Abstract
The mechanistic target of rapamycin (mTOR) kinase is a conserved regulator of cell growth, proliferation, and survival. In cells, mTOR is the catalytic subunit of two complexes called mTORC1 and mTORC2, which have distinct upstream regulatory signals and downstream substrates. mTORC1 directly senses cellular nutrient availability while indirectly sensing circulating nutrients through growth factor signaling pathways. Cellular stresses that restrict growth also impinge on mTORC1 activity. mTORC2 is less well understood and appears only to sense growth factors. As an integrator of diverse growth regulatory signals, mTOR evolved to be a central signaling hub for controlling cellular metabolism and energy homoeostasis, and defects in mTOR signaling are important in the pathologies of cancer, diabetes, and aging. Here we discuss mechanisms by which each mTOR complex might regulate cell survival in response to metabolic and other stresses.
mTOR is a signaling hub for cell metabolism. It serves as a point of convergence between a nutrient-sensing pathway and PI3K–AKT signaling (i.e., as part of mTORC1) and as a regulator of AKT itself (i.e., as part of mTORC2).
The mechanistic target of rapamycin (mTOR) is a highly conserved serine/threonine protein kinase belonging to the phosphatidylinositol kinase-related kinase (PIKK) family and in mammalian cells is a central regulator of cell growth, proliferation, and survival (for review, see Sengupta et al. 2010; Zoncu et al. 2010). As its name implies, mTOR is the target of the naturally occurring compound rapamycin, which in association with the FK506-binding protein (FKBP12) is an allosteric inhibitor of mTOR. Although rapamycin is now known to only partially inhibit mTOR activity, derivatives of the drug have important clinical applications in oncology, in preventing restenosis after angioplasty, and as an immunosuppressant following organ transplants.
THE mTOR COMPLEXES AND THEIR REGULATION
In cells, mTOR exists in two distinct complexes called mTOR complex 1 (mTORC1) and mTORC2 that have both shared and unique subunits (Fig. 1). In addition to mTOR, both complexes contain mammalian Lethal with SEC13 protein 8 (mLST8 also known as GβL) and DEP domain-containing mTOR-interacting protein (DEPTOR). Regulatory-associated protein of mTOR (RAPTOR) and 40-kDa pro-rich Akt-substrate (PRAS40) are unique to mTORC1, whereas rapamycin-insensitive companion of mTOR (RICTOR), mammalian stressed-activated map-kinase interacting protein 1 (mSIN1), and protein observed with RICTOR (PROTOR) are specific to mTORC2. Recent work is beginning to reveal insight into the structure of mTORC1 (Yip et al. 2010). A cryo-EM structure of mTORC1 at 26 Å indicates that the complex is a dimer with interlocking mTOR–RAPTOR interactions and where PRAS40 acts as a competitive inhibitor for the binding of mTORC1 substrates to RAPTOR (Wang et al. 2008; Yip et al. 2010). mLST8 associates with the mTOR kinase domain, located in the carboxyl terminus (Kim et al. 2003). In the presence of rapamycin, the mTOR–mLST8 interaction is stable, whereas the drug weakens the mTOR–RAPTOR interaction (Kim and Sabatini 2004; Yip et al. 2010). The structure of mTORC2 remains a mystery.
Figure 1.
The mTOR pathway. The mTOR kinase exists in two distinct complexes called mTOR complex 1 (mTORC1) and mTORC2. In addition to mTOR, both complexes also contain mLST8 and DEPTOR. RAPTOR and PRAS40 are unique to mTORC1, whereas RICTOR, mSIN1, and PROTOR are specific to mTORC2. mTORC1 is well known to control cell growth. Although the downstream mechanisms by which mTORC1 controls growth are still being elucidated, several direct mTORC1 substrates have now been validated. For example, mTORC1 can regulate growth by directly phosphorylating S6K1 and 4EBP1, two regulators of protein translation; by regulating autophagy through the direct phosphorylation of ULK1; by regulating lipid metabolism at least in part by phosphorylating Lipin1; and by modulating insulin signaling through Grb10. In cells, mTORC1 activity is controlled by nutrient availability, particularly that of amino acids, through a novel pathway requiring the Rag GTPases. In the presence of amino acids, the Rags deliver mTORC1 to the lysosome, which is thought to be the signaling center for mTORC1-driven growth control (not shown but described in text). Growth factor signaling through PI3K–AKT, as well as numerous stresses, such as energy deprivation and hypoxia, can also impinge on mTORC1 activity through the TSC1–TSC2 complex. TSC2 negatively regulates a small GTPase called Rheb that directly activates mTORC1 by an unknown biochemical mechanism. AKT can also activate mTORC1 by directly phosphorylating PRAS40, which relieves an inhibitory function of this subunit. mTORC2 was discovered more recently, and although it is known to be activated in response to growth factors, the mechanisms are unknown. The best described substrates of mTORC2 are AKT and SGK, which require mTORC2-dependent phosphorylation for full biochemical activity. PKC is also regulated by mTORC2.
mTORC1 is the best understood mTOR complex and is well known to control cell autonomous growth by integrating at least four growth regulatory inputs: nutrient availability, growth factor signaling, cellular energy status, and cellular stress levels (for review, see Sengupta et al. 2010; Zoncu et al. 2010). The best described mechanism by which mTORC1 controls growth is by directly phosphorylating two regulators of protein translation, p70-S6 kinase 1 (S6K1) and 4E binding protein 1 (4E-BP1). mTORC1-dependent phosphorylation of S6K at T389 activates its kinase activity toward several substrates involved in mRNA maturation and protein translation. In contrast to S6K1, 4E-BP1 represses translation, and its multisite phosphorylation by mTORC1 decreases its affinity to the translation initiation factor eIF4E, thus activating cap-dependent translation (for review, see Ma and Blenis 2009). Recently, several new substrates for mTORC1 have been identified and characterized. For example, mTORC1 also controls growth by negatively regulating autophagy through the direct phosphorylation of Unc-51-like kinase (ULK1; discussed below) (Chan 2009; Ganley et al. 2009; Hosokawa et al. 2009; Jung et al. 2009). In addition, mTORC1 directly phosphorylates the phosphatidate phosphatase Lipin1 (which regulates lipid metabolism through SREBP1) and the growth factor receptor-bound protein 10 (Grb10; part of a negative feedback loop targeting insulin receptor signaling) (Hsu et al. 2011; Peterson et al. 2011; Yu et al. 2011). In fact, two recent proteomic studies identifying Grb10 as an mTORC1 substrate further suggest that the insulin-stimulated phosphorylation cascade is largely mTOR-dependent (Hsu et al. 2011; Yu et al. 2011). A role for mTORC1 in regulating PGC1α activity and mitochondrial function has also been described (Cunningham et al. 2007; Ramanathan and Schreiber 2009).
How do upstream signals regulate mTORC1? In multicellular organisms, one mechanism by which growth factor signaling regulates mTORC1 is through the PI3K–AKT pathway (for review, see Manning and Cantley 2007). Following phosphoinositide 3-kinase (PI3K) activation, AKT (also known as PKB) is recruited to the membrane, which triggers its phosphorylation and activation. Among its many substrates, AKT directly phosphorylates and inactivates TSC2, which together in a complex with TSC1 negatively regulates mTORC1 activity (Inoki et al. 2002). The TSC1/TSC2 complex inhibits mTORC1 by suppressing the activity of its activator, a small GTPase called Ras homolog enriched in brain (Rheb) (for review, see Manning and Cantley 2003). AKT can also activate mTORC1 by directly phosphorylating PRAS40 and relieving its inhibitory effect on the complex (Sancak et al. 2007; Vander Haar et al. 2007). In addition to being inactivated by PI3K–AKT signaling, the TSC1/TSC2 complex can also be inhibited by other growth-promoting signals including mitogens or cytokines through the action of the mitogen-activated protein kinase (MAPK) ERK1/2 and the p90 ribosomal S6 kinase (RSK) (for review, see Huang and Manning 2008). In contrast, growth inhibitory signals such as energy deprivation and hypoxia can activate TSC1/TSC2 through the AMP-activated protein kinase (AMPK) and regulated in development and DNA damage responses 1 (REDD1), respectively, to suppress mTORC1 activity. In addition, Wnt-regulated glycogen synthase kinase 3β (GSK3β) coordinates with AMPK to inhibit mTOR signaling such that when Wnt signaling is active, GSK3β/AMPK-dependent activation of TSC2 is inhibited to allow mTORC1 activation (Inoki et al. 2006). Thus, the TSC1/TSC2 complex integrates numerous positive and negative growth signals and adjusts mTORC1 activity accordingly.
In contrast to growth factors, nutrient sensing by mTORC1—particularly of amino acids—is an ancient function of the complex conserved from yeast to humans. However, the mechanism by which amino acids regulate mTORC1 is just beginning to be elucidated. Amino acids (in particular, leucine) promote mTORC1 activity independently of TSC2 through a pathway that requires Rheb-dependent activation of the complex (Smith et al. 2005; Sancak and Sabatini 2009). mTORC1 activation by l-leucine is also dependent on glutamine transport into the cell, although glutamine alone has no effect on mTORC1 (DeBerardinis et al. 2007; Nicklin et al. 2009). Although the amino acid-sensing mechanism is just beginning to be revealed, it appears to require a second family of GTPases: the Rag proteins, which bind to mTORC1 and translocate it from an undefined location in the cytoplasm to the lysosome upon amino acid stimulation (Kim et al. 2008; Sancak et al. 2008, 2010). Mammals have four Rag proteins that form heterodimers: RagA or RagB (which are more closely related to each other) with RagC or RagD (which likewise are more closely related). When RagA/B is bound to GTP, RagC/D is bound to GDP and vice versa. In the presence of amino acids, for unknown reasons, RagA/B is GTP bound, and this somehow promotes the interaction and lysosomal recruitment of mTORC1. This interaction may be facilitated by the p62/sequestosome 1, which colocalizes with the Rags at the lysosome and is required for mTORC1 recruitment there (Duran et al. 2011). The Rags are tethered to the lysosome by a complex of proteins also essential for mTORC1 function called the Ragulator (formed by p18, p14, and MP1 proteins) (Sancak et al. 2010). The Ragulator forms the mTORC1 lysosomal docking site. Because a fraction of Rheb also resides at the lysosome, the current model is that the Rags/Ragulator, by an amino-acid-dependent mechanism, bring mTORC1 into close proximity to its activator Rheb-GTP at the lysosomal surface.
Why is the lysosome the site of mTORC1 activation in response to amino acid sufficiency and not the plasma membrane or some other cellular location? Although still a major question, the mystery is beginning to unravel. A complex of lysosomal proteins collectively called the v-ATPase is required for mTORC1 activity (Zoncu et al. 2011). Amino-acid-stimulated recruitment of mTORC1 to the lysosome and its ability to phosphorylate downstream substrates requires the v-ATPase, which interacts directly with the Ragulator. Interestingly, amino acids appear to accumulate inside the lysosomal lumen and signal through the v-ATPase to the Rags and Ragulator to activate mTORC1 by what has been dubbed an “inside-out” mechanism. Exactly how this happens is still under investigation but appears to require ATP hydrolysis and an associated rotation of the v-ATPase stalk. Interestingly, mTORC1 also negatively regulates lysosome biogenesis (Settembre et al. 2012), indicating that mTORC1 might also play an important role in controlling the number of lysosomes in the cell. Importantly, much of the understanding of the nutrient-sensing mechanism has been solved in vitro, and it will be important in future studies to determine the extent to which the amino acid-sensing pathway functions in various tissues and tumor cells.
Compared with mTORC1, mTORC2 was discovered more recently, and we know considerably less about its regulation and function. Growth factors activate mTORC2 at least in part through PI3K signaling, but the mechanism is unknown. Few substrates of mTORC2 have been described (Sparks and Guertin 2010). However, the discovery that mTORC2 directly phosphorylates AKT revealed a key role for mTORC2 in mediating downstream PI3K signaling in response to growth factor activation (Sarbassov et al. 2005). There are three mammalian AKT isoforms (AKT1, AKT2, and AKT3) that have both overlapping and distinct functions. In general, AKT1 is believed to have a more critical role in cell survival (Chen et al. 2001; Cho et al. 2001b), whereas AKT2 regulates glucose homeostasis (Cho et al. 2001a; Garofalo et al. 2003). AKT3 is implicated in brain development (Tschopp et al. 2005). As mentioned above, full AKT activation requires dual phosphorylation by both PDK1 (which phosphorylates AKT1 at T308 in the kinase motif) and by mTORC2 (which phosphorylates AKT1 at S473 in a carboxy-terminal hydrophobic motif). In addition to AKT, another AGC kinase family protein, Serum and glucocorticoid-induced kinase (SGK), is directly phosphorylated by mTORC2 (Garcia-Martinez and Alessi 2008).
Most signals upstream of mTOR appear to target either mTORC1 or mTORC2. However, a mechanism of regulation shared by both complexes may occur through DEPTOR, which is a common subunit of both mTOR complexes (for review, see Zoncu et al. 2010). Although the regulation of DEPTOR is complicated, it appears to be a natural inhibitor of both mTOR complexes because in its absence, S6K, AKT, and SGK activity increases (Peterson et al. 2009). DEPTOR levels are controlled though the ubiquitin-dependent degradation pathway by a mechanism requiring direct phosphorylation of DEPTOR by mTOR in an apparent positive feedback loop (Peterson et al. 2009; Duan et al. 2011; Gao et al. 2011; Zhao et al. 2011). Interestingly, DEPTOR levels are low in many cancers, which may promote mTOR-dependent cell growth, proliferation, and survival (Peterson et al. 2009). However, in a subset of multiple myelomas, DEPTOR is highly expressed. In these cells, DEPTOR overexpression inhibits mTORC1, but this relieves strong negative feedback loops to PI3K that may override its inhibitory effect on mTORC2 and consequently promote AKT-mediated cell survival (discussed below).
TSC2 PROTECTS AGAINST METABOLIC STRESS-INDUCED APOPTOSIS
As mentioned above, the TSC1/TSC2 complex integrates many growth regulatory signals to control mTORC1 activity. Many of these signals convey information regarding the metabolic state of the cell such that when nutrients are limiting, the cell will restrict mTORC1-dependent growth pathways. One of the best examples is that of AMPK-dependent phosphorylation and activation of TSC2, which is required for cell survival under glucose deprivation conditions (Inoki et al. 2003). AMPK is a major sensor of cellular energy status that is activated under conditions of metabolic stress (e.g., glucose deprivation) that decrease ATP production (for review, see Hardie 2007). AMPK is activated by increases in cellular AMP levels to promote catabolic pathways necessary to restore a critical level of ATP required for cell survival. It was initially found that AMPK activation inhibits S6K1 phosphorylation, suggesting a link between energy-sensing pathways and mTORC1 signaling (Kimura et al. 2003). It was subsequently shown that at least one mechanism by which AMPK inhibits mTORC1 is by directly phosphorylating and activating TSC2 (Inoki et al. 2003). Later work found that AMPK also directly phosphorylates RAPTOR, which can also suppress mTORC1 activity (Gwinn et al. 2008). By activating TSC2 and inhibiting mTORC1, AMPK shuts down mTORC1-dependent growth pathways from consuming cellular energy. In the absence of TSC2 function (e.g., in TSC2−/− cells), glucose deprivation results in cell death (Inoki et al. 2003). Consistent with death being driven by mTORC1 pathways, rapamycin treatment prevents the death of TSC2−/− cells when glucose is unavailable in the culture medium (Inoki et al. 2003; Choo et al. 2010).
A number of mechanisms might explain why TSC2-deficient cells are sensitive to apoptosis when deprived of glucose. One possibility is that uncontrolled mTORC1 activity in starving cells continues to drive protein translation through 4E-BP1 and S6K1, and because translation is a major consumer of cellular energy, this exhausts the ATP reserves. However, rapamycin is only a partial mTORC1 inhibitor and relatively inefficient at suppressing translation in mammalian cells, suggesting that other rapamycin-sensitive mTORC1 pathways might also be important (Feldman et al. 2009; Thoreen et al. 2009; Choo et al. 2010).
A second possibility is that mTORC1 activation during nutrient stress promotes p53 synthesis and accumulation (Lee et al. 2007). AMPK has also been shown to stabilize p53 by direct phosphorylation (Jones et al. 2005). Thus, glucose deprivation in normal cells causes AMPK to inhibit mTORC1 and stabilize p53, stalling cell growth and division; but when TSC2−/− cells are glucose deprived, AMPK stabilizes p53 whereas unrestricted mTORC1 signaling drives p53 synthesis. This synergistically results in greatly elevated levels of p53 and subsequently apoptosis. Notably, the connections between p53, mTORC1 activity, and survival are complex (Feng et al. 2005). For instance, p53 also induces the transcription of PTEN, TSC2, and REDD1, which negatively regulate mTORC1 (Stambolic et al. 2001; Ellisen et al. 2002; Feng et al. 2005). In addition, p53 also activates AMPK as well as Sestrin 1 and Sestrin 2, which are also negative regulators of mTORC1 signaling (Feng et al. 2007; Budanov and Karin 2008; Feng 2010).
A third possibility is that in the glucose-deprived state, losing TSC2 function promotes mTORC1-dependent negative feedback loops that suppress PI3K–AKT signaling, squelching critical survival signals. The best described mTORC1 negative feedback loops function through S6K1 and Grb10 (Zoncu et al. 2010; Hsu et al. 2011; Yu et al. 2011). Another mechanism of negative feedback occurs through the unfolded protein response (UPR) (Ozcan et al. 2008). The UPR senses unfolded proteins in the ER lumen and transmits that information to the cell nucleus, where it drives a transcriptional program to reestablish homeostasis (Kozutsumi et al. 1988). In this model, mTOR hyperactivation during glucose deprivation induces ER stress (presumably through increased client load) and therefore, the UPR. It was found that the UPR promotes feedback inhibition of PI3K–AKT signaling, possibly through the JNK kinase (Ozcan et al. 2008). The UPR might also directly activate apoptotic pathways in response to the overwhelming demand on the ER to faithfully regulate protein folding.
Another more recent report finds that the hypersensitivity of TSC2-deficient cells to glucose deprivation is not linked to blocking apoptosis, to p53 levels, or to activating autophagy, but rather to rapamycin’s ability to decrease metabolic consumption, maintain ATP levels, and suppress AMPK, thus preventing energetic stress (Choo et al. 2010). These investigators also find that TSC2−/− cells deprived of glucose shift to using glutamine as a carbon source and that rapamycin fails to suppress cell death in the absence of glutamine. Therefore, in this model, rapamycin’s protective effect is the result of decreasing the bioenergetic demand in order to balance cellular metabolism with the supply of nutrients and to support the shift to a glutamine-based metabolism. This response is at least partially dependent on S6K1, but not on eIF4E, consistent with rapamycin’s relative ineffectiveness at blocking 4E-BP1 phosphorylation (Feldman et al. 2009; Thoreen et al. 2009; Choo et al. 2010).
mTORC1 DIRECTLY REGULATES AUTOPHAGY
Although the mTORC1 pathways responsible for triggering apoptosis in cultured TSC2−/− cells deprived of glucose are complex, one clear and conserved connection between mTORC1 and a pathway critical for cell survival upon nutrient deprivation is the discovery that mTORC1 directly regulates autophagy. Normally cells activate autophagy (or macroautophagy) in times of nutrient deprivation to salvage critical nutrients essential for cell survival. By mechanisms still being worked out, autophagy targets proteins and organelles (such as the mitochondria) to the autophagosome, which then delivers the cargo to the lysosome for degradation and recycling of macromolecules (for review, see Chen and Klionsky 2010; Yang and Klionsky 2010; Das et al. 2012). More than 30 different autophagy genes (ATGs) have been identified that regulate autophagy induction, cargo selection, vesicle formation, autophagosome fusion, cargo degradation, and release (for review, see He and Klionsky 2009). In cells, autophagy is a critical survival mechanism under nutrient deprivation conditions, and when inhibited either genetically or pharmacologically, nutrient deprivation can result in apoptosis (Boya et al. 2005). Autophagy is also essential for the survival of newborn mice, which require autophagy to mobilize nutrient stores during a brief starvation period immediately after birth (Kuma et al. 2004). Defective autophagy is also implicated in neuronal degeneration, cancer, and aging-associated pathologies (for review, see Yang and Klionsky 2009).
Although understanding mammalian autophagy regulation is an emerging and intense area of research, it is generally accepted that mTORC1 is a major negative regulator of autophagosome formation. Studies in yeast suggested early on that TORC1 inhibits autophagy. For example, rapamycin treatment activates autophagy in yeast even when they are growing in nutrient-rich conditions (Noda and Ohsumi 1998). Yeast TORC1 directly prevents assembly of an ATG1 kinase-containing complex required for autophagy induction (Kamada et al. 2000). Rapamycin inhibits TORC1’s ability to disrupt ATG1 complex assembly, thus activating ATG1 kinase activity. It was thought that mTORC1 also controlled autophagy in mammalian cells, but until recently, the mechanism was vague. In general, rapamycin is less effective at activating autophagy in mammalian cells, although in some cases, rapamycin treatment causes accumulation of insoluble protein aggregates characteristic of those typically associated with failed autophagy (Spilman et al. 2010). In contrast to rapamycin, catalytic (ATP-competitive) inhibitors of mTOR are much more potent activators of autophagy in mammalian cells, indicating the role of mTOR in autophagy regulation is clearly conserved (Thoreen et al. 2009).
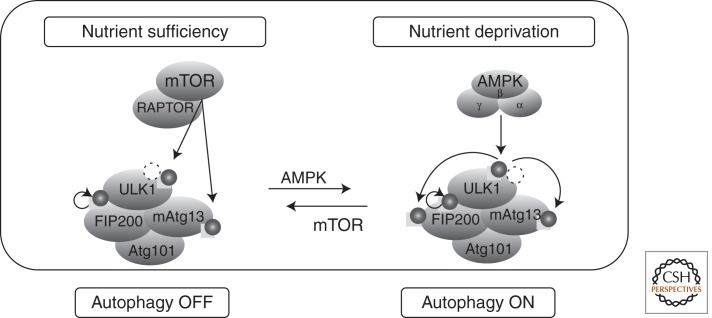
A series of reports suggests that the mechanism by which mTORC1 regulates autophagy is at least in part through direct phosphorylation of Unc-51-like kinase (ULK1), the homolog of yeast ATG1 (Chan 2009; Ganley et al. 2009; Hosokawa et al. 2009; Jung et al. 2009). In mammalian cells, ULK1 is the catalytic subunit of a complex containing mAtg13, Focal adhesion kinase-interacting protein of 200 kD (FIP200), and Atg101, all of which are essential for starvation-induced autophagy (Fig. 2). mAtg13 binds ULK1 and mediates the interaction between ULK1 and FIP200, but both FIP200 and mAtg13 appear to regulate ULK1 localization and stability. In nutrient-rich conditions, mTORC1 associates with the ULK1–mAtg13–FIP200 complex through a direct interaction between RAPTOR and ULK1, and this facilitates phosphorylation of both mAtg13 and ULK1 by mTORC1. The function of mTORC1-dependent ULK1 phosphorylation is not entirely clear, but it appears to diminish ULK1 kinase activity, thus reducing autophagic vesicle formation (Kim et al. 2011; Shang et al. 2011).
Figure 2.
Regulation of the ULK complex by mTORC1 and AMPK. The ULK complex contains ULK1, mAtg13, FIP200, and Atg101. In nutrient-rich conditions, mTORC1 associates with the complex through a direct interaction between RAPTOR and ULK1 and phosphorylates both mAtg13 and ULK1. mTORC1-dependent ULK1 phosphorylation diminishes ULK1 kinase activity, preventing autophagy induction. In the nutrient-deprived state, mTORC1 dissociates from the ULK1 complex, resulting in ULK1 dephosphorylation at the mTORC1-dependent sites and phosphorylation at distinct sites by AMPK. Under these conditions, AMPK also tightly interacts with ULK1 to promote autophagy induction.
In the nutrient-deprived state, mTORC1 dissociates from the ULK1 complex, resulting in ULK1 dephosphorylation (Kim et al. 2011; Shang et al. 2011). However, this alone does not result in autophagy activation. For this to occur, ULK1 also requires direct activating phosphorylation by AMPK, emphasizing again the interplay between nutrient and energy-sensing pathways (Egan et al. 2011; Kim and Guan 2011; Kim et al. 2011; Zhao and Klionsky 2011). In starved cells, AMPK tightly binds ULK1, and this interaction is enhanced by rapamycin and disrupted by Rheb overexpression (Behrends et al. 2010; Kim and Guan 2011). In addition, AMPK can also inhibit mTORC1 by phosphorylating and activating TSC2, and directly by phosphorylating the RAPTOR subunit, which inhibits mTORC1 activity (Krause et al. 2002; Inoki et al. 2003; Gwinn et al. 2008). Interestingly, mTORC1 can be reactivated after prolonged starvation by the autolysosomal products generated by autophagy, indicating that a minimal level of mTORC1 activity is required for survival (Liang et al. 2007; Matsui et al. 2007; Herrero-Martin et al. 2009). This latter requirement for mTORC1 might be important for recycling lysosomes (Yu et al. 2010).
The widely held view is that autophagy is downstream from mTORC1 such that when mTORC1 is “OFF” autophagy is “ON.” However, the final destination of autophagic cargo including proteins/amino acids is the lysosome, and the amino acid signal that activates mTORC1 was recently shown to emanate from within the lysosome (discussed above). Thus, autophagy should activate mTORC1, which, in fact, has been observed (Liang et al. 2007; Matsui et al. 2007; Herrero-Martin et al. 2009). How can this be reconciled? An alternative view of the relationship between mTORC1 and autophagy might explain this. In this alternative view, autophagy is actually upstream of mTORC1, and AMPK is the main regulator of autophagy that directly activates ULK1 and suppresses mTORC1. For example, nutrient and energy deprivation reduces mTORC1 activity and activates AMPK, and active AMPK promotes autophagy as described above. But without mTORC1 activity, cells would die. Therefore, by delivering amino acids to the lysosome, autophagy actually maintains mTORC1 in a minimally active state. In turn, mTORC1 inhibits autophagy as part of a negative feedback loop to prevent cells from eating themselves to death. Thus, autophagy may actually regulate mTORC1.
How long can cells survive nutrient deprivation? It would seem that prolonged starvation would, in fact, lead cells to consume themselves to death or initiate apoptosis. In fact, under certain conditions, autophagy can kill cells through a process known as autophagic cell death or cell death type II (for review, see Eskelinen 2005). However, feedback mechanisms may exist to prevent cell death caused by prolonged activation of autophagy, and one appears to require mTORC1-dependent phosphorylation of DAP1 (death-associated protein 1) (Koren et al. 2010). Under amino acid starvation (i.e., mTORC1 inhibited), DAP1 is rapidly dephosphorylated, and by an unclear mechanism, this restricts excessive autophagy. Protein phosphatases likely also work together with mTORC1 and AMPK to control autophagy. In fact, mice lacking the protein phosphatase 1 (PP1) regulatory subunit Gadd34 (growth arrest and DNA damaged protein) cannot turn off autophagy because of sustained phosphorylation of the AMPK target site in TSC2 (Uddin et al. 2011). Thus, although the regulation of autophagy is complex, by linking autophagy induction to both mTORC1 inhibition and AMPK activation, cells can tightly regulate cellular energy homeostasis and survive under conditions of metabolic stress.
mTORC2-DEPENDENT CELL SURVIVAL PATHWAYS
In addition to its role in promoting cell growth through the TSC–mTORC1 pathway, AKT has long been thought to promote cell survival directly through several mechanisms including (1) directly phosphorylating and inhibiting pro-apoptotic proteins such as BAD; (2) directly phosphorylating and inhibiting the Forkhead box O (FoxO) transcription factors, which regulate pro-apoptotic genes such as BIM and Fas ligand (also known as CD95L); (3) promoting p53 degradation by activating the murine double minute 2 (MDM2); (4) blocking the glycogen synthase kinase 3 (GSK3)-mediated inhibitory signals to the pro-survival protein Mcl-1; and (5) activating the NF-κB survival pathway via phosphorylating IκB kinase α (IKKα) (for review, see Manning and Cantley 2007). The widespread role of AKT in regulating cell survival pathways predicts that mTORC2 might control some or all of these processes. Although this seems likely, the exact role of mTORC2-mediated AKT-S473 phosphorylation in regulating cell survival is still under investigation.
One unresolved issue is whether mTORC2-dependent AKT phosphorylation is required for all AKT functions, or for only a subset of its targets. Typically, many protein kinases of the protein kinase A/protein kinase G/protein kinase C (AGC) family, such as AKT, serum, and glucocorticoid-induced protein kinase (SGK), and S6K, require prior phosphorylation in their hydrophobic motif (HM), which creates a docking site for PDK1 (Biondi et al. 2002; Frodin et al. 2002). For example, mTORC1-mediated phosphorylation on the HM site of S6K enhances the affinity for PDK1 binding and promotes full S6K activation (for review, see Jacinto and Lorberg 2008). Accordingly, acute in vitro knockdown experiments suggest that mTORC2 is required for both AKT Thr308 and Ser473 phosphorylation (Sarbassov et al. 2005). However, genetic studies indicate that Thr308 is still phosphorylated even in the complete absence of mTORC2 activity (by deleting the rictor, mlst8, or sin1 genes), arguing that in other contexts, these events might occur independently (Guertin et al. 2006; Jacinto et al. 2006; Shiota et al. 2006). The reason for this discrepancy remains unclear but may reflect a difference between acute knockdown and chronic knockout experiments, or the existence of a compensatory mechanism. Although phosphorylation at both T308 and S473 is required for maximal AKT activity in vitro, it appears that T308 phosphorylation alone empowers AKT with enough activity to phosphorylate many of its substrates in cultured cells or in tissues (Alessi et al. 1996; Guertin et al. 2006; Jacinto et al. 2006; Yang et al. 2006; Bentzinger et al. 2008; Kumar et al. 2008, 2010; Cybulski et al. 2009; Gu et al. 2011).
To date, only a select few of the predicted AKT substrates have been examined for signaling defects caused by loss of mTORC2 activity. One AKT substrate important in cell survival that appears to require mTORC2 activity is the FoxO1/3a transcription factors (Guertin et al. 2006, 2009; Jacinto et al. 2006; Yang et al. 2006). For example, FoxO1 (T24) and FoxO3a (T32) phosphorylation is decreased upon mTORC2 inactivation in both knockdown and genetic knockout studies. In the absence of AKT-mediated phosphorylation, FoxOs accumulate in the nucleus and activate metabolic and cell survival genes (Biggs et al. 1999; Nakae et al. 1999; Rena et al. 1999; Tang et al. 1999). Interestingly, phosphorylation of other AKT targets such as BAD, TSC2, and GSK3β show little to no effect upon genetic ablation of mTORC2 (Guertin et al. 2006, 2009; Shiota et al. 2006; Yang et al. 2006). Other key survival proteins downstream from AKT such as MDM2, Caspase-9 and IKKα have not yet been investigated. Because FoxOs have critical roles in cell survival, mTORC2 may regulate cell viability through Akt–FoxO pathways.
Because AKT is predicted to activate mTORC1, it is not unreasonable to predict that losing mTORC2-dependent AKT phosphorylation might also decrease mTORC1 activity and induce autophagy. However, despite most models placing mTORC2 upstream of the AKT–TSC2–mTORC1 signaling axis, evidence that this connection is important in vivo is lacking. In fact, it appears that losing mTORC2 activity has minimal effects on mTORC1 signaling in many cell types (Guertin et al. 2006; Jacinto et al. 2006; Shiota et al. 2006; Bentzinger et al. 2008; Kumar et al. 2008, 2010; Cybulski et al. 2009; Gu et al. 2011). Interestingly, mTORC2 may regulate autophagy independently of mTORC1 via the AKT–FoxO3a axis (Mammucari et al. 2007). In fasting skeletal muscle, FoxO3a positively controls transcription of several autophagy-related genes, including LC3 and Bnip3. Tamoxifen-induced activation of recombinant AKT (AKT fused with estrogen receptor) blocks FoxO3a activation and autophagy induction. Conversely, knockdown of RICTOR promotes FoxO3 nuclear retention and autophagosome formation. Another report indicates that insulin signaling also inhibits autophagy in the liver through a FoxO1-mediated mechanism (Liu et al. 2009). Thus, mTORC2 may regulate autophagic survival through AKT-dependent, mTORC1-independent mechanisms.
In vivo studies of conventional rictor, sin1, and mlst8 knockout embryos (all of which result in selective mTORC2 ablation) indicate that mTORC2 is essential for progression through mid-embryonic development (Guertin et al. 2006). Although the exact cause of lethality is unknown, increased cell death was not readily apparent in the knockout embryos. To gain further insight into the tissue-specific functions of mTORC2, conditional knockout models of rictor have been developed, including skeletal muscle, white adipose tissue, and pancreatic β cells (Bentzinger et al. 2008; Kumar et al. 2008, 2010; Cybulski et al. 2009; Gu et al. 2011). Although some metabolic defects are reported, there is no indication from these studies that mTORC2 loss—at least under otherwise normal physiological conditions—results in increased apoptosis. This may reflect the fact that in all three of these tissues, AKT Thr308 phosphorylation is largely preserved despite decreased mTORC2-dependent Ser473 phosphorylation. Thus, more studies are needed to determine exactly if and how mTORC2 might regulate cell survival.
DOES mTORC2 REGULATE CANCER CELL SURVIVAL?
Although it is not clear exactly which AKT pathways require mTORC2 under normal conditions, several lines of evidence suggest that mTORC2 may be more essential for AKT signaling in cells with oncogenic activation of PI3K activity. For example, in a PTEN-deletion-driven mouse model of prostate cancer deleting RICTOR blocks tumor formation but has no effect on normal prostate growth or function (Guertin et al. 2009). Interestingly, ablating mTORC2 activity in the PTEN-null tumor cells reduces both AKT Ser473 and Thr308 phosphorylation, perhaps suggesting a differential requirement for mTORC2 in prostate cancer cells compared with MEFs. The experiments in the prostate cancer model are reminiscent of genetic studies in Drosophila, in which dRICTOR is less essential for fly development but is required for phenotypes induced by PTEN deletion or PI3K activation (Hietakangas and Cohen 2007). Importantly, it is unclear in these models whether the rictor/mTORC2-deficient cells have survival defects.
In vitro studies using several human cancer cell lines further indicate that knocking down RICTOR is toxic to transformed cells with elevated AKT activity. For example, mTORC2 activity is elevated in gliomas and is required for anchorage-independent growth and proliferation in vitro and for tumor growth in a xenograft model (Masri et al. 2007). Moreover, mTORC2 promotes cell cycle progression and anchorage-independent growth of breast (MCF7) and prostate (PC3) cancer cell lines (Hietakangas and Cohen 2008; Guertin et al. 2009). However, regarding a specific role for mTORC2 in cell survival, only one report shows that stable knockdown of RICTOR specifically impairs survival of a cancer cell line in vitro (in this case, colorectal cancer cells) (Gulhati et al. 2009). Cell proliferation is also inhibited in these cells.
One possible explanation for the differential requirement for mTORC2 activity in normal cells versus cancer cells is that under normal conditions, basal AKT activity (Thr308 phosphorylation only) is sufficient for maintaining AKT’s essential functions, whereas in PTEN-deficient or PI3KCA (phosphatidyl inositol 3-kinase catalytic subunit) mutant transformed cells, the demand for AKT signaling is maximal, requiring both T308 and S473 phosphorylation to achieve its full activation potential. Alternatively, the difference may reflect a compensatory signal that reactivates AKT by up-regulated T308 phosphorylation upon prolonged loss of mTORC2 activity, and this pathway might not yet be functional following acute loss of mTORC2 in transient knockdown experiments. These compensatory mechanisms may exist specifically to avoid cell death. Such protective circuits may be cell type specific or only active under specific conditions, and clearly this needs further examination. Nevertheless, studies to date suggest that cancer cells with an abnormally high level of PI3K–AKT activity may have a greater requirement for mTORC2 than otherwise normal cells, and this provides rationale for developing a therapeutic strategy that selectively targets mTORC2. Importantly, however, it remains unclear to what extent mTORC2 activity is required for cancer cell survival in more advanced, therapeutically relevant stages of cancer, and whether a selective mTORC2 inhibitor (if it existed) would have an acceptable therapeutic window.
In addition to AKT, another AGC kinase family protein, SGK, is also directly phosphorylated by mTORC2 (Garcia-Martinez and Alessi 2008). It is reported that SGK can regulate cell viability through activation of MDM2-dependent p53 degradation (Amato et al. 2009). Moreover, like AKT, SGK also exists in three isoforms (SGK1, SGK2, and SGK3) in human and mouse (Brunet et al. 2001). However, in contrast to AKT, even basal SGK activity is controlled by mTORC2 because phosphorylation in its hydrophobic motif by mTORC2 is required for phosphorylation in the kinase domain by PDK1 (Garcia-Martinez and Alessi 2008). mTORC2 regulation of SGK could also have relevance in cancer cell survival as suggested by a recent study that finds SGK3 signaling (but not AKT signaling) downstream from PIK3CA mutations is essential for the survival of certain cancer cells (Vasudevan et al. 2009). Thus, mTORC2 may regulate cell survival through both AKT-dependent and AKT-independent pathways, although definitive mechanisms require further investigation.
DO mTOR INHIBITORS AFFECT CANCER CELL SURVIVAL?
Aberrant PI3K–AKT–mTOR signaling is a common feature of most cancers (Shaw and Cantley 2006; Zoncu et al. 2010; Hanahan and Weinberg 2011). Consequently, there is intense interest in developing mTOR inhibitors as cancer therapeutics. First-generation mTOR inhibitors are based on the chemical structure of rapamycin, but despite the strong rationale for using rapamycin in oncology, this class of drugs has unfortunately had limited success (O’Reilly et al. 2006; Sudarsanam and Johnson 2010; for review, see Guertin and Sabatini 2009; Benjamin et al. 2011). The best albeit modest responses to rapamycin as a therapy have been reported in renal cell carcinoma, mantle cell lymphoma, neuroendocrine tumors of the pancreas, and in treating tuberous sclerosis (caused by mutations in TSC1 or TSC2) (Benjamin et al. 2011). There are several reasons that could explain this including the fact that rapamycin is an allosteric inhibitor of mTOR that binds outside the kinase domain and only partially inhibits mTORC1 activity. For example, rapamycin universally inhibits mTORC1-dependent S6K1 phosphorylation but has only minor and acute inhibitory effects on other mTORC1 substrates including 4E-BP1 (Choo et al. 2008; Feldman et al. 2009; Thoreen et al. 2009). In addition, rapamycin relieves strong negative feedback loops to PI3K that exist downstream from mTORC1 (Fig. 3) (Choo and Blenis 2009). As mentioned above, these feedback loops can function through mTORC1 substrates including S6K1 and Grb10. S6K1 directly phosphorylates insulin receptor substrate 1 (IRS-1), mislocalizing it and targeting it for degradation. The recently discovered mTORC1 substrate Grb10 directly binds to and negatively regulates the insulin and insulin-like growth factor receptors (Hsu et al. 2011; Yu et al. 2011). mTORC1 phosphorylation of Grb10 promotes its stability. Interestingly, Grb10 levels are often decreased in cancer, suggesting that it could have tumor-suppressor functions. In both cases, rapamycin relieves feedback inhibition and promotes PI3K–AKT survival signaling. The clinical relevance of losing feedback inhibition is emphasized in human trials that find rapamycin increases AKT activation in many malignancies (O’Reilly et al. 2006; Tabernero et al. 2008; Sudarsanam and Johnson 2010). Rapamycin can also activate the MAPK pathway, providing another potential avenue to resistance (Carracedo et al. 2008). Thus, as a single agent, rapamycin may actually promote cancer cell survival; however, rapamycin may ultimately prove to be useful in combination with agents such as PI3K, AKT, or MAPK inhibitors.
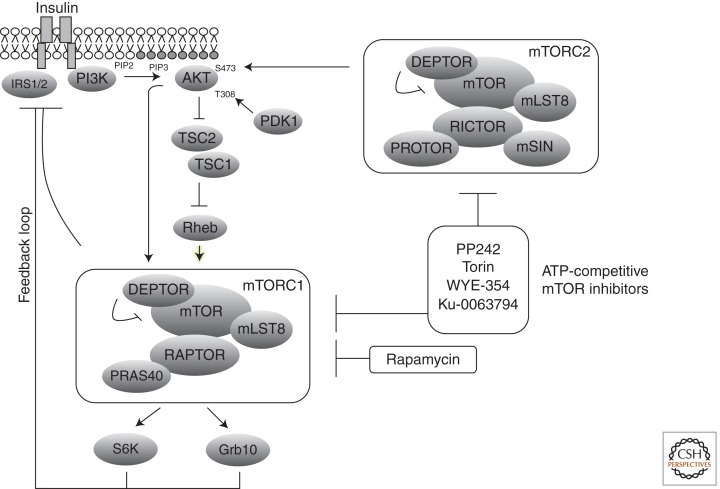
Figure 3.
Feedback inhibition of insulin signaling. Several mechanisms of negative feedback inhibition of insulin signaling exist downstream from mTORC1. The best described are mediated by S6K1, which can phosphorylate and inhibit IRS1; and by Grb10, which is stabilized by mTORC1-dependent phosphorylation and suppresses signaling from the insulin and insulin-like growth factor receptors. mTOR inhibitors such as rapamycin and the new generation of ATP-competitive inhibitors can relive these negative feedback loops, resulting in PI3K–AKT activation.
The discovery of mTORC2, which is generally rapamycin insensitive, and the widespread ineffectiveness of rapamycin as a monotherapy led to development of inhibitors that directly target the mTOR catalytic site (for review, see Guertin and Sabatini 2009). The first to be reported include Torin1, PP242, Ku-0063794, and WYE-354 (Feldman et al. 2009; Garcia-Martinez et al. 2009; Thoreen et al. 2009; Yu et al. 2009). The ATP-competitive inhibitor class more completely inhibits mTORC1 and additionally inhibits mTORC2. Of note, prolonged exposure to rapamycin can inhibit mTORC2 in a subset of cell types, and this might explain some of the clinical successes with the drug (Sarbassov et al. 2006; Gulhati et al. 2009). The mechanism is not entirely understood but may result from rapamycin blocking the assembly of new mTORC2 complexes. The mTOR ATP-competitive inhibitors are just beginning to be tested for clinical efficacy, and it is hoped that they will outperform rapamycin in the clinic. In a few preclinical studies, the mTOR catalytic inhibitors were shown to induce cell death in combination with other inhibitors (Janes et al. 2010; Sini et al. 2010).
Although the preclinical studies with mTOR catalytic site inhibitors are exciting, several questions regarding their efficacy remain. For example, will feedback activation of PI3K–AKT signaling still promote survival even though mTORC2 is also inhibited? Evidence that this could be problematic comes from studies of the natural mTOR inhibitor DEPTOR (discussed above), which emphasize the fact that losing feedback inhibition by inhibiting mTORC1 can override mTORC2 inhibition with respect to AKT activation (Peterson et al. 2009). Another potential concern is whether mTOR catalytic inhibitors will be well tolerated, although preclinical tests in rodent models are promising (for review, see Benjamin et al. 2011). The mTOR catalytic inhibitors are also more formidable activators of autophagy compared with rapamycin (Chresta et al. 2009; Thoreen et al. 2009). Because autophagy can promote cell survival in nutrient-limiting conditions, increasing autophagic activity could also promote cancer cell survival in the nutrient-deprived tumor microenvironment. In fact, in melanoma cells, inhibiting autophagy in combination with nutrient deprivation induces apoptosis, suggesting that autophagy can protect cancer cells from nutrient-limiting conditions (Sheen et al. 2011).
CONCLUSION
In this article, we review mechanisms by which cells respond to nutrient deprivation that impinge on mTORC1 signaling. We also discuss possible mechanisms by which the less-well-understood mTORC2 might regulate cell survival. Although starvation is clearly detrimental to a cell’s ability to maintain long-term homeostasis, nutrient overload also stresses cells by forcing them to elevate their metabolism, increasing damaging reactive oxygen species (ROS) and oxidative stress (for review, see Wellen and Thompson 2010). This is the case in cancer, in which oncogenic pathways drive aberrant nutrient uptake and metabolism; and diabetes, where nutrient overload promotes obesity and insulin resistance. mTOR, by functioning as a point of convergence between a nutrient-sensing pathway and PI3K–AKT signaling (i.e., as part of mTORC1) and as a regulator of AKT itself (i.e., as part of mTORC2), is central to understanding how both normal and cancer cells survive nutrient excess and is a growing area of research. In sum, mTOR integrates growth signals from diverse mechanisms that sense nutrient availability and as part of the response regulates cell survival. Pathways deregulated in many human diseases clearly impinge on mTOR signaling; thus, defining the cell and tissue-specific mechanisms through which mTOR regulates cell survival will be critical to developing therapies to treat cancer and metabolic diseases.
ACKNOWLEDGMENTS
D.A.G. is supported by grants from the NIH (R00 CA129613 and R21 CA161121), the Charles Hood Foundation, the UMass Center for Clinical and Translational Sciences, and the PEW Charitable Trusts.
Footnotes
Editors: Eric H. Baehrecke, Douglas R. Green, Sally Kornbluth, and Guy S. Salvesen
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA 1996. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J 15: 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- Amato R, D’Antona L, Porciatti G, Agosti V, Menniti M, Rinaldo C, Costa N, Bellacchio E, Mattarocci S, Fuiano G, et al. 2009. Sgk1 activates MDM2-dependent p53 degradation and affects cell proliferation, survival, and differentiation. J Mol Med (Berl) 87: 1221–1239 [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW 2010. Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin D, Colombi M, Moroni C, Hall MN 2011. Rapamycin passes the torch: A new generation of mTOR inhibitors. Nat Rev Drug Discov 10: 868–880 [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, et al. 2008. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab 8: 411–424 [DOI] [PubMed] [Google Scholar]
- Biggs WH 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC 1999. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci 96: 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi RM, Komander D, Thomas CC, Lizcano JM, Deak M, Alessi DR, van Aalten DM 2002. High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J 21: 4219–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al. 2005. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 25: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME 2001. Protein kinase SGK mediates survival signals by phosphorylating the Forkhead transcription factor FKHRL1 (FOXO3a). Mol Cell Biol 21: 952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV, Karin M 2008. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, et al. 2008. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest 118: 3065–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY 2009. mTORC1 phosphorylates the ULK1–mAtg13–FIP200 autophagy regulatory complex. Sci Signal 2: e51. [DOI] [PubMed] [Google Scholar]
- Chen Y, Klionsky DJ 2010. The regulation of autophagy—Unanswered questions. J Cell Sci 124: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, et al. 2001. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev 15: 2203–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB 3rd, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ 2001a. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ). Science 292: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ 2001b. Akt1/PKBα is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem 276: 38349–38352 [DOI] [PubMed] [Google Scholar]
- Choo AY, Blenis J 2009. Not all substrates are treated equally: Implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle 8: 567–572 [DOI] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J 2008. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci 105: 17414–17419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, Cantley LC, Blenis J 2010. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell 38: 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, Vincent JP, Ellston R, Jones D, Sini P, et al. 2009. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res 70: 288–298 [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P 2007. mTOR controls mitochondrial oxidative function through a YY1–PGC–1α transcriptional complex. Nature 450: 736–740 [DOI] [PubMed] [Google Scholar]
- Cybulski N, Polak P, Auwerx J, Ruegg MA, Hall MN 2009. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc Natl Acad Sci 106: 9902–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Das G, Shravage BV, Baehrecke EH 2012. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a008813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB 2007. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci 104: 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M 2011. mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol Cell 44: 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT 2011. 62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell 44: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA 2002. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell 10: 995–1005 [DOI] [PubMed] [Google Scholar]
- Eskelinen EL 2005. Doctor Jekyll and Mister Hyde: Autophagy can promote both cell survival and cell death. Cell Death Differ 12: 1468–1472 [DOI] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM 2009. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol 7: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z 2010. 53 regulation of the IGF-1/AKT/mTOR pathways and the endosomal compartment. Cold Spring Harb Perspect Biol 2: a001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S 2005. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci 102: 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ 2007. The regulation of AMPK β1, TSC2, and PTEN expression by p53: Stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1–AKT–mTOR pathways. Cancer Res 67: 3043–3053 [DOI] [PubMed] [Google Scholar]
- Frodin M, Antal TL, Dummler BA, Jensen CJ, Deak M, Gammeltoft S, Biondi RM 2002. A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J 21: 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X 2009. ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284: 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, et al. 2011. mTOR drives its own activation via SCF(βTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell 44: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR 2008. mTOR complex-2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum and glucocorticoid induced protein kinase-1 (SGK1). Biochem J 416: 375–385 [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR 2009. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem J 421: 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo RS, Orena SJ, Rafidi K, Torchia AJ, Stock JL, Hildebrandt AL, Coskran T, Black SC, Brees DJ, Wicks JR, et al. 2003. Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKBβ. J Clin Invest 112: 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Lindner J, Kumar A, Yuan W, Magnuson MA 2011. Rictor/mTORC2 is essential for maintaining a balance between β-cell proliferation and cell size. Diabetes 60: 827–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM 2009. The pharmacology of mTOR inhibition. Sci Signal 2: e24. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM 2006. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev Cell 11: 859–871 [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen JH, Mullholland DJ, Magnuson MA, Wu H, Sabatini DM 2009. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell 15: 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulhati P, Cai Q, Li J, Liu J, Rychahou PG, Qiu S, Lee EY, Silva SR, Bowen KA, Gao T, et al. 2009. Targeted inhibition of Mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clin Cancer Res 15: 7207–7216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hardie DG 2007. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8: 774–785 [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ 2009. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, Jaattela M 2009. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J 28: 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V 2008. TOR complex 2 is needed for cell cycle progression and anchorage-independent growth of MCF7 and PC3 tumor cells. BMC Cancer 8: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM 2007. Re-evaluating AKT regulation: Role of TOR complex 2 in tissue growth. Genes Dev 21: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. 2009. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. 2011. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332: 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD 2008. The TSC1–TSC2 complex: A molecular switchboard controlling cell growth. Biochem J 412: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 12: 12. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590 [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. 2006. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126: 955–968 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Lorberg A 2008. TOR regulation of AGC kinases in yeast and mammals. Biochem J 410: 19–37 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B 2006. SIN1/MIP1 maintains rictor–mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127: 125–137 [DOI] [PubMed] [Google Scholar]
- Janes MR, Limon JJ, So L, Chen J, Lim RJ, Chavez MA, Vu C, Lilly MB, Mallya S, Ong ST, et al. 2010. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med 16: 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18: 283–293 [DOI] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH 2009. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20: 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guan KL 2011. Regulation of the autophagy initiating kinase ULK1 by nutrients: Roles of mTORC1 and AMPK. Cell Cycle 10: 1337–1338 [DOI] [PubMed] [Google Scholar]
- Kim DH, Sabatini DM 2004. Raptor and mTOR: Subunits of a nutrient-sensitive complex. Curr Top Microbiol Immunol 279: 259–270 [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov dos D, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM 2003. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell 11: 895–904 [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Tokunaga C, Dalal S, Richardson C, Yoshino K, Hara K, Kemp BE, Witters LA, Mimura O, Yonezawa K 2003. A possible linkage between AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes Cells 8: 65–79 [DOI] [PubMed] [Google Scholar]
- Koren I, Reem E, Kimchi A 2010. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr Biol 20: 1093–1098 [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332: 462–464 [DOI] [PubMed] [Google Scholar]
- Krause U, Bertrand L, Maisin L, Rosa M, Hue L 2002. Signalling pathways and combinatory effects of insulin and amino acids in isolated rat hepatocytes. Eur J Biochem 269: 3742–3750 [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N 2004. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Kumar A, Harris TE, Keller SR, Choi KM, Magnuson MA, Lawrence JC Jr 2008. Muscle-specific deletion of rictor impairs insulin-stimulated glucose transport and enhances basal glycogen synthase activity. Mol Cell Biol 28: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Lawrence JC Jr, Jung DY, Ko HJ, Keller SR, Kim JK, Magnuson MA, Harris TE 2010. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes 59: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Inoki K, Karbowniczek M, Petroulakis E, Sonenberg N, Henske EP, Guan KL 2007. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J 26: 4812–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. 2007. The energy sensing LKB1–AMPK pathway regulates p27kip1 phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol 9: 218–224 [DOI] [PubMed] [Google Scholar]
- Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W 2009. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: Inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem 284: 31484–31492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J 2009. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. 2007. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 6: 458–471 [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci 28: 573–576 [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC 2007. AKT/PKB signaling: Navigating downstream. Cell 129: 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri J, Bernath A, Martin J, Jo OD, Vartanian R, Funk A, Gera J 2007. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res 67: 11712–11720 [DOI] [PubMed] [Google Scholar]
- Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J 2007. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 100: 914–922 [DOI] [PubMed] [Google Scholar]
- Nakae J, Park BC, Accili D 1999. Insulin stimulates phosphorylation of the Forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem 274: 15982–15985 [DOI] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273: 3963–3966 [DOI] [PubMed] [Google Scholar]
- O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al. 2006. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 66: 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, Hotamisligil GS 2008. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell 29: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM 2009. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137: 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. 2011. mTOR complex 1 regulates Lipin 1 localization to control the SREBP pathway. Cell 146: 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Schreiber SL 2009. Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci 106: 22229–22232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G, Guo S, Cichy SC, Unterman TG, Cohen P 1999. Phosphorylation of the transcription factor Forkhead family member FKHR by protein kinase B. J Biol Chem 274: 17179–17183 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Sabatini DM 2009. Rag proteins regulate amino-acid-induced mTORC1 signalling. Biochem Soc Trans 37: 289–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25: 903–915 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM 2010. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307: 1098–1101 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159–168 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM 2010. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 40: 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L, Chen S, Du F, Li S, Zhao L, Wang X 2011. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci 108: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC 2006. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441: 424–430 [DOI] [PubMed] [Google Scholar]
- Sheen JH, Zoncu R, Kim D, Sabatini DM 2011. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell 19: 613–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota C, Woo JT, Lindner J, Shelton KD, Magnuson MA 2006. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev Cell 114: 583–589 [DOI] [PubMed] [Google Scholar]
- Sini P, James D, Chresta C, Guichard S 2010. Simultaneous inhibition of mTORC1 and mTORC2 by mTOR kinase inhibitor AZD8055 induces autophagy and cell death in cancer cells. Autophagy 6: 553–554 [DOI] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG 2005. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem 280: 18717–18727 [DOI] [PubMed] [Google Scholar]
- Sparks CA, Guertin DA 2010. Targeting mTOR: Prospects for mTOR complex 2 inhibitors in cancer therapy. Oncogene 29: 3733–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, Richardson A, Strong R, Galvan V 2010. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-β levels in a mouse model of Alzheimer’s disease. PLoS ONE 5: e9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW 2001. Regulation of PTEN transcription by p53. Mol Cell 8: 317–325 [DOI] [PubMed] [Google Scholar]
- Sudarsanam S, Johnson DE 2010. Functional consequences of mTOR inhibition. Curr Opin Drug Discov Devel 13: 31–40 [PubMed] [Google Scholar]
- Tabernero J, Rojo F, Calvo E, Burris H, Judson I, Hazell K, Martinelli E, Ramon y Cajal S, Jones S, Vidal L, et al. 2008. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: A phase I tumor pharmacodynamic study in patients with advanced solid tumors. J Clin Oncol 26: 1603–1610 [DOI] [PubMed] [Google Scholar]
- Tang ED, Nunez G, Barr FG, Guan KL 1999. Negative regulation of the Forkhead transcription factor FKHR by Akt. J Biol Chem 274: 16741–16746 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp O, Yang ZZ, Brodbeck D, Dummler BA, Hemmings-Mieszczak M, Watanabe T, Michaelis T, Frahm J, Hemmings BA 2005. Essential role of protein kinase Bγ (PKBγ/Akt3) in postnatal brain development but not in glucose homeostasis. Development 132: 2943–2954 [DOI] [PubMed] [Google Scholar]
- Uddin MN, Ito S, Nishio N, Suganya T, Isobe K 2011. Gadd34 induces autophagy through the suppression of the mTOR pathway during starvation. Biochem Biophys Res Commun 407: 692–698 [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH 2007. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol 9: 316–323 [DOI] [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, et al. 2009. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell 16: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Harris TE, Lawrence JC Jr 2008. Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J Biol Chem 283: 15619–15627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB 2010. Cellular metabolic stress: Considering how cells respond to nutrient excess. Mol Cell 40: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ 2009. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr Opin Cell Biol 22: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ 2010. Eaten alive: A history of macroautophagy. Nat Cell Biol 12: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Inoki K, Ikenoue T, Guan KL 2006. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev 20: 2820–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip CK, Murata K, Walz T, Sabatini DM, Kang SA 2010. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell 38: 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Toral-Barza L, Shi C, Zhang WG, Lucas J, Shor B, Kim J, Verheijen J, Curran K, Malwitz DJ, et al. 2009. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res 69: 6232–6240 [DOI] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465: 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. 2011. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332: 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Klionsky DJ 2011. AMPK-dependent phosphorylation of ULK1 induces autophagy. Cell Metab 13: 119–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xiong X, Sun Y 2011. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell 44: 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM 2010. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]