the gypsy TE in follicle cells (Figure 1A; Olivieri et al., 2010; Sarot et al., 2004). The evolutionarily conserved maelstrom (mael) gene scored strongly in this assay (Figure 1A). This came as a surprise, as Mael levels are low in ovarian somatic cells (Findley et al., 2003) and a recent study indicated that mael is dispensable for TE silencing in ovarian somatic cells (Klenov et al., 2011). To support the gypsy-lacZ results, we induced tissue-specific mael RNAi in soma or germline and analyzed RNA levels of several marker TEs. In both cell types, mael KD resulted in desilencing of TEs to extents comparable to KD of the essential piRNA biogenesis factor Armitage (Armi) (Figures 1B and 1C). Ovaries from mael loss-of-function flies also exhibited derepression of soma and germline transposons (Figure S1A available online; note that Klenov et al. [2011] did not use mael null alleles).
Figure 1.
Maelstrom Is Required for Piwi-Mediated Silencing, but Not for piRNA Biogenesis
(A) Shown are β-gal stainings of egg chambers as readout for gypsy silencing. w[1118] flies or indicated VDRC lines for piwi or mael were crossed to tj-GAL4, gypsy-lacZ flies carrying a restrictive flamenco background.
(B and C) Displayed are fold changes in steady-state RNA levels of indicated TEs in mael or armi KD ovaries (B, soma; C, germline). Values are averages of three biological replicates (error bars represent SD) and are normalized to control knockdowns.
(D) Confocal section of a follicular epithelium stained for Piwi (magenta) and Armi (red) in which armi-RNAi has been clonally activated (marked by GFP; boundary indicated by dashed yellow line).
(E) Confocal section of a follicular epithelium stained for Piwi (magenta) and Mael (red) in which mael-RNAi has been clonally activated (marked by GFP; boundary indicated by dashed yellow line).
(F) Western blot showing protein levels of Piwi, Armi, Mael, and Tubulin in OSCs transfected with indicated siRNAs.
(G) Fold changes in steady-state RNA levels of indicated TEs from OSCs transfected with siRNAs against indicated gene (normalized to siGFP; n = 3; error bars represent SD.)
(H) Northern blots showing levels of indicated piRNAs in total RNA from OSCs transfected with siRNAs against indicated genes. To the left, an RNA size marker (nt) is shown. The miR-311 blot serves as loading control.
(I) Radiogram of polyacrylamide gel in which Piwi-bound small RNAs (CIP-kinase labeled) isolated from equal amounts of OSCs transfected with indicated siRNAs were separated. RNA size marker (nt) is indicated to the left.
(J) Shown are levels of indicated piRNA populations from ovaries of indicated genotypes (normalized to sequenced miRNAs and to respective heterozygotes). Contrasted are piRNA levels from mael−/− ovaries (blue) to piRNA levels from ovaries lacking indicated primary piRNA biogenesis factors (red).
See also Figure S1.
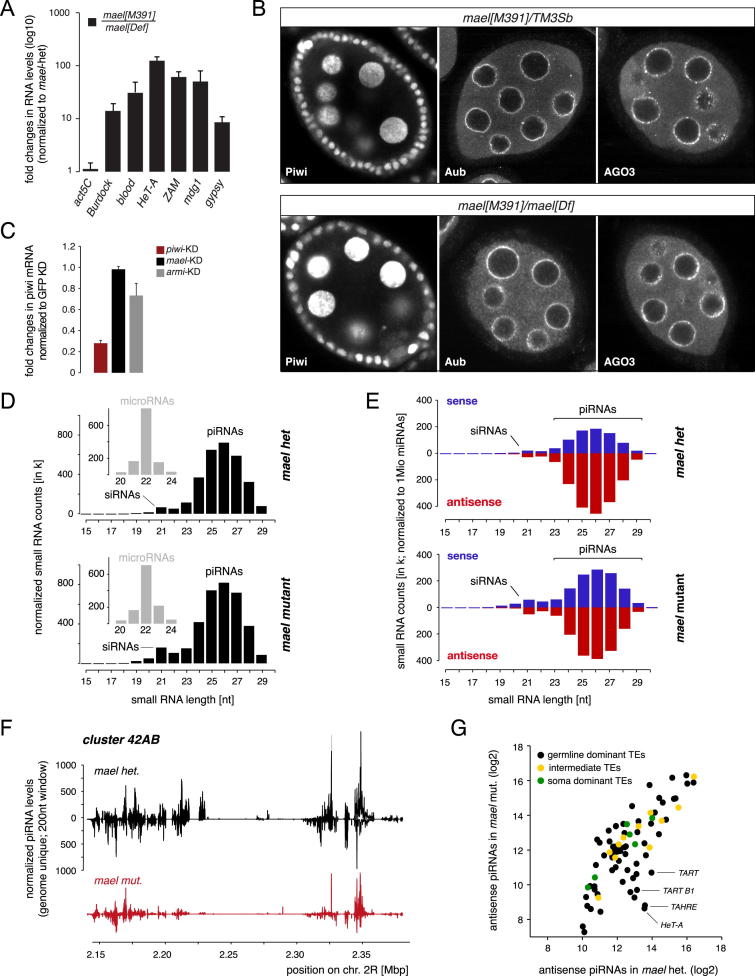
Figure S1.
Related to Figure 1
(A) Displayed are fold changes in steady state RNA levels of indicated TEs in mael null mutant ovaries (normalized to mael heterozygote siblings; values are averages of 3 biological replicates (error bars: StDev.).
(B) Confocal images of mael heterozygous (top) or mael[M391]/mael[Def] (bottom) egg chambers stained for Piwi, Aub or AGO3.
(C) Displayed are fold changes in piwi steady state mRNA levels in OSCs after transfection with indicated siRNAs. Values are averages of 3 biological replicates (error bars: StDev.) and normalized to GFP siRNA treated cells.
(D) Shown are length profiles of small RNAs (normalized to 1 million microRNAs; small insets) isolated from ovaries of mael[M391] heterozygous or mael[391]/mael[Def] flies. siRNA and piRNA populations are indicated.
(E) Shown are length profiles of repeat derived ovarian small RNAs (normalized to 1Mio miRNAs) from mael heterozygous and mael[M391]/mael[Def] flies. (red antisense; blue sense). (F) Normalized piRNA profiles (sense up; antisense down; 200nt windows) from mael het. (black) or mael mut. (red) libraries mapping uniquely to the 42AB piRNA cluster.
(G) Scatter plot (log2 scale) showing levels of antisense piRNAs mapping to soma dominant (green), intermediate (yellow) or germline dominant (black) TEs in mael het. or mael mut. libraries.
To identify the level at which Mael acts in the piRNA pathway, we monitored Piwi in clones of mael KD cells in the follicular epithelium. Defective piRNA biogenesis (e.g., armi KD) triggers loss of Piwi, presumably as unloaded Piwi is unstable (Figure 1D). In contrast, depletion of Mael had no impact on nuclear Piwi levels (Figure 1E). Similarly, levels and localizations of all PIWI proteins were unaffected in soma and germline of mael null ovaries (Figure S1B). This suggested that Mael does not act in piRNA biogenesis. To test this, we monitored TE expression and piRNA levels in OSCs upon mael KD or armi KD (Figures 1F–1I). Both KDs resulted in derepression of the TEs mdg1 and 412, but not of the germline-specific element HeT-A (Figure 1G), demonstrating an essential role for Mael in the OSC piRNA pathway. However, whereas loss of Armi resulted in reduced Piwi protein (Figure 1F; but not mRNA: Figure S1C) as well as in reduced piRNA levels (Figure 1H), loss of Mael did not. The size of Piwi-bound piRNAs was also unaffected upon mael KD (Figure 1I).
We finally sequenced piRNAs from mael mutant ovaries and compared them to heterozygous controls. In agreement with the OSC data and in contrast to known primary biogenesis factors, loss of Mael did not affect piRNAs derived from the soma-dominant flamenco cluster or the traffic jam 3′UTR (Figure 1J). For the global pool of ovarian piRNAs (soma and germline), we observed a slight shift toward sense piRNAs, probably due to abundant derepressed TE messages (Figures S1D and S1E). piRNAs derived from the germline-dominant 42AB cluster were moderately reduced (Figure S1F). At the level of most individual TEs, loss of Mael had only mild impacts on antisense piRNA populations from soma-dominant, intermediate, and many germline-dominant TEs (Figure S1G). The most notable exceptions were the telomeric TEs HeT-A, TAHRE, and TART that exhibited strong piRNA losses. We speculate that desilencing of these TEs interferes with piRNA precursor transcription at the same loci, therefore blocking piRNA production.
Taken together, Mael is not required for biogenesis or nuclear accumulation of the Piwi-RISC yet is essential for Piwi-mediated TE silencing.
Piwi/Mael-Mediated Silencing Is a Nuclear Process
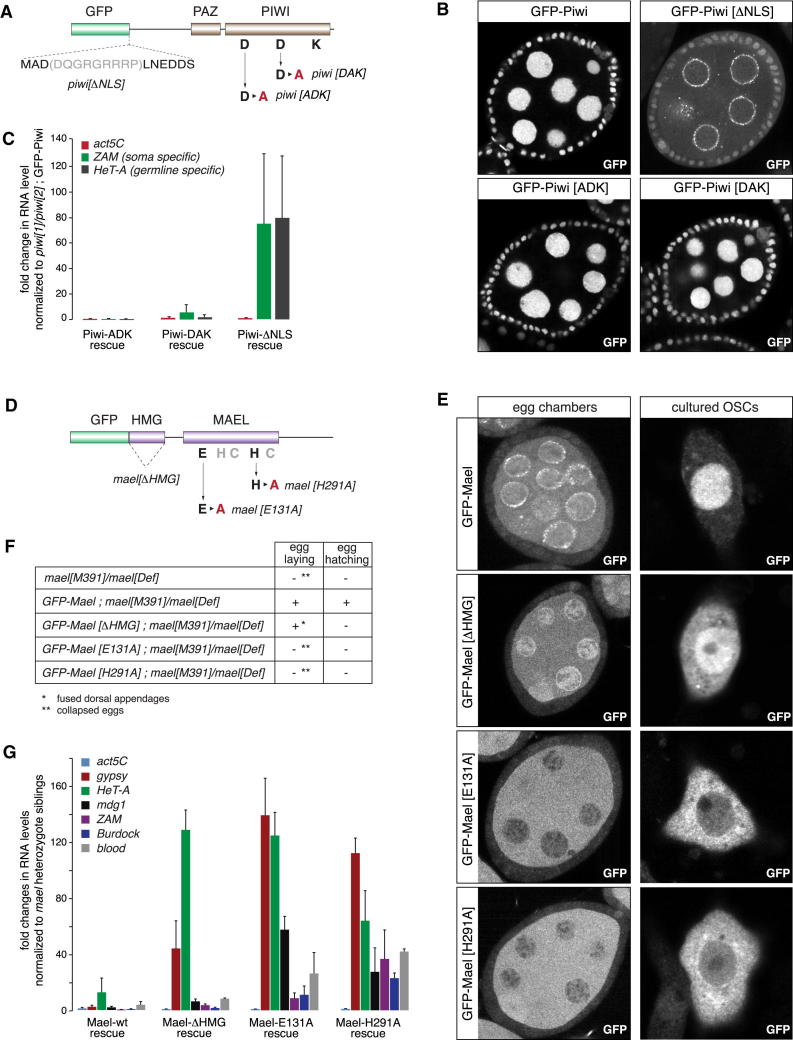
The subcellular localization of the Piwi-RISC suggests a nuclear silencing process. Indeed, experiments in OSCs indicated that Piwi's nuclear localization, but not its slicer activity, is required for silencing (Saito et al., 2010). Also in flies, N terminally truncated Piwi is cytoplasmic and piwi[ΔN] flies are defective in TE silencing (Klenov et al., 2011). We reconstructed these findings in vivo by complementing piwi[1]/piwi[2] mutant flies with various GFP-tagged genomic piwi rescue constructs. A nine-amino-acid deletion at the N terminus (ΔNLS) largely prevented nuclear accumulation of Piwi-GFP, whereas both slicer mutant GFP-Piwis (ADK or DAK) localized like wild-type GFP-Piwi to the nucleus (Figures 2A and 2B; efficient loading of all variants with piRNAs verified by IP-CIP-kinase experiments). Real-time quantitative PCR (RT-qPCR) analysis of TE RNA levels showed derepression of soma- and germline-specific TEs in piwi[ΔNLS], but not in piwi[ADK] or piwi[DAK] ovaries (Figure 2C). Moreover, both slicer mutant flies resembled wild-type flies in fertility, whereas only some eggs laid by piwi[ΔNLS] flies developed into larvae and adults.
Figure 2.
Piwi/Mael-Mediated TE Silencing Is a Nuclear Process
(A) Cartoon showing protein domains of GFP-tagged Piwi and the residues that have been altered for the mutant analysis.
(B) Confocal images of egg chambers expressing the indicated GFP-Piwi constructs in piwi[1]/piwi[2] background.
(C) Fold changes in steady-state RNA levels of actin5C and indicated TEs from ovaries expressing indicated GFP-Piwi constructs in piwi[1]/piwi[2] background (normalized to piwi[1]/piwi[2]; GFP-Piwi; n = 3; error bars represent SD).
(D) Cartoon showing protein domains of GFP-tagged Mael and the residues that have been altered for the mutant analysis.
(E) Confocal images of egg chambers (left) or OSCs (right) expressing the indicated GFP-Mael constructs (GFP) in wild-type background.
(F) Summary of the rescue experiments (egg laying and egg hatching) with indicated GFP-Mael constructs in mael[391]/mael[Def] background.
(G) Fold changes in steady-state RNA levels of actin5C and indicated TEs from ovaries expressing indicated GFP-Mael constructs in mael[391]/mael[Def] background (n = 3; error bars represent SD).
An involvement of Mael in the silencing process predicts a nuclear localization for this protein. In ovaries, endogenous Mael, as well as GFP-tagged Mael expressed under the mael control regions, is abundant in germline cells and localizes to cytoplasm, nuage, and nucleus (Figures 2D and 2E, upper left; Findley et al., 2003). As levels in follicle cells were low, we turned to OSCs in which endogenous Mael, as well as N- and C-tagged GFP-Mael, localized throughout the cell but were clearly enriched in the nucleus (Figure 2E, upper right; data not shown). Based on Mael's domain architecture, we tested the requirement of HMG and MAEL domains for nuclear localization and TE silencing in complementation assays using mael loss-of-function alleles and GFP-tagged mael rescue constructs. Whereas the wild-type construct rescued sterility and TE derepression nearly completely, two constructs harboring point mutations in conserved residues of the MAEL domain (Zhang et al., 2008a) did not (Figures 2F and 2G). In both cases, nuclear accumulation of mutant Mael was strongly reduced in ovaries and OSCs (Figure 2E). Loss of the HMG domain had only mild effects on Mael's subcellular localization (Figure 2E). mael[ΔHMG] flies did lay eggs, but these displayed defects in egg asymmetry, presumably as TE silencing was only partially rescued in these flies (Figures 2F and 2G).
We conclude that Piwi-mediated silencing is a nuclear process that is independent of Piwi's slicer activity but requires Mael and, in particular, its MAEL domain.
Piwi Silences TEs at the Transcriptional Level in a Mael-Dependent Manner
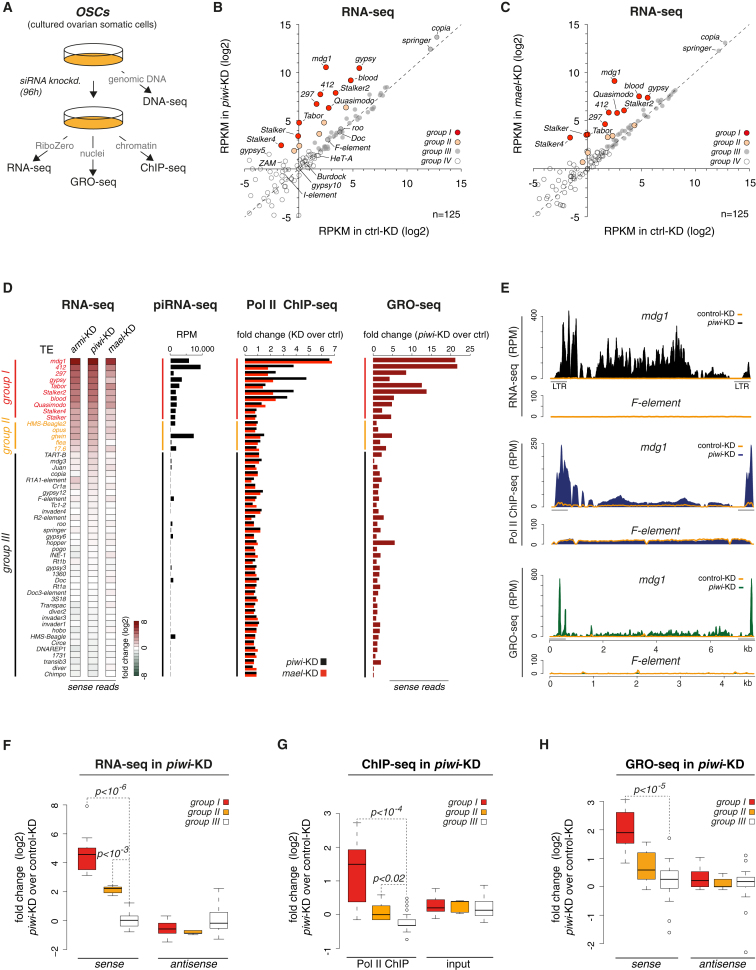
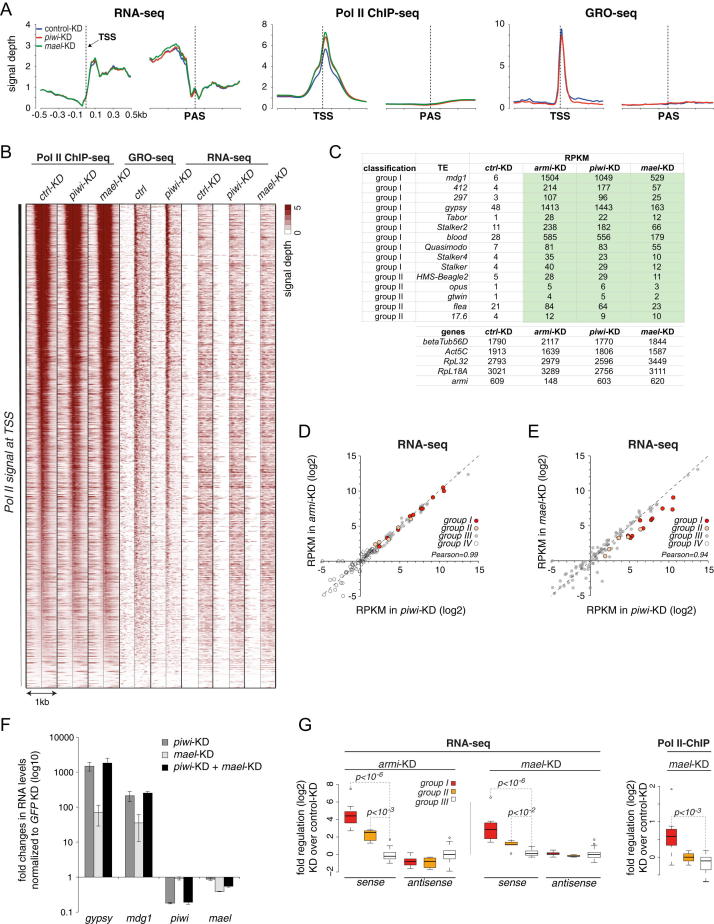
To dissect at which step of TE expression Piwi mediates silencing, we took advantage of cultured OSCs. These cells express a functional linear piRNA pathway and allow gene knockdowns using siRNAs. We profiled gene expression at three hierarchical levels in cells treated with GFP siRNAs (control KD) or with siRNAs targeting key pathway factors (piwi KD, armi KD, mael KD; Figure 3A). We first defined the set of TEs that are repressed by the piRNA pathway by comparing steady-state RNA levels (RNA-seq) between control KD and piRNA pathway KD cells. We then determined transcription rates by measuring RNA polymerase II (Pol II) occupancy (Rpb3 chromatin immunoprecipitation sequencing (ChIP-seq); Adelman et al., 2005) and nascent RNA polymerase output via global run-on sequencing (GRO-seq; Figures S2A and S2B; Core et al., 2008).
Figure 3.
The piRNA Pathway Silences TEs at the Transcriptional Level
(A) Experimental scheme of genome-wide profiling experiments performed for this study.
(B) Scatter plot of RPKM values (log2) for all TEs (n = 125) in GFP (control) or piwi knockdown samples based on RNA-seq. Four TE groups are color coded.
(C) Scatter plot of RPKM values (log2) for all TEs (n = 125) in GFP (control) or mael knockdown samples based on RNA-seq. Four TE groups are color coded.
(D) Displayed are fold changes of TE expression (groups I–III; colors as in B) in OSCs transfected with indicated siRNAs (normalized to control cells) at the level of steady-state sense RNA (RNA-seq; heatmap), Pol II occupancy (ChIP-seq), or nascent sense RNA (GRO-seq). The piRNA-seq diagram indicates Piwi-bound piRNA levels mapping antisense to indicated TEs.
(E) Density profiles of normalized reads from RNA-seq (top), Pol II ChIP-seq (middle), and GRO-seq (bottom) experiments on mdg1 (group I) and F-element (group III). Orange line indicates levels in control cells, and solid signal indicates levels in piwi KD cells.
(F–H) Box plots showing fold changes (log2) in the expression of group I, group II, and group III TEs based on RNA-seq (F), Pol II ChIP-seq (G), or GRO-seq (H) upon piwi KD (compared to control; p values based on Wilcoxon rank-sum test). Box plots show median (line), 25th–75th percentile (box) ± 1.5 interquartile range; circles represent outliers. Contrasted are sense and antisense reads (RNA-seq and GRO-seq) and IP versus input (Pol II ChIP-seq).
See also Figure S2.
Figure S2.
Related to Figure 3
(A) Metagene profiles of normalized RNA-seq, Pol II ChIP-seq and GRO-seq reads around (±0.5 kb) transcriptional start site (TSS) and polyadenylation site (PAS) from indicated siRNA mediated knockdowns (GFP: blue; piwi: red; mael: green). Only genes meeting the following criteria were used: RNA-seq RPKM ≥ 5, length > 1 kb, no overlaps with flanking gene up to 200 nt upstream of annotated TSS; n = 2628; note the strong TSS bias for Pol II occupancy and GRO-seq; note also that only RNA-seq shows the expected drop at the PAS, while Pol II occupancy and GRO-seq do not as expected as they continue to transcribe downstream of the PAS.
(B) Heatmap showing TSS data presented in (A) at single gene-resolution. Profiles were sorted for decreasing signal of Pol II ChIP-seq in control knockdowns.
(C) Table listing RNA-seq RPKM values for indicated TEs (upper part) or a set of highly expressed genes (lower part) upon indicated siRNA knockdowns in OSC.
(D) Scatter plot showing RNA-seq RPKM values (log2) of group I-IV TEs in armi KD versus piwi KD OSCs.
(E) Scatter plot showing RNA-seq RPKM values (log2) of group I-IV TEs in mael KD versus piwi KD OSCs.
(F) Displayed are fold changes in steady state RNA levels of indicated TEs and genes upon piwi KD or mael KD or piwi+mael KD in OSCs. Note that the gypsy primer pair spans a splice junction, which explains the higher de-repression values compared to the RNA-seq data. Values are averages of 3 biological replicates (error bars: StDev.) and normalized to GFP siRNA treated cells.
(G) Box plot analysis indicating fold changes (log2) in RNA-seq RPKM values (left) or Pol II occupancy values (right) for indicated TE groups in armi KD or mael KD cells; for the RNA-seq analysis, reads mapping sense or antisense to TEs were contrasted; p-values were computed with Wilcoxon rank-sum test. Box plots show median (line), 25th–75th percentile (box) ± 1.5 interquartile range; circles represent outliers.
To determine steady-state RNA levels, we sequenced total RNA after removal of ribosomal RNA. Reads per kilobase per million mapped reads (RPKM) values for annotated genes were highly correlated between piwi KD and control KD cells (Pearson correlation coefficient 0.95). In contrast, several out of the 125 annotated D. melanogaster TE families showed strong increases in RNA levels (Figure 3B). For example, the LTR elements mdg1 or gypsy increased by >200- or >30-fold upon piwi KD, respectively. With RPKM values of >1,000, both TEs were among the most abundant coding transcripts in OSCs (Figure S2C). Almost identical results were obtained upon knockdown of Armi (Figure S2D; Pearson correlation coefficient piwi KD/armi KD 0.99). Thus, loss of the Piwi-RISC led to highly reproducible increases in the RNA levels of a subset of TEs.
Based on these data, we classified TEs into four groups (Figure 3B). Group I elements exhibited RNA increases >10-fold; group II elements. 3- to 10-fold; group III elements, <3-fold; and group IV elements were expressed below an RPKM cutoff of five in any of the analyzed libraries (no or very low expression and not further analyzed). Knockdown of Mael resulted in slightly weaker but otherwise very similar derepression of TEs at the RNA level (Figures 3C and S2E). This strongly supports the notion that Mael is an integral piRNA pathway factor. Indeed, piwi+mael double-KD cells exhibited TE derepression similar to piwi KD cells, indicating that both proteins act in the same pathway (Figure S2F).
Strikingly, changes in steady-state RNA levels were highly correlated with changes in RNA Pol II occupancy (Pol II ChIP-seq; piwi KD and mael KD), as well as with changes in nascent RNA levels (GRO-seq; piwi KD; Figure 3D). Upon piRNA pathway KD, both measures indicative of active transcription were strongly increased in the LTR regions (containing the TSS), as well as in the internal portions of regulated TEs, but not of nonregulated TEs (Figure 3E). For example, overall Pol II occupancy and nascent RNA levels for mdg1 increased 6.5-fold and 22-fold, respectively. For all three assays, the respective controls (antisense reads for RNA-seq; input for Pol II ChIP-seq; antisense reads for GRO-seq) showed no difference among the three TE groups (Figures 3F–3H and S2G). This argues against a general increase in repeat or heterochromatin transcription upon pathway loss. Moreover, an analysis of the RNA-seq, Pol II ChIP-seq, and GRO-seq data on genes indicated no global alterations in gene expression upon piRNA pathway loss (Figure S2B).
Of note, TE antisense piRNA levels were typically very high for the set of regulated elements (group I and II) but mostly absent for the nonregulated yet expressed TEs (group III) (Figure 3D). Despite being expressed, group III elements can therefore not be regulated by the pathway, simply because no guide RNAs exist in Piwi-RISC.
We conclude that Piwi/Mael-mediated silencing acts predominantly or even exclusively at the transcriptional level by blocking Pol II recruitment and therefore transcription.
Piwi Mediates Transcriptional Silencing of TEs In Vivo in a piRNA-Dependent Manner
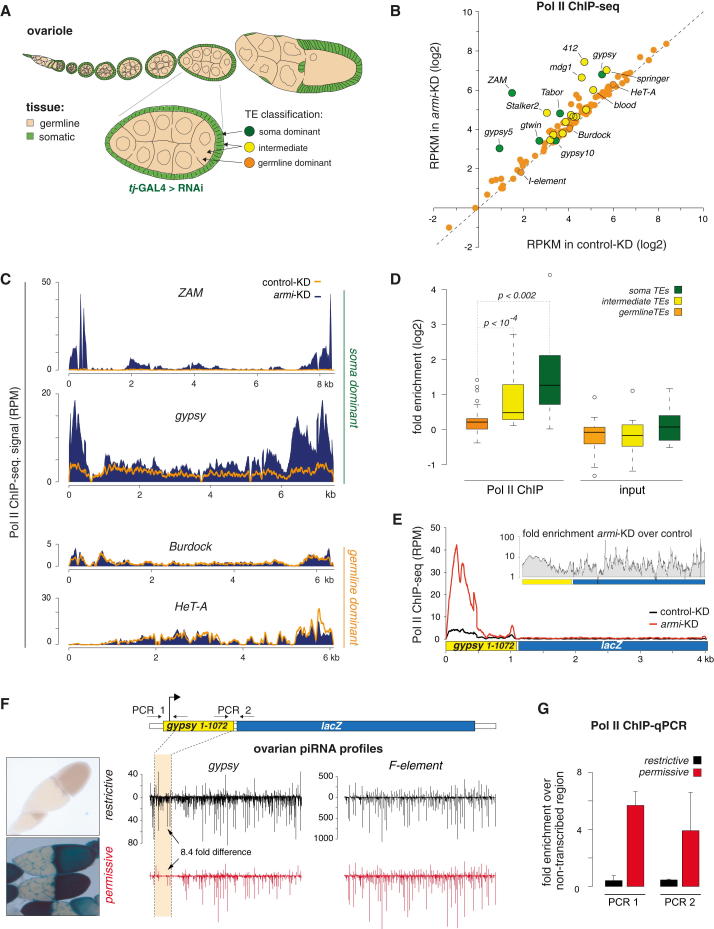
OSCs have been cultured for several years in the laboratory, and we therefore wanted to test whether Piwi guides transcriptional silencing (TGS) also in flies. We knocked down the piRNA pathway specifically in ovarian somatic cells (Figure 4A) and determined Pol II occupancy on chromatin from dissected ovaries. The tested genotypes were armi KD (piwi KD results in rudimentary ovaries) and tejas KD (control; dispensable for the somatic piRNA pathway; Patil and Kai, 2010). The Pearson correlation coefficient between both knockdowns on annotated genes was 0.99, indicating that the piRNA pathway has no general impact on gene transcription.
Figure 4.
Piwi-RISC Mediates TGS of TEs in Ovarian Somatic Cells
(A) Scheme of a Drosophila ovariole and an individual egg chamber (somatic cells in green, germline cells in beige). Indicated is the classification of TEs according to Malone et al. (2009).
(B) Scatter plot of Pol II ChIP-seq RPKM values (log2) for all TEs (n = 125; color code as in A) from control KD ovaries (tj-GAL4 > RNAi tej) versus armi KD ovaries (tj-GAL4 > RNAi armi).
(C) Density profiles of normalized Pol II ChIP-seq reads on ZAM and gypsy (soma dominant) and Burdock and HeT-A (germline dominant). Orange line indicates levels in control, and solid signal indicates levels in armi KD ovaries.
(D) Box plots indicating fold enrichments (log2) of Pol II ChIP-seq reads on TEs belonging to the indicated classes. Contrasted are IP (Pol II) versus input (p values based on Wilcoxon rank-sum test). Box plots are as in Figure 3.
(E) Normalized Pol II ChIP-seq read density on the gypsy-lacZ reporter in control ovaries (black line) versus armi KD ovaries (red line). Small inset displays the fold change (armi KD versus control) of Pol II occupancy on the reporter.
(F) Shown to the left are β-gal stainings of egg chambers from gypsy-restrictive ovaries (top) and gypsy-permissive ovaries (bottom) harboring the gypsy-lacZ reporter (Sarot et al., 2004). In the center, piRNA levels (black, restrictive strain; red, permissive strain) mapping to the indicated TEs (sense up, antisense down; normalized to 1Mio miRNAs) are displayed, and the portion of gypsy present in the gypsy-lacZ reporter (cartoon at top) is indicated.
(G) Shown is the Pol II ChIP-qPCR analysis on the gypsy-reporter (primers 1 and 2 indicated in F) in ovaries from restrictive versus permissive strains (enrichments calculated over intergenic region; n = 3; error bars represent SD.).
See also Figure S3.
In contrast, Pol II occupancy increased significantly for several TE families upon armi KD (Figure 4B). These were nearly exclusively classified as soma-dominant or intermediate TEs (Malone et al., 2009) that are targeted by ovarian somatic piRNAs (Figures 4A and 4B; note that the TE expression pattern differs between follicle cells and OSCs). With more than 20-fold, the retroelement ZAM showed the strongest increase, in agreement with it being one of the most strongly repressed elements in follicle cells (Figure 4C; Olivieri et al., 2010). In contrast, Pol II occupancy was unchanged on TEs that are strongly repressed by the pathway in germline cells (Figures 4C and 4D).
The fly strain used for the soma knockdown also harbored a gypsy-lacZ reporter, allowing us to analyze regulation at a single genomic locus (Figure 4E). Pol II occupancy at the gypsy TSS (multicopy), as well as across the lacZ ORF (single copy), increased ∼10 fold upon armi KD. The gypsy-lacZ reporter was integrated into a euchromatic landing site, and its repression depends on flamenco-derived piRNAs antisense to the ∼750 bp gypsy RNA segment (Sarot et al., 2004). Hence, Piwi-RISC mediates TGS in trans.
Loss of the piRNA pathway leads to derepression of many TEs and results in DNA damage and developmental aberrations. To exclude that these defects underlie the loss of TE repression, we took advantage of two fly strains that carry the identical gypsy-lacZ reporter but differ in their silencing ability due to differences in their gypsy piRNA repertoire (Figure 4F; Sarot et al., 2004). Both strains are devoid of active gypsy TEs and are fertile. We confirmed strongly reduced piRNA levels (8.4-fold) antisense to the gypsy portion of the reporter in the permissive versus the restrictive strain by small RNA sequencing from ovaries (Figure 4F; miRNA normalized; F-element piRNAs serve as control). Based on ChIP-PCR experiments, Pol II occupancy on the reporter was 4- to 6-fold higher in permissive versus restrictive strains (Figure 4G; primers specific to the reporter construct).
Taken together, Piwi represses transcription of TEs in flies, and piRNAs antisense to the target are required for this silencing process.
Piwi-Mediated TGS Induces Formation of H3K9me3 Heterochromatin
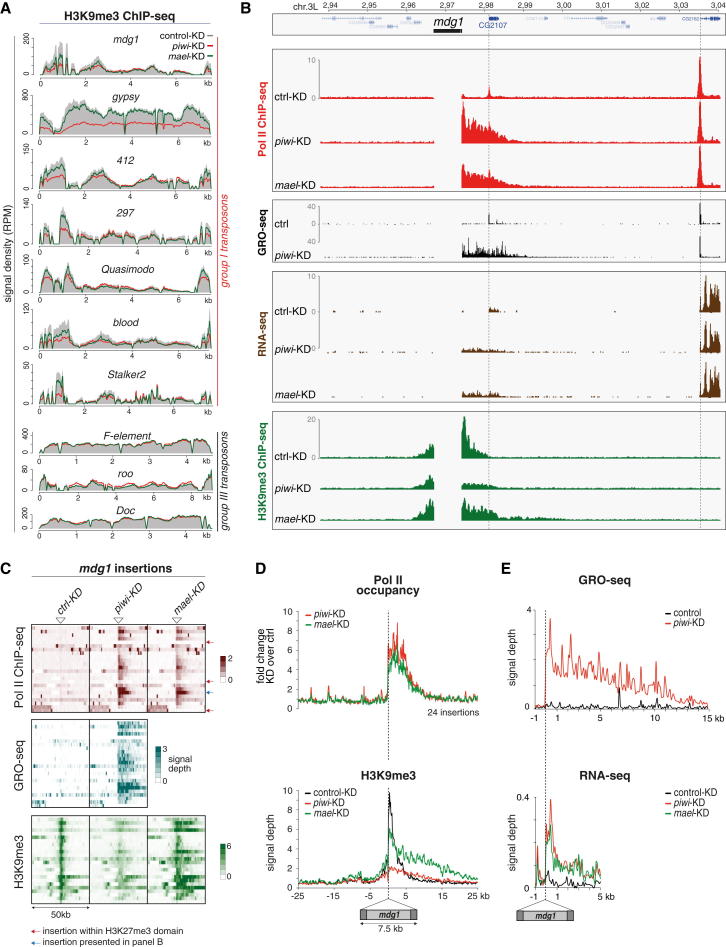
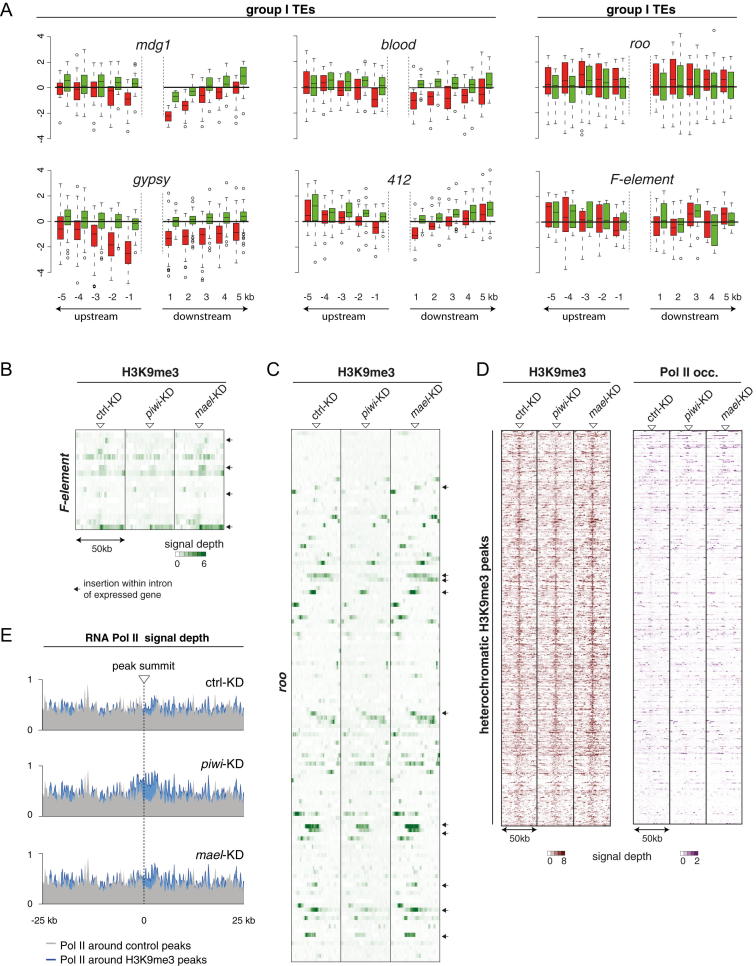
To explore the basis of Piwi-mediated TGS, we reasoned that alterations in TE promoter accessibility via chromatin modifications are the most likely scenario. We focused on the major repressive histone marks H3 lysine 9 trimethylation (H3K9me3) and H3 lysine 27 trimethylation (H3K27me3). Using ChIP-qPCR, we found lower levels for H3K9me3 on mdg1 or gypsy upon piwi KD. We therefore profiled H3K9me3 patterns genome-wide in OSCs upon control KD, piwi KD, or mael KD. ChIPseq reads mapping to TE consensus sequences indicated a significant decrease in H3K9me3 marks upon Piwi loss for group I TEs (repressed elements), but not for group III TEs (Figure 5A). Of note, H3K9me3 levels did drop to a much milder extent upon mael KD. It is important to note that this analysis combines all genomic instances of a certain TE, irrespective of whether they are full length or not, active or inactive, and irrespective of their insertion into hetero- or euchromatin. To allow a more meaningful analysis of the data, we therefore explored the possibility of whether individual TE insertions could be analyzed by virtue of their flanking and therefore unique genomic surroundings.
Figure 5.
Loss of Piwi, but Not Mael, Leads to Decreased H3K9me3 on TEs
(A) Profiles of normalized H3K9me3 signals on indicated TE consensus sequences (group I versus group III) in piwi KD cells (red line), mael KD cells (green line), and GFP KD cells (solid gray).
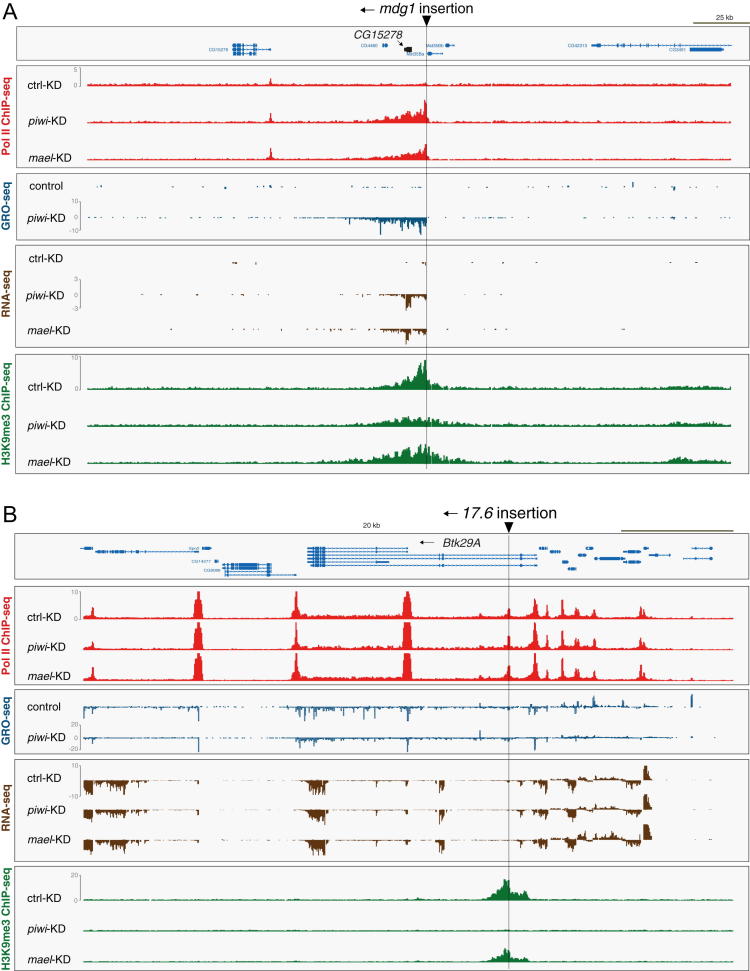
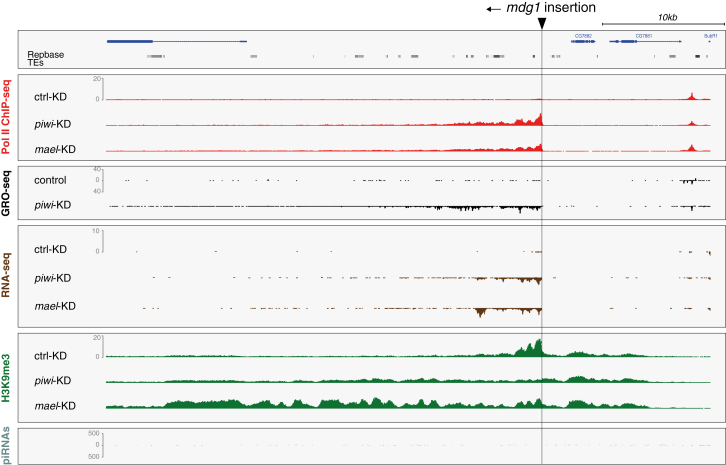
(B) Profiles of transcriptional activity (Pol II ChIP-seq, GRO-seq, RNA-seq) and H3K9me3 density of OSCs after indicated knockdowns (left) in a 100 kb window (chr. 3L; 63E) that contains an mdg1 insertion (only genome unique reads are displayed).
(C) Heatmaps presenting Pol II occupancy, GRO-seq signal, and H3K9me3 ChIP-seq signal within 50 kb windows centered on all 24 euchromatic mdg1 insertions (oriented 5′ to 3′) in control, piwi, or mael KD cells. Red arrows mark insertions within H3K27me3 domains; the insertion marked by the blue arrow is the one displayed in (B).
(D and E) Metaplots for the genomic regions flanking the average euchromatic mdg1 insertion; displayed are changes in Pol II occupancy (D), normalized H3K9me3 signal (D), normalized GRO-seq signal (E), and normalized RNA-seq signal (E) in control KD (black), piwi KD (red), or mael KD cells (green).
See also Figure S4.
Indeed, an annotated mdg1 insertion at position 63E showed remarkable patterns in the regions flanking the insertion (Figure 5B). The insertion is in the genomic sense orientation and ∼7 kb away from the next expressed gene (CG2107). Strikingly, upon piwi or mael KD, Pol II occupancy (red), nascent RNA levels (black), and steady-state RNA levels (brown) were all strongly increased, exclusively downstream of the insertion site. Thus, derepression of this mdg1 insertion leads to pronounced bleeding of Pol II into the flanking ∼15 kb.
Remarkably, in control KD cells, the mdg1 insertion was flanked by a ∼12 kb domain of strong H3K9me3 signal with summits right at the insertion site, very indicative of heterochromatin spreading (Figure 5B). Whereas piwi or mael KDs increased transcription in a very similar manner, changes in H3K9me3 patterns differed significantly between the two knockdowns. Loss of Piwi triggered a strong reduction of the H3K9me3 signal downstream of the insertion. Loss of Mael instead led to only a modest reduction at the insertion site but resulted in increased H3K9me3 spreading downstream of it.
We wanted to extend this analysis to the entire set of mdg1 insertions. The assembled D. melanogaster genome contains 17 full-length mdg1 insertions. But aside from the 63E insertion, only two others displayed the above-described impacts on flanking regions. We speculated that differences in TE insertion sites between the OSC genome and the reference genome might underlie this discrepancy and therefore sequenced OSC genomic DNA to call TE insertions de novo. This identified 24 mdg1 insertions within euchromatic areas, only three of which were shared with the reference genome (for definition of euchromatin, see Table S6). Nearly all OSC mdg1 insertions exhibited the characteristic changes in transcription and H3K9me3 patterns upon piRNA pathway loss (Figure 5C). Based on all 24 insertions, we calculated signals for Pol II occupancy, GRO-seq, RNA-seq, and H3K9me3 in the regions flanking the average mdg1 insertion (Figures 5D and 5E). This “meta-mdg1” analysis confirmed that loss of Piwi or Mael resulted in highly directional bleeding of transcription up to ∼15 kb downstream of the insertion. It also re-emphasized the different impacts that piwi or mael KDs have on H3K9me3 patterns: Piwi loss resulted in severely reduced H3K9me3 signals, whereas Mael loss triggered a modest decrease only right downstream of the insertion site but led to increased H3K9me3 spreading. Similar results were obtained by calculating changes in H3K9me3 for all 24 insertions in 1 kb bins around the insertion site (Figure S3A). Given that extent and asymmetry of H3K9me3 spreading mirrored the increases in transcription, we propose that Pol II activity facilitates H3K9me3 spreading. Indeed, even upon Piwi loss, the slope of the residual H3K9me3 signal was shallower than in control cells. This likely also explains the asymmetric H3K9me3 distribution around mdg1 insertions in control cells, as even here, a low level of Pol II occupancy is detected right downstream of the insertion site (e.g., Figure 5B).
Figure S3.
Related to Figure 4
(A) Box-plots showing the changes of the H3K9me3 ChIPseq levels in 1kb bins for the flanking genomic regions (upstream and downstream 5 kb) of mapped insertions of the indicated TEs in piwi (red) or mael (green) knockdowns compared to GFP control KDs. All TEs except F-element and roo (group III) belong to the piRNA-regulated group I.
(B) Heatmap showing distribution of H3K9me3 density in genomic regions (50kb windows) around all euchromatic insertions of F-element. Black arrows indicate insertions located in sense orientation within a transcriptionally active unit.
(C) Heatmap showing distribution of H3K9me3 density in genomic regions (50kb windows) around all euchromatic insertions of roo. Black arrows indicate insertions located in sense orientation within a transcriptionally active unit.
(D) Heatmaps showing signal of H3K9me3 (left) or Pol II occupancy (right) in 50kb windows centered on heterochromatic H3K9me3 peaks (n = 655) in control KD, piwi KD or mael KD cells. Only genome unique reads are displayed.
(E) Shown are average profiles of normalized Pol II ChIP-seq signals in the 50kb regions around all euchromatic peaks of H3K9me3 (blue, n = 466) or randomly chosen peaks (gray, n = 466) in OSCs upon indicated knockdowns. Pol II occupancy is not lower in the vicinity of H3K9me3 peaks.
Taken together, an analysis of individual TE insertions strongly supports the TGS model, argues for an integral role of H3K9me3 in the silencing process, and suggests that Mael acts in a silencing process downstream of Piwi and H3K9me3 establishment.
The Widespread Impact of TEs and the piRNA Pathway on H3K9me3 Domains
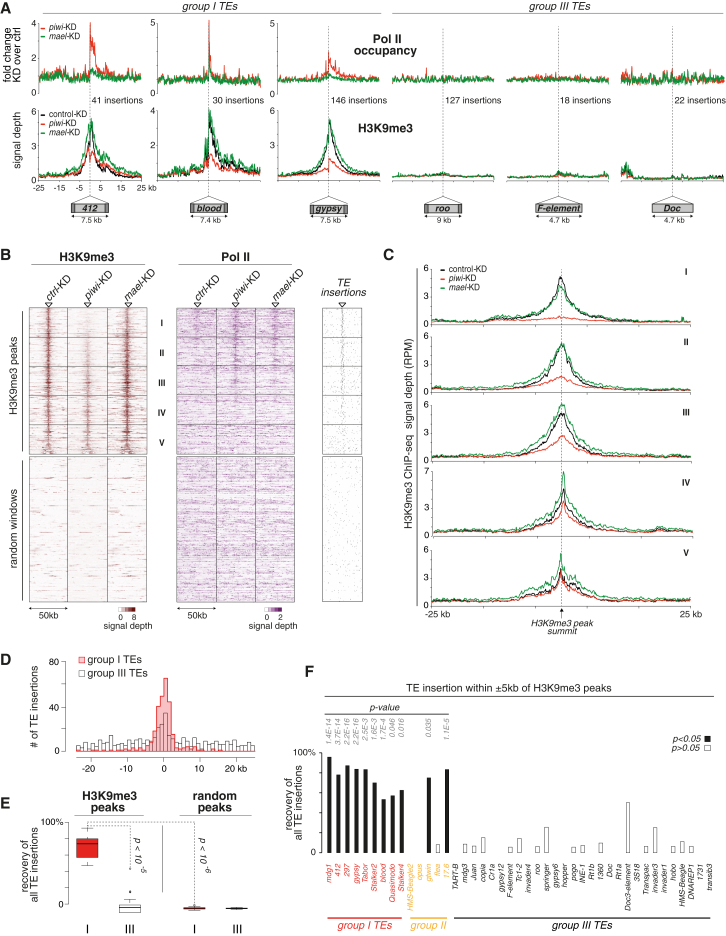
We extended the metatransposon analysis to other group I (Piwi-repressed) and group III TEs (not repressed) (Figure 6A). Similar to mdg1, transcription increased significantly downstream of 412, blood, or gypsy insertions. Increases were stronger in piwi KD than in mael KD cells, and the extent of “bleeding” was element specific. Importantly, no changes were observed downstream of roo, F-element, or Doc insertions (all group III).
Figure 6.
Euchromatic H3K9me3 Islands Correlate with TE Insertions and Depend on Piwi
(A) Metaplots for the genomic regions flanking the average euchromatic insertion of indicated TEs (copy number indicated) belonging to group I or III; displayed are changes in Pol II occupancy (compared to control KD) and normalized H3K9me3 signal in control KD (black), piwi KD (red), or mael KD cells (green).
(B) Heatmaps showing signal of H3K9me3 (left) or Pol II occupancy (right) in 50 kb windows centered on euchromatic H3K9me3 peaks (n = 466) and in a random set of euchromatic 50 kb windows (n = 466) in control KD, piwi KD, or mael KD cells. Windows with euchromatic H3K9me3 peaks were sorted according to the loss of signal in piwi KD cells, and five equally sized bins were defined (I–V). To the right, positions of all TE insertions within the euchromatic H3K9me3 windows are displayed.
(C) Metaplots of H3K9me3 signals (individual plots for the five bins defined in B) for the 50 kb window flanking the average euchromatic H3K9me3 peak in control (black), piwi KD (red), or mael KD cells (green).
(D) Distribution of group I (red) or III (white) TE insertions in the 50 kb windows centered on all H3K9me3 peaks (n = 466).
(E) Percentages of all group I (red) or all group III (white) TE insertions found within 5 kb of euchromatic H3K9me3 summits in comparison to an average and randomly selected set of control regions. Box plots are as in Figure 3.
(F) Percentages of insertions of indicated individual TEs found within 5 kb of euchromatic H3K9me3 summits (p values based on binominal test using random control areas).
All analyzed group I TEs induced H3K9me3 spreading into flanking genomic regions, and these H3K9me3 domains were dependent on Piwi (Figure 6A). Upon mael KD, H3K9me3 at the insertion site was not or was only marginally reduced yet showed again increased asymmetric spreading (Figures 6A and S3A). In contrast, none of the analyzed group III elements triggered formation of H3K9me3 domains in euchromatin (Figure 6A). The average F-element, however, had per copy number an H3K9me3 signal comparable to mdg1 (data not shown). The OSC genome contains >100 F insertions, and only 18 map to euchromatin. We speculate that euchromatic F insertions are transcriptionally inactive (supported by very low RNA-seq RPKM levels) and devoid of H3K9me3 marks (hence no H3K9me3 spreading) and that the high H3K9me3 levels for the average F-element stem from the abundant heterochromatic insertions. A few euchromatic F and roo insertions did, however, trigger H3K9me3 spreading in a Piwi-dependent manner (Figures S3B and S3C). Interestingly, these were typically in sense orientation within introns of transcribed genes and thus provide targets for antisense piRNAs that do exist in OSCs. Thus, Piwi-guided H3K9me3 depends on transcription.
Intrigued by the strong correlation between H3K9me3 and group I TE insertions, we determined all euchromatic H3K9me3 peaks (n = 466; FDR 10−4) and displayed H3K9me3 signals, as well as Pol II occupancy, in a 50 kb window centered on the peak summit in control, piwi, or mael KDs (Figures 6B and 6C). We sorted these peaks according to their loss in H3K9me3 signal upon piwi KD and divided them into five equally sized bins (I–V). Strikingly, piwi KD led to significant reductions in the H3K9me3 signal for most peaks, whereas mael KD did not and instead resulted often in a broadened H3K9me3 domain (Figures 6B and 6C). At the same time, piwi or mael KDs triggered increased Pol II occupancy in proximity to the H3K9me3 summit for those peaks that depended on Piwi (Figure 6B). We compared these results to randomly chosen euchromatic 50 kb windows (Figure 6B) and to all heterochromatic H3K9me3 peaks (n = 655; Figure S3D; analysis based on genome unique reads). Though the repetitive nature of heterochromatin complicates the analysis, Piwi seemingly impacted H3K9me3 patterns predominantly in euchromatic areas. Of note, Pol II occupancy was highly similar in euchromatic H3K9me3 windows and random control windows (Figure S3E), and it was not reduced within H3K9me3 domains compared to their surroundings. Thus, H3K9me3—a mark typically implicated in condensed chromatin state—is compatible with transcription, at least within euchromatic domains.
The data in Figures 6B and 6C suggested that most euchromatic H3K9me3 islands are linked to the piRNA pathway. We investigated whether this correlated with TE insertions or whether this indicated a TE-independent role of Piwi. Strikingly, nearly all (88%) H3K9me3 peaks had a TE insertion within 5 kb up- or downstream of the summit (Figure 6B, right; random expectation: 14%). Sixty-one percent of these insertions belonged to group I TEs (random expectation 11%), and these exhibited a very pronounced enrichment at H3K9me3 summits (Figure 6D). On average, ∼80% of all group I TE insertions were found within 5 kb of H3K9me3 summits compared to only ∼5% of group III TE insertions (Figure 6E; p < 10−5). For example, 95% of mdg1 insertions (n = 24), 87% of gypsy insertions (n = 146), but only 7% of roo insertions (n = 127) were within 5 kb of H3K9me3 summits (Figure 6F). Within random windows, percentages of group I and group III TE insertions were identical (Figure 6E). We noted that group III elements were also slightly enriched at H3K9me3 summits (Figure 6D). As noted above, the underlying insertions mapped often in sense orientation to introns of expressed genes and therefore are targets for piRNA-mediated H3K9 trimethylation.
Taken together, TE-derived transcripts targeted by Piwi-RISC are a major trigger of H3K9 trimethylation in euchromatin. Within constitutive heterochromatin, additional or alternative pathways seem to dominate H3K9me3 patterns, though the repetitive sequence nature complicates data interpretation in these domains.
The Impact of TE Insertions and the piRNA Pathway on Gene Expression
Gene bodies constitute nearly half of D. melanogaster euchromatin, and therefore many piRNA-repressed TE insertions are expected in proximity to expressed genes. By browsing TE insertions in the genome, we found several examples in which piRNA pathway loss led to remarkable changes in the expression of nearby genes.
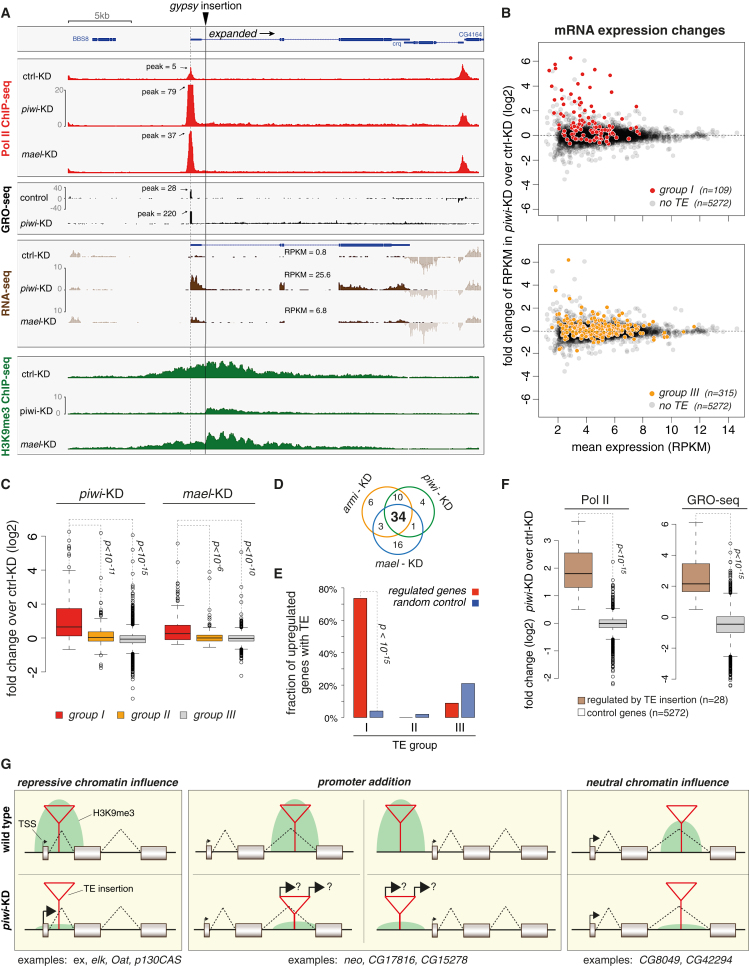
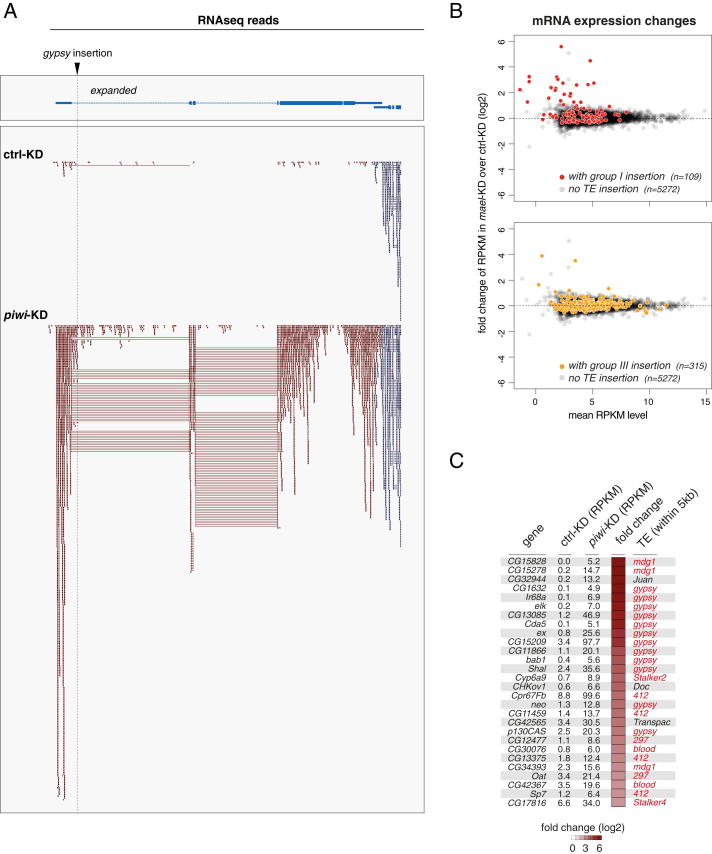
Figure 7A shows the locus of expanded (ex), which harbors an OSC-specific gypsy insertion in its first intron, ∼1.2 kb downstream of the TSS. In control cells, this gypsy insertion triggered H3K9me3 spreading into the surrounding 10–12 kb, covering also the ex TSS. Pol II occupancy at the TSS was detectable albeit weak, and based on RNA-seq, ex was hardly expressed (RPKM <1). Remarkably, upon piwi or mael KD, Pol II occupancy at the ex TSS (as well as within the gene body), nascent RNA levels, and steady-state ex mRNA levels were all strongly increased. An analysis of the RNA-seq reads indicated that the first intron was faithfully spliced despite the gypsy insertion (Figure S4A). Thus, derepression of the ex promoter was causative for the increased expression. Loss of Piwi resulted in a complete loss of H3K9me3 marks upstream of the gypsy insertion exposing the ex TSS (Figure 7A). In contrast, loss of Mael—despite strong increases in ex transcription—had only minor impacts on H3K9me3 patterns. Once again this indicated that Piwi-RISC triggers H3K9me3 independent of Mael, that a Mael-dependent silencing step exists downstream of H3K9me3, and that Pol II recruitment and activity are compatible with a substantial H3K9me3 level.
Figure 7.
The Impact of TEs and the piRNA Pathway on Gene Expression
(A) Transcriptional activity (Pol II ChIP-seq, GRO-seq, RNA-seq) and H3K9me3 density at the expanded locus. OSC-specific insertion site of gypsy indicated with gray line; ex TSS, with dashed line.
(B) Changes in mRNA abundance for the set of expressed genes (RPKM >5 in one library) between piwi KD and control KD cells. The upper plot contrasts genes with no insertion (gray) and genes with an insertion of a group I TE in sense orientation (red). The lower plot contrasts genes with no insertion (gray) and genes with no group I TE but with a group III TE insertion (yellow).
(C) Box plot showing the changes (log2) in RNA-seq RKPM levels for the three gene groups defined in (B) upon knockdown of Piwi (left) or Mael (right).
(D) Venn diagram displaying the number of upregulated genes upon depletion of the indicated piRNA pathway components (criteria: armi and piwi KDs >4-fold, mael KD >2-fold; compared to GFP KD).
(E) Shown is the percentage of all upregulated genes (red) with TE insertions belonging to class I, II, or III in close proximity (± 5 kb) in comparison to random control genes (blue).
(F) Box plots displaying fold changes (log2) in Pol II occupancy (left) or GRO-seq signal (right) upon piwi KD versus GFP KD for upregulated genes with TE insertion (n = 28) versus all expressed genes with no TE insertion (n = 5272; RPKM >5). Box plots are as in Figure 3.
(G) Cartoon showing the three categories of how TE insertions impact gene loci. For category A (repressive chromatin influence), the TE insertion is close to the gene's TSS and results in TSS repression via H3K9me3 spreading. For category B (promoter addition), the TE insertion serves as an ectopic promoter through transcriptional bleeding. For category C (neutral chromatin influence), the TE insertion does not dampen gene expression but triggers local H3K9me3 spreading. Examples for each category are given below.
See also Figure S5.
Figure S4.
Related to Figure 5
(A) Depicted are individual RNA-seq reads (including spliced reads) from GFP or piwi KD cells mapping to the expanded locus (reads mapping to the flanking transcription unit in blue); note that the first intron is faithfully spliced despite the inserted gypsy element.
(B) Changes in mRNA abundance for the set of expressed genes (RPKM > 5 in one library) between mael KD and control KD cells. The upper plot contrasts genes with no insertion (gray) and genes with an insertion of a group I TE in sense orientation (red). The lower plot contrast genes with no insertion (gray) and genes with no group I TE, but with a group III TE insertion (yellow).
(C) List of all upregulated genes (4x in piwi and armi KDs, 2x in mael KDs) with mapped TE insertions either in the gene body or in close proximity (up to 5kb). Given are also the RNA-seq RPKM values in control and piwi KD cells.
To globally determine the impact of the piRNA pathway on gene expression, we split the set of genes with RPKM >5 in any of the RNA-seq libraries into three groups: genes with no TE insertion, genes with a group I TE insertion, and genes with no group I but with a group III TE insertion. We plotted their mean RPKM levels versus their fold RPKM change upon piwi KD (Figure 7B) or mael KD (Figure S4B). Strikingly, whereas genes with no insertion (gray) and those with group III insertions (yellow) were distributed symmetrically around the baseline, the population of genes with group I insertions (red) was significantly skewed to increased RNA levels upon Piwi or Mael loss. As a whole, the set of genes with group I TE insertions had significantly increased RNA-seq RPKM values over genes with group III TE insertions, as well as over genes with no TE insertions (Figure 7C).
Reciprocally, we selected the set of genes with the most consistent changes upon piRNA pathway KD (4-fold in piwi and armi KD and 2-fold in mael KD cells). Thirty-four genes were upregulated, with no gene being downregulated (Figure 7D). Eighty percent of these genes were associated with a TE insertion within 5 kb, and 85% of these TEs belonged to group I (Figure 7E). In comparison to an average set of random control genes, group I elements were highly enriched in derepressed genes (p value < 10−15), wheras group III elements were not (Figure 7E). Importantly, the 28 genes with TE insertions (Figure S4C) that exhibited increased steady-state RNA levels also showed increased Pol II occupancy (p < 10−15) and increased nascent RNA output (p < 10−15; Figure 7F). Many TE insertions therefore have profound impacts on the expression of nearby genes.
After inspecting dozens of TE insertions in the vicinity of genes, we defined three major categories of how TEs and the piRNA pathway impact expression or chromatin status of gene loci (Figure 7G; see also Feschotte, 2008). For category A genes (repressive chromatin influence), TEs are inserted close to the TSS of a per se transcribed gene. By nucleating spreading of silencing marks such as H3K9me3, the TE represses accessibility of Pol II to the gene's promoter and dampens transcription. Upon loss of the piRNA pathway, the repressive chromatin environment is lost and Pol II gains access to the gene's promoter. For category B genes (promoter addition), the TE insertion does not affect the gene's promoter directly, either as the insertion is too distant or as the promoter is anyways off in OSCs. Upon loss of the pathway, an increase in gene expression results from Pol II bleeding out of the TE body into the downstream transcription unit (e.g., Figure S5A). RNA-seq patterns indicated that this often leads to accumulation of spliced mature or 5′ truncated mRNAs, dependent on the location of the TE insertion. TE insertions belonging to category C (neutral chromatin influence) trigger H3K9me3 in a Piwi-dependent manner but do not impact gene expression (e.g., Figure S5B). In most cases, the involved TEs are themselves not active or are only mildly active but are still targeted by OSC piRNAs (e.g., 17.6, roo). As mentioned above, these insertions depend on the host gene's transcription for being able to trigger H3K9me3.
Figure S5.
Related to Figure 7
(A and B) Shown are normalized density profiles of Pol II ChIP-seq (red), GRO-seq (black), RNA-seq (brown) and H3K9me3 ChIP-seq (green) for the indicated OSC knockdowns (left).
(A) Shown is the ∼140kb area with an mdg1 insertion upstream of the typically non-expressed gene CG15278. Upon loss of the piRNA pathway transcriptional bleeding from the TE insertion into the CG15278 locus leads to accumulation of RNA reads.
(B) Shown is a ∼120kb area with a 17.6 insertion in sense orientation into an intron of the Btk29A transcription unit. This insertion triggers H3K9me3 spreading, which depends on Piwi but only weakly on Mael. Loss of the piRNA pathway does not lead to upregulation of the host gene, classifying this insertion.
Taken together, our data illustrate the profound impact of TEs on gene expression patterns and show how loss of the piRNA pathway triggers derepression of dozens of genes simply by a loss of local TE repression.
Discussion
Piwi-RISC Guides Transcriptional Silencing
Small-RNA-guided transcriptional gene silencing (TGS) is widespread in fungi and plants (Grewal, 2010; Moazed, 2009; Slotkin and Martienssen, 2007). In animals, evidence for TGS guided by small RNAs is sparse, with the exception of studies in C. elegans (Burton et al., 2011; Ashe et al., 2012; Buckley et al., 2012; Gu et al., 2012; Shirayama et al., 2012; Luteijn et al., 2012).
Our data demonstrate that Drosophila Piwi-RISC silences TEs via TGS. Loss of Piwi triggered increased Pol II occupancy and increased nascent RNA output for repressed TEs, and this paralleled increases in steady-state RNA levels. It therefore seems that TGS is the major route through which Piwi represses TEs, although an additional layer of PTGS cannot be excluded.
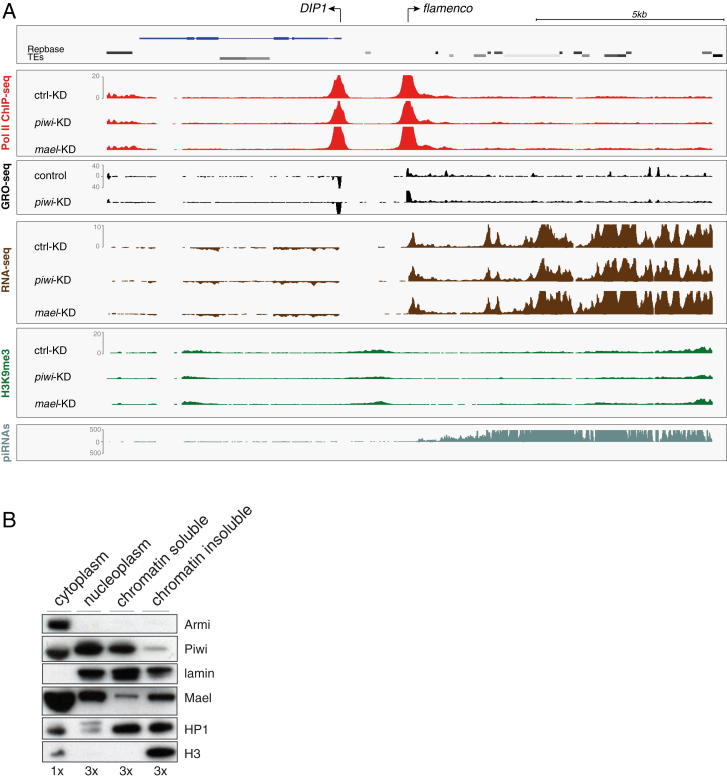
TGS by Piwi-RISC shows several features. (1) Silencing cannot be maintained in the absence of Piwi-RISC. Thus, any chromatin modification installed by Piwi-RISC is lost relatively quickly, either at particular cell-cycle phases or due to overriding activity of transcriptional activators. (2) Piwi silences TEs in trans. piRNAs are processed from piRNA clusters such as flamenco (Brennecke et al., 2007; Pélisson et al., 2007), and Figure S6A shows that transcription at the flamenco locus is unaffected in piwi or mael KD cells. Thus, piRNAs originating from clusters are loaded into Piwi, and subsequently, Piwi-RISC silences TEs throughout the genome in trans. (3) Piwi-mediated silencing requires transcription of the target (Pélisson et al., 2007; Sarot et al., 2004). Strong support for this stems from intronic insertions of TEs that are transcriptionally inactive but against which antisense OSC piRNAs exist. Only if inserted in sense into an active transcription unit do these TEs nucleate H3K9me3 spreading into the flanking genome. Thus, nascent RNA at the target locus must recruit Piwi to chromatin via complementary piRNAs. Indeed, careful fractionation experiments reproducibly indicate that significant levels of Piwi as well as Mael are present in chromatin fractions (Figure S6B). We were, however, not able to map Piwi specifically to target loci using ChIP experiments. Possibly, the distance between nascent RNA and DNA template prevents efficient crosslinking.
Figure S6.
Related to Discussion
(A) Shown are normalized density profiles of Pol II ChIP-seq (red), GRO-seq (black), RNA-seq (brown), H3K9me3 ChIP-seq (green) and piRNA-seq (light green) for the indicated OSC knockdowns (left). Shown is the ∼20kb area around the transcriptional start site of the flamenco cluster. Shown are only reads mapping uniquely to the genome but we note that nearly all areas in this window are genome-unique.
(B) Western blot showing protein levels of Armi, Piwi, Lamin, Mael, HP1 and Histone 3 (H3) in cytoplasmic, nucleoplasmic, soluble and insoluble chromatin fractions of OSCs. The relative amount of each fraction loaded per lane (based on fraction volume) is given below. The following antibodies were used: α-Lamin (ADL67.10, DSHB), α-HP1 (C1A9, DSHB) and α-H3 (Abcam, ab1791).
The Mechanism of Piwi-Mediated TGS
The molecular events that occur downstream of Piwi-RISC recruitment to a target RNA are unclear. Our data show that one key step is trimethylation of H3K9, a major heterochromatic mark. So-called “readers” of this mark are HP1 proteins (Vermaak and Malik, 2009). In Drosophila, the prototypical HP1 member implicated in heterochromatin biology is Su(var)205, and its knockdown leads to TE desilencing in the germline (Wang and Elgin, 2011). HP1 proteins are known to interact with H3K9 methyltransferases (Schotta et al., 2002). Whether Piwi instructs H3K9 trimethylation via recruitment of HP1 (as suggested in Brower-Toland et al., 2007, but see Wang and Elgin, 2011) or via recruitment of an H3K9 methyltransferase like in S. pombe (Zhang et al., 2008b) remains to be determined.
A central finding of our study is that H3K9me3 itself cannot be the final silencing mark, as we observed strong TE derepression upon loss of Mael despite no or only very modest changes in H3K9me3. This places Mael in an unknown silencing step downstream of Piwi and downstream of or in parallel to H3K9me3. Interestingly, this resembles RNAi-guided TGS in S. pombe, which requires the activity of histone deacetylation and histone chaperone complexes downstream of H3K9me3 (Reyes-Turcu and Grewal, 2012). Mael is evolutionarily conserved, and its mouse homolog is required for TE silencing in testes (Aravin et al., 2009). Also, mice express a nuclear PIWI protein (MIWI2), and this has been linked to DNA methylation of targeted TE insertions (Aravin et al., 2008). As DNA methylation is absent in Drosophila, it will be interesting to test whether the murine piRNA pathway also induces H3K9me3 and whether DNA methylation is the stable manifestation of an initially histone-specific silencing mark.
Implications for Heterochromatin Biology
Heterochromatin has long been viewed as the highly condensed and inert part of the genome that is refractory to transcription. Recent insights are challenging this view and attest a dynamic interplay between transcription and establishment/maintenance of the heterochromatic character (Grewal, 2010; Moazed, 2009; Slotkin and Martienssen, 2007). In agreement, our data illustrate that establishment of the heterochromatic H3K9me3 mark on dispersed TE insertions depends on transcription. More surprisingly, we found that the H3K9me3 mark is compatible with ongoing transcription and that Pol II activity enhances H3K9me3 spreading. The most notable example of increased H3K9me3 spreading was an mdg1 insertion in pericentromeric heterochromatin (Figure S7). Upon loss of Mael, this insertion nucleated H3K9me3 spreading over >30 kb. Maybe H3K9me3 spreading is generally more pronounced in heterochromatic areas, as local concentrations of required factors are higher in these chromatin environments.
Figure S7.
Related to Discussion
Shown are normalized density profiles of Pol II ChIP-seq (red), GRO-seq (black), RNA-seq (brown), H3K9me3 ChIP-seq (green) and piRNA-seq (light green) for the indicated OSC knockdowns (left). Shown is a ∼60kb area that resides in the peri-centromeric heterochromatin of chromosome 2R (cytological position 42A); the position of the mdg1 insertion (minus strand) is indicated; note the absence of piRNAs mapping to this region and the massive spreading of H3K9me3 in mael KD cells.
A key feature of the OSC system is that hundreds of TEs and H3K9me3 domains can be monitored simultaneously upon loss of silencing. This strengthens experimental conclusions but also allows unexpected discoveries. For example, three euchromatic mdg1 insertions did not show increased Pol II bleeding upon Piwi loss and nucleated only low H3K9me3 levels in their vicinity. These insertions might simply lack essential promoter features. However, we found that all three insertions reside in H3K27me3 domains (red arrows in Figure 5C; G.S. and J.B., unpublished data), suggesting that H3K27me3 is dominant over Piwi mediated TGS.
The Impact of TEs and the piRNA Pathway on Gene Expression
Many links have been made between TEs and the regulation of genes (Feschotte, 2008; Slotkin and Martienssen, 2007). The unique aspect of our work is that hundreds of TE insertions can be studied upon loss of their repression. This shed considerable light on two key modes of how TEs impact gene expression. On the one hand, TEs act positively on gene expression by providing transcriptional competence to certain genomic regions (promoter addition). On the other hand, TEs act negatively on flanking genes if they are silenced transcriptionally as the “silencing character” spreads into flanking domains (repressive chromatin influence). In both cases, loss of TE silencing will often lead to increased expression of the neighboring gene.
Considering the promoter addition model, we were surprised to see how far Pol II transcription can bleed into sequences flanking the insertion. All piRNA-repressed TEs in OSCs are LTR elements, and it is unclear whether the 3′ LTR serves as an independent TSS or whether transcriptional bleeding implies a large transcript encompassing the entire TE plus flanking sequences. Transcriptional bleeding often traversed genic transcription units and triggered legitimate splice events, suggesting synthesis of stable hybrid mRNAs.
Whether a TE insertion dampens transcription of a nearby gene via the chromatin influence model seems to depend on the distance between TE insertion and gene TSS, as well as on the strength of the gene's promoter (we rarely observed impacts on highly expressed genes). Our data therefore show that piRNA target sequences within introns can impact host gene transcription. Whether cotranscriptional splicing lowers the impact of intronic target sequences remains to be determined.
All in all, the piRNA pathway does considerably impact gene expression via TGS of TEs. An important open question is to what extent TE insertions display transcriptional bleeding or nucleate chromatin changes also in nongonadal cells in which the piRNA pathway is thought to be not active.
Experimental Procedures
Drosophila Stocks
Fly stocks are listed in Table S1.
Cell Culture
OSCs were cultured as described (Niki et al., 2006) and transfected with Cell Line Nucleofector kit V (Amaxa Biosystems; program T-029).
Antibodies
α-Piwi and α-Armi (rabbit) were described in Olivieri et al. (2010) and mouse α-Piwi and mouse α-Armi in Saito et al. (2010). Rabbit α-Mael was raised against the SDNDFSVNGADGKLKK peptide. α-Rpb3 was described in Adelman et al. (2005), and α-H3K9me3 was from Abcam (ab8898).
CIP-Kinase Labeling of Small RNAs
Cells lysates were prepared from respective knockdowns. Piwi-RISC was isolated with α-Piwi. Small RNAs were extracted, dephosphorylated (CIP), and radioactively labeled (T4 PNK). For details, see the Extended Experimental Procedures.
Extended Experimental Procedures.
X-Gal Staining
Ovaries from 5-7 day old flies were dissected into ice cold PBS (max 30 min), fixed in 0.5% Glutaraldehyde/PBS (RT, 15 min), and washed with PBS. The staining reaction was performed with staining solution (10mM PBS, 1mM MgCl2, 150 mM NaCl, 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, 0.1% Triton, 0.1% X-Gal) at room temperature over night.
CIP-Kinase RNA Labeling
Cells were transfected twice with respective siRNAs. Cells were lysed in Lysis buffer (20mM HEPES-NaOH (pH7.0), 150mM NaCl, 2.5mM MgCl2, 250mM sucrose, 0.05% NP40, 0.5%Triton). 4mg total protein was used from each sample for immunoprecipitation. Immunoprecipitation was performed using Dynabeads Protein G (Invitrogen) and rabbit anti- Piwi antibody (Brennecke et al., 2007). Lysates were incubated 2 hr at 4°C with antibody crosslinked to beads and washed 3 times with 300mM NaCl and 0.2%NP-40 and 3 times without detergent. Beads were treated with Proteinase K (Roche) and RNA was isolated with TRIzol (Invitrogen). Isolated RNA was dephosphorylated using CIP (Alkaline Phosphatase, Calf Intestinal), phosphorylated using PNK and radio-labeled gamma-ATP. RNAs were separated on a 15% PAA-Urea Gel.
Transposon QPCR Analysis
cDNA was prepared via random priming of 1μg total RNA isolated from ovaries of 5-7 day old flies. Quantitative PCR was performed using BioRad IQ SYBR Green Super Mix. Each experiment was performed in biological triplicates with technical duplicates. Relative RNA levels were calculated by the 2-ΔΔCT method (Livak and Schmittgen, 2001) and normalized to rp49 levels. Fold enrichments were calculated in comparison to respective RNA levels obtained from heterozygous flies or from flies not harboring a knockdown hairpin. For ChIP-qPCR experiments, enrichments were calculated over input and normalized to an intergenic region.
Northern Blot
Total RNA was isolated from respective knockdowns and separated on a 15% polyacrylamide urea gel. RNA was transferred to Amersham Hybond-NX (RPN303T) membrane and crosslinked by EDC (1-ethyl-3-(3- dimethylaminoprophy) carbodiimide) for 1 hr. The membrane was pre-hybridized in Church Buffer and hybridized to probes overnight at 37°C. The membrane was washed 3 times 10 min with 2xSSC, 0.1% SDS and exposed.
Small RNA Cloning
Small RNA cloning and sequencing was performed as described in (Brennecke et al., 2007). In brief, 20 ug of total RNA was isolated from maelM391/maelDf or respective heterozygous ovaries by TRIzol and Phenol/Chloroform extraction, was resolved on a denaturing polyacrylamide gel and RNAs corresponding to 18-28 nt were isolated and subjected to ligations of 3′-, and 5′-adaptors followed by reverse transcription and PCR amplification; libraries were sequenced on GAII or HiSeq2000 platforms (Illumina). For sequencing of piRNAs (Figure 3D) we purified Piwi-bound piRNAs from OSCs and followed the protocol described above.
GRO-Seq
10 millions of OSC nuclei were isolated per experiment and subjected to the nuclear run-on reaction in the presence of Br-UTP for 5min at 30 degrees according to (Larschan et al., 2011). The reaction was terminated with Trizol LS (Invitrogen) followed by extraction of total RNA. RNA was fragmented by base hydrolysis with 1M NaOH on ice to ∼20-150nt and remaining DNA was removed with DNaseI (Promega). Br-UTP-containing RNA fragments were enriched and purified using anti-deoxyBrU beads (Santa Cruz Biotech). RNA was then end-repaired and ligated to 3′- and 5′-adaptor used in the small RNA cloning procedure. Each ligation step was followed by purification with anti-deoxyBrU beads. Cloned library of nascent RNA fragments was reverse transcribed, PCR amplified and sequenced on HiSeq2000 (Illumina).
ChIP-Seq
10 million OSCs were fixed with 1% formaldehyde for 10 min followed by quenching with glycine. For ChIP from tissue, 50 ul of ovaries were dissected into ice cold PBS, washed and crosslinked with 1,8% formaldehyde for 10 min, quenched and dounced to disrupt the tissue. Nuclei were isolated, washed and lysed; isolated chromatin was fragmented using tip sonicator (Omni-Ruptor) to fragment sizes of 200-400nt. Immuno-precipitation was done overnight with specific antibodies. Intensive washing steps removed non-specific background and beads were eluted with 1% SDS. For de-crosslinking of protein-DNA complexes eluates were incubated 6h at 65 degrees and remaining proteins were digested with proteinase K and RNA with RNase A. DNA fragments were extracted with phenol/chloroform and used as template either for qPCR or library preparation. Libraries were cloned with NEBNext ChIP-Seq Library Prep Reagent Set for Illumina (NEB) and sequenced on HiSeq2000 (Illumina).
DNA-Seq
OSCs were lysed in RIPA buffer (Tris 50mM pH7.5, NaCl 150mM, SDS 0.1%, sodium deoxycholate 0.5%, Triton X-100 1%) followed be o/n incubation with proteinase K (Roche). RNA was removed by digestion with RNase A (Fermentas) and genomic DNA was extracted with phenol/chloroform. DNA was fragmented, cloned with NEBNext ChIP-Seq Library Prep Reagent Set for Illumina (NEB) and sequenced on HiSeq2000 (Illumina).
Cellular Fractionation
OSCs were resuspended in buffer I (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM MgCl2, 0.1mM EDTA, 0.1mM DTT, 0,1% NP40) and incubated on ice followed by mechanical isolation of nuclei with syringe and centrifugation. Supernatant was pre-cleared and taken as cytoplasmic fraction. Pellet was washed twice with buffer I and washed twice in lysis buffer (10 mM Tris (pH 5 7.5), 2 mM MgCl2, 3 mM CaCl2, 10% glycerol, 0.5% NP40). Nuclei were next pre-cleaned in buffer F (50mM Tris-Cl (pH 5 8.3), 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA) and opened with triton lysis buffer (50 mM Tris pH 7.5, 0.5% Triton, 137.5 mM NaCl, 5mM EDTA, 10% glycerol). Supernatant was collected as nuclear fraction. Remaining pellet was washed twice with the same buffer and afterward mechanically disrupted by extensive pipetting (saved as soluble chromatin). Remaining pellet was collected as insoluble chromatin.
Computational Analyses
Mapping of Short Reads
All the Illumina short reads were quality controlled (filtering out N-containing reads, sequencing artifacts) and mapped to the Drosophila melanogaster genome (dm3) excluding chromosome Uextra with bowtie 0.12.7 (Langmead et al., 2009). For piRNA-seq as well as for GRO-seq we allowed up to 1 mismatch due to short fragments, whereas up to 3 mismatches were allowed for RNA-seq and ChIP-seq reads. Genome-aligned reads were mapped to transposon consensus sequences (obtained from BDGP (http://www.fruitfly.org/data/p_disrupt/datasets/ASHBURNER/D_mel_transposon_sequence_set.fasta) and Repbase (Jurka, 1998) and available upon request) allowing up to 1 mismatch for GRO-seq and up to 3 mismatches for the other methods used in this study.
To avoid cross-contamination between highly similar TE sequence stretches, only reads mapping to one transposon in our list were retrieved. For reads mapped twice within one entry (as the TE consensus sequences contain always two LTRs for LTR-containing TEs) we applied a weighting scheme. Number of reads mapped to each TE was normalized to its length and total number of genome-aligned reads (RPKM value, Reads Per Kilobase of exon model per Million mapped reads) (Mortazavi et al., 2008).
Processing of Small RNA Sequencing Reads and Global Nuclear Run-On Sequencing Reads
Our RNA cloning strategy introduces 4 random nucleotides at 3′ end of 5′ linker and 5′ end of 3′ linker, which reduces ligation biases (Jayaprakash et al., 2011). Reads were first stripped of the 3′ adaptor and then the introduced 4 random nucleotides at each end of the read were removed. Only reads larger than 22 nt were selected to increase mapping specificities. Potential contaminants and degradation products of abundant cellular RNAs were removed (reads mapping to rRNA, mitochondrial RNA, microRNAs (all from Flybase) and Drosophila C virus (highly expressed virus present in OSCs)). Next, reads were mapped to Drosophila genome release 5 (excluding Uextra) with bowtie 0.12.7 (Langmead et al., 2009).
RNA Sequencing
We sequenced rRNA-depleted total RNA in a strand-specific manner from OSCs upon different siRNA-mediated knockdowns. This yielded ∼14-32 million genome- and transcriptome-mappable reads. For the computational analyses, we first extracted high quality bases from every read (6-56 nt) and mapped these to the Drosophila genome as well as to the FlyBase transcriptome. Uniquely aligned reads were used for quantification of gene expression levels according to coordinates in the Flybase gene annotation (r5.31) by calculating RPKM values. For computing TE expressions we used genome-mapped reads, aligned them to the TE consensus and proceeded as described above.
Chromatin Immunoprecipitation followed by Sequencing
We performed ChIP-seq analysis of Pol II, histone H3K9me3 and their inputs in two replicates. Based on the high correlations between replicates we pooled them together and used these pooled libraries throughout this study. The ChIP-seq reads were mapped to the genome and TE consensus sequence to compute RPKM values.
H3K9me3 Peak Calling
For calling H3K9me3 peaks we used findPeaks from HOMER software (Heinz et al., 2010). Enrichments of H3K9me3 signal were calculated against input (false discovery rate (FDR) of 10−4). We merged all the euchromatic peaks within 8kb distance from each other, which yielded in total 1121 peaks (466 euchromatic and 655 heterochromatic).
Identification of TE Insertions
For TE insertion calling, we used genomic DNA-seq reads (single-end, 100bp) from OSCs. We took advantage of reads spanning boarders between TE insertions and neighboring genomic regions. First, we removed all the reads, which map to the Drosophila C-virus genome or human rRNA (both 0 mismatches), which we expect to be contaminants. Second, to validate the presence of already annotated insertions, we filter out reads aligning to the assembled Drosophila melanogaster genome with masked repeats, then aligned separately to genomic repeats only and finally to TE consensus sequences (all up to 3 mismatches). Next, we took 25nt of 5′- as well as 3′-end of each remaining read (∼17.8% of total reads) and used these for further analyses. We examined if one part of those reads maps to a TE consensus sequence (up to 2 mismatches) while the other part maps to the genome (uniquely; up to 1 mismatch; ∼0.06% of all reads). To bias the identification of insertions toward full-length elements, we required the TE mapping part to map to the LTR of LTR-elements and to the first 500nt of non-LTR elements. Only insertions covered by ≥ 2 reads were used further (1852 in euchromatin).
Data Visualization
For preparation of meta-plots representing an average signal distribution around H3K9me3 peaks or TE insertions we identified the summit of every peak and used ± 25kb windows to display signal of H3K9me3 or Rpb3 ChIP-seq in 100nt bins. All heatmaps generated in this study were prepared with Java Treeview (Saldanha, 2004). For visualization of sequencing tracks we used the UCSC Genome Browser (Kent et al., 2002).
Coordinates of Heterochromatin
The borders for heterochromatin used throughout this study are depicted in Table S6. These were mostly informed by H3K9me3 densities and the location of piRNA clusters, which map typically at the borders between euchromatin and heterochromatin (Brennecke et al., 2007).
Statistical Analysis
We used statistical packages implemented in R 2.15.0 for all calculations and plots in this study. For data visualization in box plot format we used the standard features: horizontal bar represents median, the box depicts 25th and 75th percentile (lower and upper quartile respectively), whiskers represent sample minimum (lower) and maximum (upper); outliers are shown as circles. Statistical significance in Figures 3–7 (Figures 3F–H, 4D, 5A, 6E, 7E) was computed with Mann-Whitney U test, whereas p-values in Figures 6E and 7E, F were calculated over random control using binomial test. Random control for peaks in Figure 6E and genes in Figure 7E was generated by calculating an average transposon recovery in random peaks from 100 simulation (Figure 6E); or by selecting a random set of genes and checking for presence TE insertion within ± 5kb (an average of 100 simulations, Figure 7E).
Northern Blot
Total RNA was isolated from respective knockdowns and separated on a 15% Urea-PAA gel. After transfer onto a membrane, radioactively labeled probes were hybridized overnight. Probe sequences are shown in Table S3, and details are provided in the Extended Experimental Procedures.
Small RNA Cloning
Small RNA cloning and sequencing was performed as in Brennecke et al. (2007).
RT-qPCR Analysis
Primer sequences and details are given in the Extended Experimental Procedures.
RNA-Seq
Total RNA from siRNA-treated OSCs was rRNA depleted using RiboZero (Epicenter), fragmented, and reverse transcribed with random hexamers. Strand-specific libraries were prepared using the UDG-digestion-based strategy, cloned with NEBNext ChIP-Seq Library Prep Reagent Set for Illumina (NEB), and sequenced on HiSeq2000 (Illumina).
GRO-Seq
Global nuclear run-on procedure was according to Core et al. (2008). In brief, ∼10 million OSC nuclei were isolated per experiment and subjected for nuclear run-on in the presence of Br-UTP followed by purification of fragmented RNA with anti-deoxyBrU beads. RNA fragments were cloned and sequenced on HiSeq2000 (Illumina). Detailed description is provided in the Extended Experimental Procedures.
ChIP-Seq
Chromatin immunoprecipitation (ChIP) was carried out according to Lee et al. (2006). In brief, ∼10 million OSCs or 50 ul dissected ovaries were fixed with 1% or 1.8% formaldehyde, respectively. Prepared chromatin was sonicated and used for respective immunoprecipitation followed by decrosslinking and purification of DNA. Recovered DNA fragments were cloned with NEBNext ChIP-Seq Library Prep Reagent Set for Illumina (NEB) and sequenced on HiSeq2000. Detailed description is provided in the Extended Experimental Procedures.
DNA-Seq
Genomic DNA of OSCs was fragmented and cloned with NEBNext ChIP-Seq Library Prep Reagent Set for Illumina (NEB) followed by sequencing on HiSeq2000 (Illumina). Detailed description is provided in the Extended Experimental Procedures.
Computational Analyses
Detailed information is provided in the Extended Experimental Procedures.
Acknowledgments
We thank K. Adelman for α-Rbp3; M. Siomi for OSCs and α-Armi; M. Madalinski for antibody purifications; S. Lopez for fly injections; the VDRC; A. Pelisson and H. Ruohola-Baker for fly strains; D. Jurczak, A. Stark, D. Gerlach, R. Maier and R. Sachidanandam for help with bioinformatics; and the CSF NGS unit for deep sequencing. We thank S. Grewal and D. Moazed for inspiring discussions and comments on the manuscript. The Brennecke laboratory is supported by the Austrian Academy of Sciences, the European Community's 7th Framework Programme (FP7/2007-2013; ERC grant # 260711EU), and the Austrian Science Fund (FWF; Y 510-B12). G.S. is supported by the Boehringer Ingelheim Fonds. G.S., D.D., and J.B. designed the experiments. G.S. and D.D. performed the experiments. G.S. performed the computational analysis. G.S., D.D., and JB analyzed the data and wrote the paper.
Published online: November 15, 2012
Footnotes
Supplemental Information includes Extended Experimental Procedures, seven figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.10.040.
Accession Numbers
All Illumina data sets (Table S5) were deposited at GEO (GSE41729).
Supplemental Information
References
- Adelman K., Marr M.T., Werner J., Saunders A., Ni Z., Andrulis E.D., Lis J.T. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Bourc'his D., Schaefer C., Pezic D., Toth K.F., Bestor T., Hannon G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A.A., van der Heijden G.W., Castañeda J., Vagin V.V., Hannon G.J., Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E.M., Mitchell J., Bagijn M.P., Cording A.C., Doebley A.L., Goldstein L.D., Lehrbach N.J., Le Pen J. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B., Findley S.D., Jiang L., Liu L., Yin H., Dus M., Zhou P., Elgin S.C., Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B.A., Burkhart K.B., Gu S.G., Spracklin G., Kershner A., Fritz H., Kimble J., Fire A., Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton N.O., Burkhart K.B., Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourt J., Brasset E., Desset S., Pouchin P., Vaury C. Polycomb group-dependent, heterochromatin protein 1-independent, chromatin structures silence retrotransposons in somatic tissues outside ovaries. DNA Res. 2011;18:451–461. doi: 10.1093/dnares/dsr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley S.D., Tamanaha M., Clegg N.J., Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Grewal S.I. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S.G., Pak J., Guang S., Maniar J.M., Kennedy S., Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat. Genet. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H.H., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Klenov M.S., Sokolova O.A., Yakushev E.Y., Stolyarenko A.D., Mikhaleva E.A., Lavrov S.A., Gvozdev V.A. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl. Acad. Sci. USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Johnstone S.E., Young R.A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn M.J., van Bergeijk P., Kaaij L.J., Almeida M.V., Roovers E.F., Berezikov E., Ketting R.F. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.D., Hannon G.J. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R., Hannon G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N., Lei E.P. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki Y., Yamaguchi T., Mahowald A.P. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc. Natl. Acad. Sci. USA. 2006;103:16325–16330. doi: 10.1073/pnas.0607435103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D., Sykora M.M., Sachidanandam R., Mechtler K., Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V.S., Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- Pélisson A., Sarot E., Payen-Groschêne G., Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Grewal S.I. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr. Opin. Genet. Dev. 2012;22:156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., Sakota E., Kotani H., Asai K., Siomi H., Siomi M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Saito K., Ishizu H., Komai M., Kotani H., Kawamura Y., Nishida K.M., Siomi H., Siomi M.C. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E., Payen-Groschêne G., Bucheton A., Pélisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Krauss V., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti K.A., Brennecke J. The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet. 2010;26:499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H.C., Gu W., Ishidate T., Conte D., Jr., Mello C.C. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S., Olovnikov I., Sergeeva A., Lavrov S., Abramov Y., Savitsky M., Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011;39:8703–8711. doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M.C., Sato K., Pezic D., Aravin A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Vermaak D., Malik H.S. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu. Rev. Genet. 2009;43:467–492. doi: 10.1146/annurev-genet-102108-134802. [DOI] [PubMed] [Google Scholar]
- Wang S.H., Elgin S.C. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. USA. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Xiong H., Shan J., Xia X., Trudeau V.L. Functional insight into Maelstrom in the germline piRNA pathway: a unique domain homologous to the DnaQ-H 3′-5′ exonuclease, its lineage-specific expansion/loss and evolutionarily active site switch. Biol. Direct. 2008;3:48. doi: 10.1186/1745-6150-3-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Mosch K., Fischle W., Grewal S.I. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Heinz, S., Benner, C., Spann, N., Bertolino, E., Lin, Y.C., Laslo, P., Cheng, J.X., Murre, C., Singh, H., and Glass, C.K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed]
- Jayaprakash, A.D., Jabado, O., Brown, B.D., and Sachidanandam, R. (2011). Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 39, e141. [DOI] [PMC free article] [PubMed]
- Jurka, J. (1998). Repeats in genomic DNA: mining and meaning. Curr. Opin. Struct. Biol. 8, 333–337. [DOI] [PubMed]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed]
- Langmead, B., Trapnell, C., Pop, M., and Salzberg, S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed]
- Larschan, E., Bishop, E.P., Kharchenko, P.V., Core, L.J., Lis, J.T., Park, P.J., and Kuroda, M.I. (2011). X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471, 115–118. [DOI] [PMC free article] [PubMed]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed]
- Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. [DOI] [PubMed]
- Saldanha, A.J. (2004). Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.