Abstract
The cDNA sequence for CAP160, an acidic protein previously linked with cold acclimation in spinach (Spinacia oleracea L.), was characterized and found to encode a novel acidic protein of 780 amino acids having very limited homology to a pair of Arabidopsis thaliana stress-regulated proteins, rd29A and rd29B. The lack of similarity in the structural organization of the spinach and Arabidopsis genes highlights the absence of a high degree of conservation of this cold-stress gene across taxonomic boundaries. The protein has several unique motifs that may relate to its function during cold stress. Expression of the CAP160 mRNA was increased by low-temperature exposure and water stress in a manner consistent with a probable function during stresses that involve dehydration. The coding sequences for CAP160 and CAP85, another spinach cold-stress protein, were introduced into tobacco (Nicotiana tabacum) under the control of the 35S promoter using Agrobacterium tumefaciens-based transformation. Tobacco plants expressing the proteins individually or coexpressing both proteins were evaluated for relative freezing-stress tolerance. The killing temperature for 50% of the cells of the transgenic plants was not different from that of the wild-type plants. As determined by a more sensitive time/temperature kinetic study, plants expressing the spinach proteins had slightly lower levels of electrolyte leakage than wild-type plants, indicative of a small reduction of freezing-stress injury. Clearly, the heterologous expression of two cold-stress proteins had no profound influence on stress tolerance, a result that is consistent with the quantitative nature of cold-stress-tolerance traits.
Although the concept that cold acclimation in plants involves altered gene expression during the induction of freezing tolerance is more than 20 years old, it has only been in recent years that most of the information concerning the influence of low temperature on gene expression has become available (Thomashow, 1994; Hughes and Dunn, 1996). Similar findings with cold-water fishes (Lin and Gross, 1981), bacteria (Goldstein et al., 1990), and yeast (Kondo and Inouye, 1991) parallel the growing body of information on low-temperature-regulated genes in plants. In plants a significant portion of the genes demonstrated to be regulated in response to low temperature are, as yet, without a known function. Some, based on sequence homologies (Hahn and Walbot, 1989; Gilmour et al., 1992; Guo et al., 1992; Houde et al., 1992; Sutton et al., 1992; Neven et al., 1993), belong to a superfamily of developmentally and environmentally regulated proteins linked with reduced water activities (Dure, 1993; Close, 1996). Others have properties in common with this superfamily, such as boiling solubility, regulation by ABA, or responsiveness to water stress (Cattivelli and Bartels, 1990; Kurkela and Franck, 1990; Lin and Thomashow, 1992; Orr et al., 1992; Nordin et al., 1993; Yamaguchi-Shinozaki and Shinozaki, 1993a, 1993b).
Freezing stress is a desiccative physical stress (Pearce, 1988), and during slow equilibrium freezing in plants, unfrozen water is withdrawn from the inside of the cell by the lower vapor pressure of extracellular ice (Mazur, 1963). This causes progressive dehydration, which leads to a drastic loss of cellular volume and an increase in the concentration of intracellular solutes. The extreme dehydration that can result from freezing has major effects on cellular components such as membrane structure (Steponkus, 1984; Steponkus et al., 1988), which in turn reduces the ability of the cell to survive a freeze/thaw cycle. Consequently, during the course of evolution plants have acquired adaptive mechanisms to help ensure their survival in situations in which freezing occurs (Sakai and Larcher, 1987; Guy et al., 1992). The idea that many of these proteins might function not only during water stress but also during freezing stress to afford some manner of protection is appealing but largely untested. Previous work with spinach (Spinacia oleracea L.) in our laboratory has supported a relationship between drought stress and cold-acclimation responses (Guy et al., 1992; Neven et al., 1993).
The major question yet unanswered is this: By what mechanisms do plants become freezing tolerant? Undoubtedly, the process is one of great complexity (Steponkus, 1984; Sakai and Larcher, 1987). The finding that many cold-stress proteins are related to proteins linked to water stress or anhydrous conditions is pivotal because it points to a specific facet of freezing stress that can be more critically examined. In spinach two cold-stress proteins, CAP85 and CAP160, have been shown to share features in common with proteins associated with water stress (Guy et al., 1992; Neven et al., 1993). Both of these proteins exist in soluble form in spinach cells, and are actively synthesized and accumulated in leaf tissue exposed to 5°C (Guy and Haskell, 1987, 1988) or subjected to water stress at 25°C (Guy et al., 1992).
One approach to determining the role of a stress protein of known or unknown function is to modify its natural expression (Artus et al., 1996) or to insert its gene into another organism in which it does not naturally exist (Georges et al., 1990; Hightower et al., 1991; McKown and Warren, 1991; Kenward et al., 1993). Here we report the cloning and characterization of cDNAs and genomic sequences for one of these proteins, CAP160, and the influence of expression of both CAP160 and CAP85 proteins on freezing-stress injury in tobacco (Nicotiana tabacum).
MATERIALS AND METHODS
Isolation of CAP160 cDNAs and Genomic Clones
Using a monoclonal antibody (2H8) raised against purified CAP160, cDNAs were isolated by immunoscreening a Uni-Zap (Stratagene) library prepared with poly(A+) RNA from spinach (Spinacia oleracea L.) leaf tissue that had been exposed to 5°C for 2 d (Neven et al., 1993). Sixteen clones were selected by the immunoscreening. Four different clones were partially sequenced and all appeared identical. Clone IIa contained the largest insert and was sequenced in its entirety using the Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems) on an automated sequencer (Applied Biosystems). Subclones were prepared by cutting at internal restriction sites, followed by cloning into pBluescript (Stratagene). All regions of the cDNA were sequenced several times and, where necessary, synthetic oligonucleotide primers were used to complete sequencing. Spinach DNA was partially restricted with EcoRI and cloned into EMBL4. The library was screened with CAP160 cDNA clone IIa. Overlapping clones representing the 5′ and 3′ regions were used to construct the complete gene sequence. Database searches for homology to CAP160 DNA and protein sequences were performed with BLAST (Altschul et al., 1990).
Nucleic Acid Isolation and Analyses
RNA was isolated using a phenol-LiCl procedure (Ausubel et al., 1991), and portions were separated onto a 1.2% agarose gel containing formaldehyde. Gels were stained with ethidium bromide to visualize rRNA bands for verification of equal loading, photographed, and then blotted onto a nylon membrane (Hybond-N, Amersham) and fixed with UV light (Stratalinker 1800, Stratagene). Blots were hybridized with the random-primed 32P-labeled cDNA clone IIa, then washed in 2× SSC, 0.2% SDS at room temperature, 0.5× SSC, 0.2% SDS at 68°C, and 0.1× SSC, 0.2% SDS at 68°C. Autoradiography was generally performed for 24 h at −80°C. DNA was isolated by the method of Gordon-Kamm et al. (1990), cut with restriction enzymes as indicated, and analyzed by DNA-blot hybridization.
CAP Expression in Escherichia coli
Fusion protein expression in E. coli XL1-Blue cells harboring pBluescript containing a cDNA insert was induced with IPTG. XL1-Blue cells containing pBluescript without an insert were included in all of the experiments as a control. Induced cells were pelleted by centrifugation at 1,600g for 15 min in a JA20 rotor (Beckman), resuspended in lysis buffer (50 mm Tris, pH 8.0, 1 mm EDTA, 1 mm PMSF, 10% Suc, 1 mg/mL lysozyme), and incubated for 10 min on ice. Triton X-100 was added to 0.1% final concentration and incubated for 10 min on ice. The solution was centrifuged at 48,000g in a JA20 rotor for 1 h, and the supernatant was removed and mixed with 2× SDS buffer. Protein sequestered in inclusion bodies was extracted from the pellet with SDS buffer. Approximately 5 μg of total protein was fractionated by SDS-PAGE in 10% acrylamide gels, blotted to PVDF membranes, and probed with anti-CAP160 or anti-CAP85 antibodies (Guy et al., 1992).
Plant Transformation and Growth Conditions
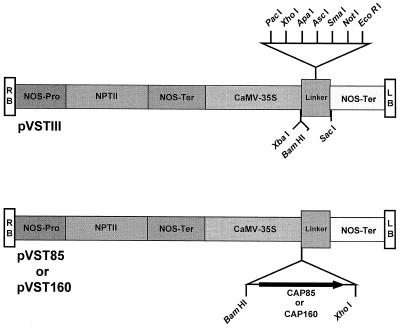
CAP160 and CAP85 (Neven et al., 1993) cDNA sequences were put under the control of the cauliflower mosaic virus 35S promoter by subcloning a 2.7-kb BamHI-XhoI fragment of CAP160 and a 1.8-kb BamHI-XhoI fragment of CAP85 into the broad-host-range vector pVSTIII (Malik and Wahab, 1993), creating plasmids pVST-160 and pVST-85, respectively (Fig. 1). These two plasmids were transferred separately into Agrobacterium tumefaciens strain LBA4404 by direct transformation (Hofgen and Willmitzer, 1988).
Figure 1.
Constructs used in the tobacco transformation experiments (Malik and Wahab, 1993). The right (RB) and left (LB) borders are indicated by the open boxes at the ends of the constructs.
Leaf discs from surface-sterilized young leaves of greenhouse-grown tobacco (Nicotiana tabacum cv Xanthi) plants were infected with A. tumefaciens harboring pVST-160, pVST85, or pVSTIII. Successful transformants were selected on regeneration medium containing 4.3 g/L MS salts (Murashige and Skoog, 1962), 1 mL/L 1000× B5 vitamin mixture, 0.3 mg/L IAA, 10 mg/L dimethylallylaminopurine, 30 g/L Suc, 100 mg/L kanamycin, and 250 mg/L cefotaxime or carbenicillin. Young shoots were transferred to this medium with no hormones and no antibiotic for rooting. Rooted plants were transferred to soil and maintained either in a growth chamber at 25°C or in the greenhouse. Seeds were collected from all primary transformants and sown on solid medium containing MS salts and 200 mg/L kanamycin. Plantlets that remained green were transferred to soil and grown under greenhouse conditions.
Protein Analysis
Leaf material was frozen in liquid nitrogen and then powdered using a mortar and pestle. The frozen powder was transferred directly to SDS buffer (Neven et al., 1993) and boiled for 5 min. Insoluble proteins were removed by spinning the samples for 10 min in a microcentrifuge. The supernatant was transferred to fresh tubes and stored at −20°C. Total protein content was determined by the Bradford method (1976). The presence of CAP160 and CAP85 were determined by protein-blot analysis of crude protein extracts using monoclonal antibodies 2H8 and 5A10, respectively. Equal loading of protein-blot lanes was verified by Coomassie brilliant blue staining of a duplicate gel.
Partial sequencing of CAP160 purified from spinach tissue was accomplished after cleavage with cyanogen bromide as previously described (Neven et al., 1993).
Tissue prints were made from flower, stem, and pedicel tissue from cold-acclimated and nonacclimated transformed tobacco and control, mock-transformed tobacco (Cassab and Varner, 1987). The resulting membranes were probed with CAP85 monoclonal antibody 5A10 (Neven et al., 1993), CAP160 monoclonal antibody 2H8 (Haskell et al., 1993), or dehydrin antibody (Close et al., 1993).
Freeze Tests
Leaves were harvested from either greenhouse- or growth-chamber-grown plants at the temperatures indicated. Leaf discs (three per tube) were frozen at a cooling rate of 0.5 or 1°C/h, as indicated. All samples were equilibrated for 1 h at 0°C, and ice nucleation was induced at −1°C by placing a small ice chip in contact with the tissue. Frozen samples were thawed at 4°C in a refrigerator overnight. The leaf discs were placed in 5 mL of double-distilled water and incubated for 2 h. Electrolyte loss was measured with a conductivity meter. After the initial conductivity measurement, the relative electrolyte loss was determined by refreezing the tissue at −80°C, thawing, and measuring the conductivity a second time.
Drought Tolerance
Young plants at the three-leaf stage were transferred to vermiculite and watered every other day with 5% PEG in one-tenth MS salts. Control plants received one-tenth MS salts alone. After 2 weeks, the concentration of PEG was raised to 10%. Six weeks later the plants were evaluated visually and fresh weights were determined.
RESULTS
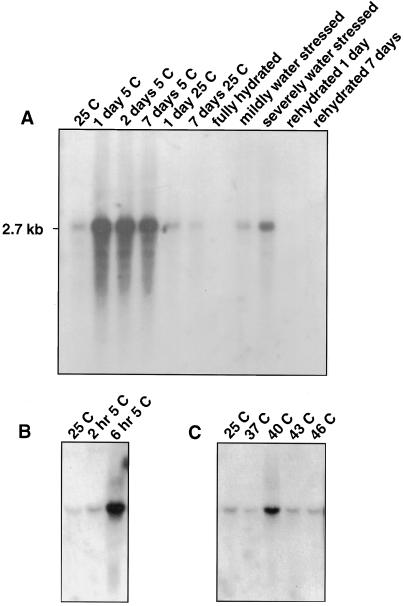
The CAP160 protein is known to be actively synthesized and/or accumulated in leaf tissue exposed to 5°C (Guy and Haskell, 1987, 1988) or during water stress at 25°C (Guy et al., 1992). Therefore, any cDNAs for this protein selected by the antibody screening should hybridize with mRNA that was up-regulated during low-temperature exposure or water stress. Several of the clones hybridized strongly to a 2.7-kb mRNA that was up-regulated during exposure to 5°C and was down-regulated upon return to 25°C (Fig. 2A). Similarly, the cDNAs hybridized to a 2.7-kb mRNA that was abundant in water-stressed leaf tissue, but was low or absent in nonstressed tissue (Fig. 2A). The mRNA for CAP160 reached a maximum abundance in leaf tissue within 24 h of 5°C exposure and remained high for up to 7 d of low-temperature exposure. The transcript abundance was slightly increased at 2 h of 5°C and by 6 h had accumulated to a high level (Fig. 2B). Expression of CAP160 was moderately stimulated by a heat-shock treatment of 40°C for 2 h, but not by higher or lower heat-shock temperatures (Fig. 2C).
Figure 2.
RNA-blot analyses of the influence of cold acclimation, deacclimation, water stress, and heat shock on the CAP160 steady-state mRNA abundance. A, Acclimation, deacclimation, and water-stress responses. B, Short-term response to low temperature. C, Response to heat shock. The cDNA hybridizes to a major 2.7-kb mRNA. Each lane was loaded with 5 μg of total RNA, and equal loading was verified by ethidium-bromide staining before blotting. Similar results were observed in several hybridization experiments. Details regarding the physiologic parameters for the water-stressed samples have been published elsewhere (Guy et al., 1992; Neven et al., 1993).
Sequence analysis of several cDNAs indicated an open reading frame encoding an acidic protein of 780 amino acids (Fig. 3). The position of the first ATG in the sequence has a similarity to a translation initiation site consensus (Joshi, 1987), and the open reading frame is broken by an in-frame stop codon 20 bp upstream from the first ATG, suggesting that the cDNA contains the entire coding sequence. The deduced protein has a molecular mass of 83.9 kD and a pI of 4.6. Whereas the calculated pI is in good agreement with the pI of 4.5 estimated from two-dimensional gels for spinach CAP160, the molecular mass of the deduced protein is little more than one-half of that estimated by SDS-PAGE (Guy and Haskell, 1987).
Figure 3.
Nucleotide and deduced amino acid sequence of CAP160 clone IIa. Amino acids in boldface and underlined correspond to cyanogen bromide-generated peptide sequences from purified CAP160. The dark-shaded box denotes a motif rich in negatively charged residues, and the lighter-shaded box denotes a repeat sequence. An imperfectly tandem repeated sequence is double underlined, and a repeating four-residue motif of P(I/S)(T/K)(G/W) is underlined.
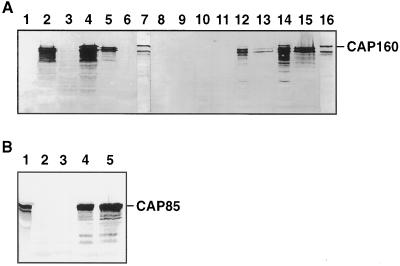
Protein-blot analyses of E. coli possessing pBluescript without an insert showed no immunoreactivity with anti-CAP160 antibodies, whereas cells containing the CAP160 cDNA expressed immunoreactive proteins with or without induction with IPTG (Fig. 4A). Expression was higher with IPTG, and the level of CAP160 accumulation in E. coli appeared to be similar to that found in cold-acclimated spinach leaves (Guy et al., 1992). Two major bands (120 and 140 kD) and a number of minor bands were present, each of which migrated faster than leaf CAP160 (Fig. 4A, upper band in lanes 7 and 16). The lower doublet bands in the leaf extracts of 115 and 110 kD are thought to be proteolytic breakdown products of CAP160 because they generally increase while the 160-kD band decreases with time after extraction. Induced bacterial cells contained a minor band at 160 kD (Fig. 4A, lanes 2, 4, 12, and 14) that comigrated with leaf CAP160 (Fig. 4A, lanes 7 and 16). Because an in-frame stop codon is present before the first ATG, it is probable that the immunoreactive recombinant protein of 160 kD in induced cells is not a β-galactosidase fusion protein, but an authentic-size CAP160 protein resulting from the reinitiation of translation at the first ATG site. The proteins of < 160 kD may be proteolytic products or may result from alternative reinitiation of translation at ATG sites farther downstream. Nevertheless, a small percentage of the protein produced appeared to be full length. This was not unexpected because eukaryotic proteins are often found to be unstable when expressed in E. coli (Ausubel et al., 1991).
Figure 4.
CAP160 and CAP85 protein expression in E. coli XL1-Blue cells. A, Lane 1, pBluescript without an insert; lane 2, CAP160 clone IIa; lane 3, clone IIb; lane 4, clone VIII; lane 5, clone VII; lane 6, clone IX; lane 7, spinach leaf CAP160; lane 8, soluble fraction of pBluescript without an insert minus IPTG; lane 9, insoluble fraction of pBluescript without an insert minus IPTG; lane 10, soluble fraction of pBluescript without an insert plus IPTG; lane 11, insoluble fraction of pBluescript without an insert plus IPTG; lane 12, soluble fraction of CAP160 IIa minus IPTG; lane 13, insoluble fraction of CAP160 IIa minus IPTG; lane 14, soluble fraction of CAP160 IIa plus IPTG; lane 15, insoluble fraction of CAP160 IIa plus IPTG; and lane 16, spinach leaf CAP160. B, Lane 1, Spinach leaf CAP85; lanes 2 and 3, CAP85 out-of-frame clone S42a plus IPTG; and lanes 4 and 5, CAP85 in-frame clone S42aE plus IPTG.
Whether the truncated products retained biological activity remains unknown. CAP160 protein products were present in both soluble and insoluble forms, and it appeared that a significant portion of the expressed protein was not sequestered in inclusion bodies. The 160-kD form was observed only in the soluble fraction (Fig. 4A, lanes 12 and 14). The reason for the low mobility of CAP160 in SDS-PAGE is unknown, but might result from the high content of Ser, Ala, and negatively charged residues (Kondo and Inouye, 1991). The CAP85 cDNA, in which the deduced protein sequence is smaller than the apparent size estimated by SDS-PAGE (Neven et al., 1993), also produced a protein that comigrated with CAP85 from leaf tissue (Fig. 4B).
Additional evidence linking the cDNA of clone IIa with CAP160 is provided by protein sequence information. Two amino acid sequences from peptide fragments prepared by cyanogen bromide cleavage of purified CAP160 closely matched the deduced sequences at residues 83 to 94 and 174 to 192 (Fig. 3).
CAP160 has several unusual motifs that may have a functional significance. It has an acidic region toward the N terminus in which 18 of 26 residues are either aspartic acid or Glu, including a central core where five aspartic acids are followed by four Glu residues (Fig. 3). Two imperfect repeats of 29 residues each are found in the N-terminal half of the protein. They are separated from each other by 133 residues. These two repeats are rich in Thr (24%), Gly (14%), and Asp (14%). Another two large, imperfect repeats of 58 and 56 amino acids, separated by 1 residue, are found beginning at residue 460. Thr (approximately 13%) is the single most abundant residue in these repeats, followed by Val (10.5%). Immediately following the second large repeat is a series of seven 4-residue repeats with the consensus of P(S/I)TG. Preceding the first large repeat is one copy of the 4-residue repeat PSTG. Given the presence of the Pro and Gly in this repeating sequence, this region might be a loop or a solvent-exposed region of the protein. The Ser and Thr residues both bearing hydroxyls might be potential phosphorylation sites on the protein. All of these characteristic features of CAP160 were individually used to do BLAST searches of the databases for matches that could provide insight into its function. In addition, the CAP160 sequence was searched for known protein motifs found in the Prosite library (release 12.2, 3/95) of the Genetics Computer Group (Madison, WI) sequence analysis software package, but no convincing matches were found that would give a clue to its function or identity.
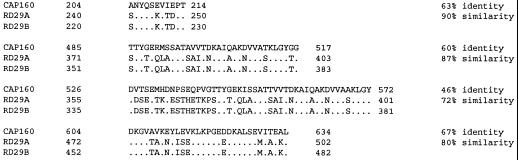
A search of the GenBank database revealed no compelling sequence matches for the entire protein. Although it was suspected that CAP160 was likely the homolog of a pair of Arabidopsis thaliana proteins with several names (cor78, lti140, rd29A, rd29B, and lti65), the total protein sequence similarity was less than 20%. Nevertheless, four discrete regions within the CAP160 sequence did have high homology with the Arabidopsis proteins (Fig. 5), leading us to believe that CAP160 is a probable homolog to these proteins. The two Arabidopsis proteins also have an acidic region (EEDDDDDELEPE or DVEDDDDEYDEQDPE) with similarities to the acidic region of CAP160, which could have some importance to a common function.
Figure 5.
Alignment of CAP160 protein sequence with the Arabidopsis rd29A and rd29B proteins. Identical residues are indicated by dots.
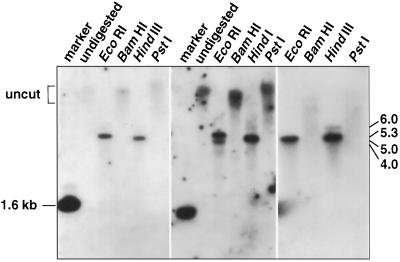
Replicated blots of genomic DNA using cDNA probes showed some blot-to-blot variation, but overall produced the predicted banding pattern based on the DNA sequence from genomic clones for CAP160 (Fig. 6). The banding pattern was relatively simple even when stringency was reduced, indicating that CAP160 is not a member of a large, high-homology gene family.
Figure 6.
Genomic DNA blot for CAP160. Results of three different experiments are shown. Clone IIa was used to make the hybridization probe.
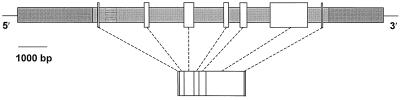
The isolation of genomic clones for CAP160 permitted the characterization of the organization of the gene. The CAP160 protein is encoded on seven exons extending over 8 kb. The exons range from 71 to 1254 bp in length, whereas the introns tend to be larger than the exons, ranging from a minimum of 440 bp to a maximum of 1437 bp (Fig. 7). Characterization of genomic clones with differences in restriction fragment length revealed the existence of a 352-bp deletion in intron 1 (Fig. 7). Aside from this deletion, the nucleotide sequences in both intron and exon regions of all isolated clones were virtually identical, suggesting the possibility that if it does not represent a second gene sequence, this clone represents an allelic sequence.
Figure 7.
Structure of the CAP160 gene. Flanking sequences are indicated by dark shading, noncoding and intron sequences are lightly shaded, and exons are shown as unshaded boxes. The hatched area in intron 1 shows the position of a large deletion present in some sequences.
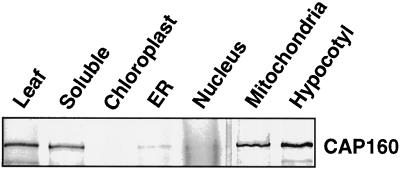
Subcellular-localization studies were conducted with leaf and hypocotyl tissue to determine where in the cell CAP160 is found. These studies were performed at the same time as studies of another spinach CAP, the results of which have been previously reported (Neven et al., 1993). This previous report includes the marker protein analysis for relative fraction contamination. The major organellar fractions were prepared, and these extracts were probed with 2H8, a monoclonal antibody specific for CAP160, using protein-blot methods (Fig. 8). A major portion of CAP160 was present in a freely soluble fraction, which would be consistent with a protein present in the cytosol and not tightly associated with any macromolecular structure that would be pelleted at relatively low g forces. However, an equally large proportion was found in the mitochondrial fraction. Whether the protein was inside or outside of the mitochondria was not determined. Protein blots using antibodies for marker proteins for all of the fractions shown in Figure 8 indicated that the presence of the strong CAP160 reactivity in the mitochondrial fraction was well in excess of the documented cross-contamination (Neven et al., 1993), and thus was likely to reflect an actual cofractionation.
Figure 8.
Subcellular localization of CAP160. The presence of CAP160 was detected by protein blot using a monoclonal antibody, 2H8, which is specific for CAP160. For marker protein analysis and relative contamination for each subcellular fraction, see Neven et al. (1993).
Expression of CAP160 and CAP85 in Transgenic Tobacco
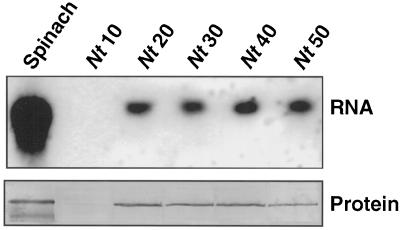
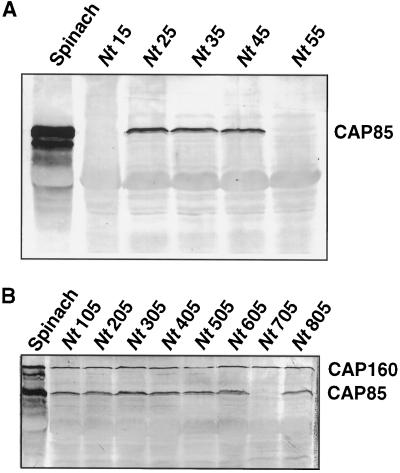
Constructs using the cDNAs for CAP160 and CAP85 (Neven et al., 1993) were placed under the control of the 35S promoter and used in A. tumefaciens-based transformation experiments. Putative transformants were assayed for expression of CAP160 and CAP85 by protein-blot analyses using highly specific monoclonal antibodies. In these experiments protein was isolated from plants grown under either growth-chamber conditions (25°C) or greenhouse conditions. Some of the leaves (as indicated) were taken from plants that were grown under sterile conditions in sealed containers with a very high humidity level. Several transformants were found to express the mRNA for each introduced gene (Fig. 9). No cross-reacting protein was found in either wild-type tobacco (not shown) or in tobacco transformed with the vector alone. CAP85 was resolved as a doublet consisting of an intense 85-kD band and a smaller and less-intense 80- to 83-kD band in spinach (Neven et al., 1993), and its expression also resulted in a doublet in transgenic tobacco. R0 CAP85 plants (Nt25 and Nt35) were crossed with R0 CAP160 plants (Nt40), and the progeny were evaluated for expression of the two proteins. A large number of the plants expressed both proteins, a few expressed only CAP160, but no plants were found that expressed only CAP85 or neither of the two proteins (Fig. 10).
Figure 9.
Analysis of spinach CAP160 RNA and protein expression in transgenic lines of tobacco. Spinach RNA and protein are shown for reference. Nt10 was transformed with the vector only. Hybridizations were done with radiolabeled cDNA clone IIa, and protein detection was with monoclonal antibody 2H8. RNA blots were loaded with 5 μg of total RNA per lane. Protein blots were loaded with 20 μg of total buffer-soluble protein per lane.
Figure 10.
Analysis of spinach CAP85 expression in tobacco (A) and coexpression of CAP85 and CAP160 in progeny from a cross of lines expressing individually either CAP85 or CAP160 (B). Cross-reacting proteins were detected with a mixture of monoclonal antibody 2H8 for CAP160 and monoclonal antibody 5A10 for CAP85.
Tissue prints were done on nitrocellulose with cold-acclimated or nonacclimated stem, pedicel, and flower tissue taken from transformed tobacco plants to assess expression patterns at the tissue level. The resulting membranes were exposed to CAP85, CAP160, or both antibodies. Both proteins were expressed in all tissues examined, and no difference was seen between cold-acclimated and nonacclimated tissue (not shown). When the same tissues were analyzed by tissue printing and probed with a dehydrin antibody (Close et al., 1993), greater staining was seen in the cold-acclimated tissue than in the nonacclimated tissue. Tissue prints from control, mock-transformed tobacco did not react with either of the two monoclonal antibodies for the spinach proteins, but did react with the anti-dehydrin antibody.
Seeds were collected from all of the R0 plants, regardless of whether they expressed the introduced CAP genes. Seeds were sown on kanamycin and the number of green plants (kanamycin resistant) versus white plants was evaluated. The number of white plants versus green plants was somewhat lower than the expected Mendelian ratio of 1:3 (1:6.6 for CAP160, 1:5 for CAP85), indicating the possibility that more than one, but probably only a few, copies of the transferred genes integrated into the genome. Selected green plants were evaluated by protein-blot analysis, and all expressed either the expected CAP160 or CAP85. Green, 1-month-old plantlets were transferred to soil, and at the three-leaf stage they were transferred into either cold (3°C) or warm (20°C) conditions. After 14 d, protein was isolated and analyzed by protein blot. In all cases the quantities of CAP85 and CAP160 protein were lower in the plants grown in the cold (data not shown).
Freezing Tests
The first test for increased hardiness was an electrolyte-leakage test performed on leaf discs of transformed and mock-transformed plants. Initial tests were conducted on R0 plants grown at 25°C. Only one transformant, CAP160 Nt40, exhibited any evidence of a decrease in electrolyte leakage, but even that was not statistically significant (Table I). A sample from each plant was taken each time the electrolyte-leakage test was performed to assay for CAP160 or CAP85 expression.
Table I.
Freezing tolerance of an Ro transgenic tobacco line expressing the spinach CAP160 protein
| Temperature | Electrolyte Leakage
|
|
|---|---|---|
| Transformed controls | CAP160 (Nt40) | |
| °C | ||
| 0.0 | 26 ± 1 | 30 ± 7 |
| −1.0 | 30 ± 2 | 31 ± 3 |
| −1.5 | 31 ± 3 | 34 ± 4 |
| −2.0 | 50 ± 6 | 43 ± 11 |
| −2.5 | 60 ± 6 | 52 ± 12 |
The data are presented as the arcsin-transformed percentage of the relative electrolyte leakage. Two different mock-transformed lines were used and the data were combined. The experiment was repeated five times.
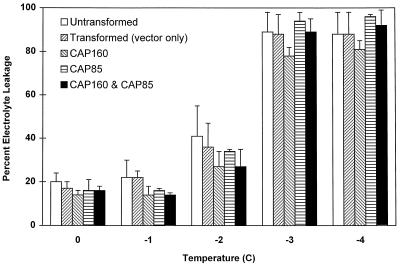
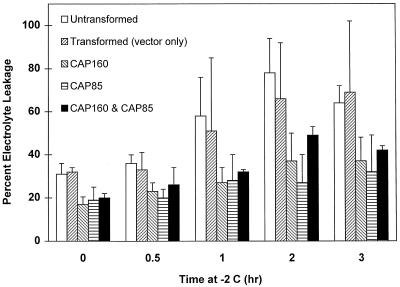
The second set of experiments was performed on the R1 generation. In this case, two sets of experiments were run. The first was a determination of LT50 as described above (Fig. 11). A slight improvement was seen in the hardiness of plants expressing the spinach protein only at −1°C, but overall the LT50 values were not different. The second test, a “kinetics-of-freezing” analysis, was designed to reveal more subtle differences between the plants. In this test, samples were placed at the approximated LT50 of −2°C and removed at various times. At all times, the plants expressing the spinach proteins exhibited less electrolyte leakage than the untransformed control plants. Because of the large variability in the mock-transformed plants, no significant difference was found between it and either the untransformed plants or those expressing the spinach proteins (Fig. 12).
Figure 11.
Influence of CAP85 or CAP160 expression or coexpression of both proteins on the severity of injury after freezing. Error bars represent the sd of two to four experiments.
Figure 12.
Differential sensitivity of tobacco expressing spinach cold-stress proteins in response to a time-course freeze/thaw regime. Error bars represent the sd of five experiments.
Whole plants expressing CAP160 or CAP85, mock-transformed (pVSTIII, vector only) plants, or control cv Xanthi plants (nontransformed) were transferred at approximately the three-leaf stage into a cold room. These plants were incubated for 1 month at 5°C. Visual inspection revealed no obvious difference in either the amount of leaf damage or plant size. The temperature was reduced to approximately 0°C, and although all of the plants suffered minor leaf damage, all survived (data not shown). Protein analyses indicated no differences in the levels of either spinach protein over that of similar plants kept at 20°C.
DISCUSSION
The function of CAP160 in spinach and its role in plant cold stress remains obscure. Homology searches indicate that CAP160, in its entirety, is a novel protein, the biological function of which cannot be identified by shared homology with proteins of known function. CAP160 does have limited similarity to the Arabidopsis lti78, or COR78 protein, a low-temperature-induced (Horvath et al., 1993; Nordin et al., 1993) and desiccation-responsive protein, also known as rd29A and rd29B (Yamaguchi-Shinozaki, 1993a, 1993b). The shared homology is enough for us to consider it a spinach homolog to these proteins. The primary structure of CAP160 shows no similarity to antifreeze or ice-nucleation proteins, the function of which is to alter the freezing of water (Green and Warren, 1985; Davies and Hew, 1990). However, several noteworthy features of the primary structure of CAP160 (Fig. 3) could provide helpful clues to its functional role in low-temperature-stress response mechanisms. These include a highly negatively charged motif at residues 61 to 86, which includes a run of nine consecutive aspartates and glutamates and several repeat motifs, a large tandem imperfect repeat, and a four-residue imperfect-repeating motif present in eight copies. All of these features were used to search the databases for matching sequences that might provide a clue to CAP160's function. Only the acidic residue tract 61 to 86 yielded a noteworthy similarity.
A variety of proteins possess extensive tracts of negatively charged residues, including nucleolins (Lapeyre et al., 1987), some transcription factors (Hisatake et al., 1991), Na/Ca,K exchanger (Reilander et al., 1992), spermine-binding protein (Chang et al., 1987), calsequestrin (Yazaki et al., 1990), and ubiquinol-Cyt c reductase protein (Van Loon et al., 1984). Of these, the relationship to that of nucleolin seems most notable. Nucleolin is a major nucleolar phosphoprotein in eukaryotic cells (Fang and Yeh, 1993) that has sequence-specific RNA-binding activity that regulates, or participates in, rRNA processing (Hanakahi et al., 1997). CAP160 lacks many of the structural features of plant nucleolin, such as nuclear-localization sequences (Bogre et al., 1996), and we do not consider it to be a nucleolin. However, it has some unusual similarities, including the fact that it is a phosphoprotein (Guy and Haskell, 1989) and is extremely sensitive to proteolytic cleavage even in purified form (D. Haskell and C. Guy, unpublished data). Nucleolin has a self-cleaving activity that resides in the C-terminal domain and that apparently recognizes repeated motifs within the molecule (Fang and Yeh, 1993). CAP160 has repeats, although not as many as are found in nucleolins.
Because one of the major effects of low-temperature exposure is a significant reduction in translation rate, perhaps CAP160's function is to participate as an accessory protein in the assembly of ribosomes or to help accelerate translation in some way. Evidence exists to suggest that the capacity of the translation apparatus is enhanced during cold acclimation (Guy et al., 1985) during a time when CAP160 is accumulating. CAP160's subcellular localization appears to be predominately cytosolic, which would be consistent with such a function. However, given its apparent cofractionation with mitochondria, alternative functions are also a possibility.
Analysis of CAP160 gene structure revealed further differences from that of the Arabidopsis cold- and drought-responsive rd29 genes. CAP160 has more and much larger introns than either of the two Arabidopsis genes, six versus three each. In addition, the two Arabidopsis genes are within 2 kb of each other, but we could find no evidence from genomic clones that such a close tandem arrangement exists for CAP160 in spinach.
The coding sequences from cDNA clones for two genes that are up-regulated during cold acclimation in spinach were placed under the control of the nearly constitutive cauliflower mosaic virus promoter and transformed separately into tobacco, a freezing-sensitive plant. These two genes were expressed in the transformed tobacco as determined by protein-blot analysis, whereas no cross-reacting proteins to the monoclonal antibodies were found in nontransformed tobacco. However, both of the proteins were expressed in tobacco at an apparently lower level than under low-temperature-induction conditions in spinach. Tests designed to determine whether their expression conferred increased tolerance to low-temperature stress indicated little overall benefit to tobacco. Freezing tolerance is considered to be a quantitative trait (Hayes et al., 1993); therefore, it is not surprising that the heterologous expression of one or two genes induced during cold acclimation would significantly alter the LT50 of freezing-sensitive tobacco. This result is similar to that when a cold-regulated, osmotin-like protein was overexpressed in potato and had no effect on freezing tolerance (Zhu et al., 1996), but it is in contrast to the constitutive overexpression of the COR15a protein in Arabidopsis (Artus et al., 1996).
In the work with Arabidopsis, overall cell survival was only subtly enhanced after a freeze/thaw cycle for overexpressing cells, but a more dramatic, positive benefit of overexpression of COR15a was the preservation of chloroplast electron-transport function by this chloroplast stromal protein. Other examples of this approach include several attempts to enhance yeast and plant freezing tolerance through the introduction of cold-water fish antifreeze proteins (Georges et al., 1990; Hightower et al., 1991; McKown and Warren, 1991; Kenward et al., 1993). Expression of fish antifreeze proteins has not significantly altered freezing of plant tissues (Hightower et al., 1991). The results of the expression of spinach cold-stress proteins in tobacco do not rule out the possibility that major enhancements in the tolerance of freezing-sensitive plants might be achieved through the expression of one or a few stress proteins, but instead further support the view of the quantitative nature of plant freezing-stress tolerance (Hayes et al., 1993).
CAP85 and CAP160 are continuously and highly expressed during cold acclimation and under continuing cold treatment, making them unlikely candidates for regulatory genes, which in all likelihood would be rare messages expressed very early during cold acclimation. A large number of genes are induced in the process of cold acclimation (Kaye and Guy, 1995). Although many of these genes fall into one of a few multigene families, particularly the LEA/dehydrin family, their function remains unknown. Most are probably involved in the metabolic adjustment of the plant during cold acclimation. The two clones used in this study, CAP85 and CAP160, are both members of the LEA/dehydrin superfamily. LEA and dehydrin proteins are known to accumulate during the later stages of seed development or during water stress and are thought to play a role during seed dry-down (Close et al., 1993). Recent evidence suggests that at least one of the spinach proteins may have cryoprotective properties (Kazuoka and Oeda, 1992). Purified CAP85 was shown to protect lactate dehydrogenase in vitro from a freeze/thaw inactivation (Kazuoka and Oeda, 1992). This opens the door to the possibility that many low-temperature-regulated genes may have cryoprotective properties (Artus et al., 1996). More likely, CAP160's role, given its subcellular localization and its induction during drought stress, will be stabilization of supramolecular structures such as membranes, ribosomes, or cytoskeletal elements; acting as that of a proteinaceous compatible solute that reduces the toxic effects of cellular solutes at a high concentration during dehydration; or that of an enzyme that synthesizes a product with compatible solute properties. Regardless of its mode of action, CAP160 represents a new class of intracellular, low-temperature-stress proteins with a function correlated with enhanced freezing tolerance only in spinach.
ACKNOWLEDGMENTS
We are grateful to J. Anderson, C. Zhang, D. Sung, and an anonymous reviewer for their constructive comments during the preparation and review of the manuscript, and T. Close for the gift of the anti-dehydrin antibody. Facilities and assistance for protein and DNA sequencing, antibody production, and computer services were provided by the core laboratories of the Interdisciplinary Center for Biotechnology Research. We also thank the Florida High Tech Council for support.
Abbreviations:
- CAP
cold-acclimation protein
- IPTG
isopropyl-1-thio-β-galactoside
- LEA
late embryogenesis abundant
- LT50
killing temperature for 50% of the cells
- MS
Murashige and Skoog
Footnotes
This research was supported in part by the U.S. Department of Agriculture/National Research Initiative Competitive Grants Program (grant no. 93-37100-8906). This is Florida Agricultural Experiment Station journal series no. R-06081.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Artus NN, Uemura M, Steponkus P, Gilmour SJ, Lin C, Thomashow MF. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA. 1996;93:13404–13409. doi: 10.1073/pnas.93.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1991) Current Protocols in Molecular Biology, Vol 2. Wiley, New York
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bogre L, Jonak C, Mink M, Meskiene I, Traas J, Ha DT, Swoboda I, Plank C, Wagner E, Heberle-Bors E and others. Developmental and cell cycle regulation of alfalfa nucMs1, a plant homolog of the yeast Nsr1 and mammalian nucleolin. Plant Cell. 1996;8:417–428. [PMC free article] [PubMed] [Google Scholar]
- Cassab GI, Varner JE. Immunocytolocalization of extensin in developing soybean seed coats by immunogold-silver staining and by tissue printing on nitrocellulose paper. J Cell Biol. 1987;105:2581–2588. doi: 10.1083/jcb.105.6.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattivelli L, Bartels D. Molecular cloning and characterization of cold-regulated genes in barley. Plant Physiol. 1990;93:1504–1510. doi: 10.1104/pp.93.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Saltzman AG, Hiipakka RA, Huang IY, Liao SS. Prostatic spermine-binding protein: cloning and nucleotide sequence of cDNA, amino acid sequence, and androgenic control of mRNA level. J Biol Chem. 1987;262:2826–2831. [PubMed] [Google Scholar]
- Close TJ. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant. 1996;97:795–803. [Google Scholar]
- Close TJ, Fenton RD, Moonan F. A view of plant dehydrins using antibodies specific to the carboxy terminal peptide. Plant Mol Biol. 1993;23:279–286. doi: 10.1007/BF00029004. [DOI] [PubMed] [Google Scholar]
- Davies PL, Hew CL. Biochemistry of fish antifreeze proteins. FASEB J. 1990;4:2460–2468. doi: 10.1096/fasebj.4.8.2185972. [DOI] [PubMed] [Google Scholar]
- Dure L., III A repeating 11-mer amino acid motif and plant desiccation. Plant J. 1993;3:363–369. doi: 10.1046/j.1365-313x.1993.t01-19-00999.x. [DOI] [PubMed] [Google Scholar]
- Fang SH, Yeh NH. The self-cleaving activity of nucleolin determines its molecular dynamics in relation to cell proliferation. Exp Cell Res. 1993;208:48–53. doi: 10.1006/excr.1993.1221. [DOI] [PubMed] [Google Scholar]
- Georges F, Saleem M, Cutler AJ. Design and cloning of a synthetic gene for the flounder antifreeze protein and its expression in plant cells. Gene. 1990;91:159–165. doi: 10.1016/0378-1119(90)90083-4. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Artus NN, Thomashow MF. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol Biol. 1992;18:13–21. doi: 10.1007/BF00018452. [DOI] [PubMed] [Google Scholar]
- Goldstein J, Pollitt NS, Inouye M. Major cold shock protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Kamm WJ, Spencer TM, Mangano ML, Adams TR, Daines RJ, Start WG, O'Brien JV, Chambers SA, Adams WR, Jr, Willets NG. Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell. 1990;2:603–618. doi: 10.1105/tpc.2.7.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RL, Warren GJ. Physical and functional repetition in a bacterial ice nucleation gene. Nature. 1985;317:645–648. [Google Scholar]
- Guo W, Ward RW, Thomashow MF. Characterization of a cold-regulated wheat gene related to Arabidopsis thaliana cor47. Plant Physiol. 1992;100:915–922. doi: 10.1104/pp.100.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C, Haskell D, Neven L, Klein P, Smelser C. Hydration-state-responsive proteins link cold and drought stress in spinach. Planta. 1992;188:265–270. doi: 10.1007/BF00216823. [DOI] [PubMed] [Google Scholar]
- Guy CL, Haskell D. Induction of freezing tolerance in spinach is associated with the synthesis of cold acclimation induced proteins. Plant Physiol. 1987;84:872–878. doi: 10.1104/pp.84.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL, Haskell D. Detection of polypeptides associated with the cold acclimation process in spinach. Electrophoresis. 1988;9:787–796. doi: 10.1002/elps.1150091115. [DOI] [PubMed] [Google Scholar]
- Guy CL, Haskell D. Preliminary characterization of high molecular mass proteins associated with cold acclimation in spinach. Plant Physiol Biochem. 1989;27:777–784. [Google Scholar]
- Guy CL, Niemi KJ, Brambl R. Altered gene expression during cold acclimation of spinach. Proc Natl Acad Sci USA. 1985;82:3673–3677. doi: 10.1073/pnas.82.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Walbot V. Effects of cold-treatment on protein synthesis and mRNA levels in rice leaves. Plant Physiol. 1989;91:930–938. doi: 10.1104/pp.91.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanakahi LA, Dempsey LA, Li MJ, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci USA. 1997;94:3605–3610. doi: 10.1073/pnas.94.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskell DW, Anderson JV, Guy CL. Antigen binding of a mouse monoclonal IgG1 is inactivated by heating but not by freeze/thaw cycling. Cryobiology. 1993;30:532–535. doi: 10.1006/cryo.1993.1054. [DOI] [PubMed] [Google Scholar]
- Hayes PM, Blake T, Chen THH, Tragoonrung S, Chen F, Pan A, Liu B. Quantitative trait loci on barley (Hordeum vulgare L.) chromosome 7 associated with components of winter hardiness. Genome. 1993;36:66–71. doi: 10.1139/g93-009. [DOI] [PubMed] [Google Scholar]
- Hightower R, Baden C, Penzes E, Lund P, Dunsmuir P. Expression of antifreeze proteins in transgenic plants. Plant Mol Biol. 1991;17:1013–1021. doi: 10.1007/BF00037141. [DOI] [PubMed] [Google Scholar]
- Hisatake K, Nishimura T, Maeda Y, Hanada K, Song CZ, Muramatsu M. Cloning and structural analysis of cDNA and the gene for mouse transcription factor UBF. Nucleic Acids Res. 1991;19:4631–4637. doi: 10.1093/nar/19.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, McLarney BK, Thomashow MF. Regulation of Arabidopsis thaliana L. (Heynh) cor78 in response to low temperature. Plant Physiol. 1993;103:1047–1053. doi: 10.1104/pp.103.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M, Danyluk J, Laliberté JF, Rassart E, Dhindsa RS, Sarhan F. Cloning, characterization, and expression of a cDNA encoding a 50-kilodalton protein specifically induced by cold acclimation in wheat. Plant Physiol. 1992;99:1381–1387. doi: 10.1104/pp.99.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Dunn MA. The molecular biology of plant acclimation to low temperature. J Exp Bot. 1996;296:291–305. [Google Scholar]
- Joshi CP. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye C, Guy CL. Perspectives of plant cold tolerance: physiology and molecular responses. Sci Prog. 1995;78:271–299. [PubMed] [Google Scholar]
- Kazuoka T, Oeda K. Heat-stable COR (cold-regulated) proteins associated with freezing tolerance in spinach. Plant Cell Physiol. 1992;33:1107–1114. [Google Scholar]
- Kenward KD, Altschuler M, Hildebrand D, Davies PL. Accumulation of type I fish antifreeze protein in transgenic tobacco is cold-specific. Plant Mol Biol. 1993;23:377–385. doi: 10.1007/BF00029012. [DOI] [PubMed] [Google Scholar]
- Kondo K, Inouye M. TIP 1, a cold shock-inducible gene of Saccharomyces cerevisiae. J Biol Chem. 1991;266:17537–17544. [PubMed] [Google Scholar]
- Kurkela S, Franck M. Cloning and characterization of a cold- and ABA-inducible Arabidopsis gene. Plant Mol Biol. 1990;15:137–144. doi: 10.1007/BF00017731. [DOI] [PubMed] [Google Scholar]
- Lapeyre B, Bourbon H, Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CL, Thomashow MF. DNA sequence analysis of a complementary DNA for cold-regulated Arabidopsis gene cor15 and characterization of the COR15 polypeptide. Plant Physiol. 1992;99:519–525. doi: 10.1104/pp.99.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Gross JK. Molecular cloning and characterization of winter flounder antifreeze cDNA. Proc Natl Acad Sci USA. 1981;78:2825–2829. doi: 10.1073/pnas.78.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik VS, Wahab SZ. Versatile vectors for expressing genes in plants. J Plant Biochem Biotechnol. 1993;2:69–70. [Google Scholar]
- Mazur PJ. Kinetics of water loss from cells at subzero temperatures and the likelihood of intracellular freezing. Gen Physiol. 1963;47:347–369. doi: 10.1085/jgp.47.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown RL, Warren GJ. Enhanced survival of yeast expressing an antifreeze gene analogue after freezing. Cryobiology. 1991;28:474–482. doi: 10.1016/0011-2240(91)90057-u. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Neven LG, Haskell DW, Hofig A, Li QB, Guy CL. Characterization of a spinach gene responsive to low temperature and water stress. Plant Mol Biol. 1993;21:291–305. doi: 10.1007/BF00019945. [DOI] [PubMed] [Google Scholar]
- Nordin K, Vahala T, Palva ET. Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol. 1993;21:641–653. doi: 10.1007/BF00014547. [DOI] [PubMed] [Google Scholar]
- Orr W, Lu B, White TC, Robert LS, Singh J. Complementary DNA sequence of a low temperature-induced Brassica napus gene with homology to the Arabidopsis thaliana kin1 gene. Plant Physiol. 1992;98:1532–1534. doi: 10.1104/pp.98.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RS. Extracellular ice and cell shape in frost-stressed cereal leaves: a low temperature scanning-electron microscopy study. Planta. 1988;175:313–324. doi: 10.1007/BF00396336. [DOI] [PubMed] [Google Scholar]
- Reilander H, Achilles A, Friedel U, Maul G, Lottspeich F, Cook NJ. Primary structure and functional expression of the Na/Ca,K-exchanger from bovine rod photoreceptors. EMBO J. 1992;11:1689–1695. doi: 10.1002/j.1460-2075.1992.tb05219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Larcher W (1987) Frost Survival of Plants: Responses and Adaption to Freezing Stress. Springer-Verlag, Berlin, pp 138–153
- Steponkus PL. Role of the plasma membrane in freezing injury and cold acclimation. Annu Rev Plant Physiol. 1984;35:543–584. [Google Scholar]
- Steponkus PL, Uemura M, Balsamo RA, Arvinte T, Lynch DV. Transformation of the cryobehavior of rye protoplasts by modification of the plasma membrane lipid composition. Proc Natl Acad Sci USA. 1988;85:9026–9030. doi: 10.1073/pnas.85.23.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton F, Ding X, Kenefick DG. Group 3 LEA gene HVA1 regulation by cold-acclimation and deacclimation in two barley cultivars with varying freezing resistance. Plant Physiol. 1992;99:338–340. doi: 10.1104/pp.99.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Arabidopsis thaliana as a model for studying mechanisms of plant cold tolerance. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 807–834. [Google Scholar]
- Van Loon AP, De Groot RJ, De Haan M, Dekker A, Grivell LA. The DNA sequence of the nuclear gene coding for the 17-kd subunit VI of the yeast ubiquinol-cytochrome c reductase: a protein with an extremely high content of acidic amino acids. EMBO J. 1984;3:1039–1043. doi: 10.1002/j.1460-2075.1984.tb01924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol. 1993a;101:1119–1120. doi: 10.1104/pp.101.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet. 1993b;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- Yazaki PJ, Salvatori S, Dahms AS. Amino acid sequence of chicken calsequestrin deduced from cDNA: comparison of calsequestrin and aspartactin. Biochem Biophys Res Commun. 1990;170:1089–1095. doi: 10.1016/0006-291x(90)90504-g. [DOI] [PubMed] [Google Scholar]
- Zhu B, Chen TH, Li PH. Analysis of late-blight disease resistance and freezing tolerance in transgenic potato plants expressing sense and antisense genes for an osmotin-like protein. Planta. 1996;198:70–77. doi: 10.1007/BF00197588. [DOI] [PubMed] [Google Scholar]