Abstract
Within the last decade, multiple novel congenital human disorders have been described with genetic defects in known and/or novel components of several well-known DNA repair and damage response pathways. Examples include disorders of impaired nucleotide excision repair, DNA double-strand and single-strand break repair, as well as compromised DNA damage-induced signal transduction including phosphorylation and ubiquitination. These conditions further reinforce the importance of multiple genome stability pathways for health and development in humans. Furthermore, these conditions inform our knowledge of the biology of the mechanics of genome stability and in some cases provide potential routes to help exploit these pathways therapeutically. Here, I will review a selection of these exciting findings from the perspective of the disorders themselves, describing how they were identified, how genotype informs phenotype, and how these defects contribute to our growing understanding of genome stability pathways.
Many human disorders are associated with genetic defects in DNA repair and damage response pathways. For example, ataxia telangiectasia is caused by defects in ATM, which is responsible for repairing double-stranded breaks in DNA.
The link between DNA damage, mutagenesis, and malignant transformation is long established. A logical extension is that a congenital defect in a fundamental DNA repair pathway, such as nucleotide excision repair (NER), would be anticipated to be associated with a pronounced cancer predisposition syndrome. Indeed this is well known to be the case considering xeroderma pigmentosum (XP) (Cleaver 1968, 1969, 1970). In most XP subtypes, the devastatingly overt >1000-fold elevated risk of developing basal and squamous cell carcinomas on sun-exposed areas of the skin is directly attributable to a failure to remove highly mutagenic solar ultraviolet (UV) radiation-induced DNA photoproducts from the genome. In this sense XP represents a paradigm of a DNA repair disorder with a clear pathological link between genotype and phenotype (Cleaver et al. 2009).
As our knowledge of the complexity of genome stability pathways has evolved, coupled with the explosive technical advances in molecular and cellular biology, more and more human disorders caused by defects in components that constitute the genome stability network continue to be described. At the most fundamental level, identification of these conditions enables future accurate molecular diagnosis (Raffan et al. 2011). This has relevance for associated comorbidities and is vital for informed counseling of the parents, not just for family planning and recurrence risk analysis, but can assuage destructive feelings of maternal/paternal guilt and bring to an end what is often a protracted and extremely stressful journey to ascertain a clinical diagnosis (Raymond et al. 2010; Evans et al. 2011; Baker et al. 2012). These disorders can also help inform the biology of genome stability. Their diverse and often unanticipated clinical features can provide evidence for previously unappreciated biological connections (Griffith et al. 2008; Rauch et al. 2008; Huang-Doran et al. 2011). Furthermore, these insights can help optimize therapeutic strategies for these conditions along with more common conditions such as cancer. For example, the application of nonmyeloablative conditioning protocols for bone marrow transplantation to combat the severe anemia and lymphoid malignancy in Fanconi anemia, or the use of reduced intensity radiotherapy strategies to treat lymphoma in ataxia telangiectasia patients have both been developed directly from our understanding the inherent sensitivity of cells from these patients to specific forms of DNA damage (Gennery et al. 2005; Resnick et al. 2005; Lavin 2008). The interest in developing specific inhibitors toward “drug-able” targets that play key roles in DNA repair and the DNA damage response, to increase the selective sensitivity of tumor cells toward conventional DNA-damaging therapies such as radiotherapy and certain chemotherapies is currently an active area of interest (Bryant and Helleday 2004; Helleday et al. 2008; Evers et al. 2010; Helleday 2010).
Since 2005, there have been several notable descriptions of novel congenital disorders caused by defects in known, or more importantly, novel components of several DNA repair and DNA damage response pathways. These conditions have been identified via a combination of approaches: candidate gene approaches coupled to educated guesswork based on known biology of a particular pathway; by classical homozygosity linkage analysis using consanguineous families; and in recent years, by the growing influence of next-generation whole exome sequencing. The latter approach, in particular, offers the tantalizing prospect of being able to identify additional potential genetic defects using single affected patients. Here, I will review examples of some of the novel disorders that have been described since 2005. I will first review disorders of DNA repair pathways, including those of nonhomologous DNA end joining (NHEJ), base excision repair (BER)–single-strand break repair (SSBR), NER, and homologous recombination (HR)–interstrand cross-link (ICL) repair, before highlighting disorders associated with defects in the DNA damage-induced signal transduction responses (phosphorylation and ubiquitination).
NOVEL DISORDERS OF NHEJ
The mechanics of the NHEJ pathway are outlined in Chiruvella et al. (2013). Two disorders, one caused by deficiency of a novel component, the other by mutation of a well-known component of NHEJ, have been described recently. Cells from both disorders show pronounced defects in DNA double-strand break repair (DSBR).
Cernunnos/XLF-SCID
NHEJ represents an important means of directly repairing DNA double-strand breaks (DSBs) by a resealing process not dependent on the availability of a homologous DNA strand. One of the primary functions of NHEJ is the repair of programmed DSBs in the immunoglobin (Ig) and T-cell receptor (TCR) gene loci during the process of V(D)J recombination to form the complete Ig and TCR repertoire of the immune system (Lieber 2010). Indeed the known human conditions defective in a core component of NHEJ, DNA ligase IV (LIG4) causing LIG4 syndrome and Artemis (DCLRE1C) causing Art-SCID (severe combined immunodeficiency), are associated with pronounced T- and B-cell deficiencies (Moshous et al. 2001; O’Driscoll et al. 2001, 2004; O’Driscoll and Jeggo 2006). These patients suffer frequent infections from an early age, invariably presenting initially in the immunology clinic. Because of the DSB repair defect and consequent ionizing radiation sensitivity of cells from these patients, these conditions are denoted as radiosensitive (RS)-SCID, distinguishing them from other more common causes of SCID such as RAG1/2 or adenosine deaminase deficiency (Riballo et al. 2004).
Understanding the cellular and clinical spectrum of RS-SCID enabled the subsequent identification of defects in what transpired to be a novel NHEJ component: Cernunnos/XRCC4-like factor (XLF), caused by mutations in NHEJ1. Using a functional cDNA library-based complementation cloning strategy, Buck and colleagues used fibroblasts from a series of patients characterized by pronounced T- and B-cell-minus SCID, growth retardation, and microcephaly (phenotypes reminiscent of LIG4 syndrome), identifying multiple mutations in a novel gene they called Cernunnos (Buck et al. 2006). In a complementary approach, Ahnesorg and colleagues showed that a previously undescribed XRCC4 interactant they had identified (XLF; XRCC4-like factor) was defective in a well-characterized NHEJ-defective cell line, 2BN (Ahnesorg et al. 2006). This line was derived from an RS-SCID patient that did not harbor a defect in any of the known NHEJ factors (Dai et al. 2003). Subsequent functional analysis suggests that Cernnunos/XLF may function in stimulating the adenylation of DNA ligase IV, an essential step in the ligation reaction thereby facilitating DSB ligation (Riballo et al. 2009).
DNA-PKcs-SCID
The DNA-dependent protein kinase (DNA-PK) complex is composed of the DNA-PK catalytic subunit (DNA-PKcs), a phosphatidyl inositol-3 kinase-like protein kinase, and the KU70/80 heterodimer. KU70/80 has a very high affinity for double-stranded DNA ends and recruits DNA-PKcs to DSBs as the initial step of NHEJ (Lees-Miller 1996; Smith and Jackson 1999; Meek et al. 2008). Although the exact physiologically relevant substrates of DNA-PK are not well defined, DNA-PKcs autophosphorylation in trans at a DSB is essential for NHEJ, particularly for recruitment of Artemis during V(D)J recombination (Chan et al. 2002; Ding et al. 2003; Cui et al. 2005; Meek et al. 2007). Artemis-endonuclease activity plays an essential role in opening the hairpin-sealed coding ends formed by RAG1/2 endonuclease-mediated cleavage at the Ig and TCR loci. Failure to process these coding ends effectively results in SCID (Moshous et al. 2001; Ma et al. 2002).
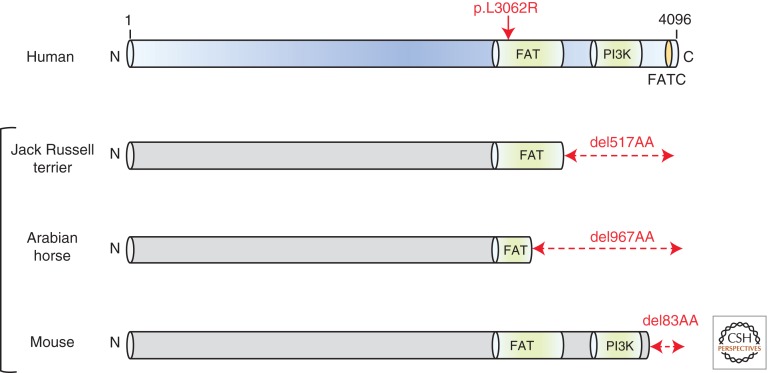
Compound heterozygous mutations in PRKDC, the gene encoding DNA-PKcs, were recently identified in a single case of RS-SCID without associated developmental features such as the microcephaly and growth delay seen in LIG4 syndrome and Cernunnos/XLF-SCID (van der Burg et al. 2009a,b). It is noteworthy in this context that Artemis deficiency similarly is not associated with developmental abnormalities (Moshous et al. 2001; Li et al. 2002). The identification of this novel and long-predicted genetic defect (spontaneous DNA-PKcs defects occur in Jack Russell terrier dogs, Arabian horses, and mice [Fig. 1]) is owing to a thorough analysis of the immunological profile of the affected case (Bosma et al. 1983; Peterson et al. 1995; Wiler et al. 1995; Meek et al. 2001; van der Burg et al. 2009b). Analysis of coding joints from bone marrow precursor cells from the affected patient showed an overrepresentation of elongated P elements (palindromic sequences) indicative of a failure to cleave hairpin-sealed coding ends (van der Burg et al. 2009b). This is a feature of Artemis deficiency, yet no pathogenic mutations were detected in DCLRE1C, or indeed in LIG4 and NHEJ1. Because Artemis activity during V(D)J is dependent on DNA-PKcs autophosphorylation and long P elements are a feature of the DNA-PKcs-mutant SCID mouse, van der Burg and colleagues focused their attention on PRKDC (Schuler et al. 1991). Compound heterozygous mutations in PRKDC were identified (p.delG2113 and p.L3062R), although unexpectedly, these did not appear to impact on DNA-PKcs stability, expression, kinase activity, or autophosphorylation capacity (van der Burg et al. 2009a). Complementation-based analysis indicated that p.L3062R, found in the highly conserved FAT domain of DNA-PKcs, alone impacted V(D)J and was the likely pathogenic hypomorphic allele (Fig. 1) (van der Burg et al. 2009a,b). These unexpected findings are not reflected by any of the known animal models for DNA-PKcs deficiency, which all lack kinase activity (Fig. 1) (van der Burg et al. 2009a,b). Although it is possible that other PRKDC mutations that do impair DNA-PKcs-dependent kinase activity may also occur in SCID patients yet to be identified, these findings indicate that assaying DNA-PK activity on patient-derived cells may not capture all potential defects.
Figure 1.
Different genetic defects in DNA-PKcs. Schematic representation of human DNA-PKcs (in purple) showing the relative positioning of the FAT, FATC, and PI3K-calalytic kinase domains. The spontaneous deletions (del) observed within DNA-PKcs in Jack Russell terriers, Arabian horse, and mouse are shown in gray, all of which involve loss of the catalytic PI3K-active site.
NOVEL DISORDERS OF BER AND SSBR PATHWAYS
BER and SSBR constitute a vital defense not only against the cytotoxic and mutagenic consequences of endogenously generated reactive oxygen species (ROS), but these pathways also repair DNA breaks and nicks induced by the programmed physiologically important function of topoisomerase I (Top I), to relieve torsional tension within the double helix, which is a normal by-product of transcription and DNA replication (Caldecott 2008). These pathways are reviewed in Krokan and Bjørås (2013).
Hyper-IgM Syndrome and Juvenile Polyposis
The first step of BER involves the action of the glycosylases, a diverse group of enzymes that act to remove the damaged/modified base, generating an apurinic/apyrimidinic (AP) abasic site. Over the last decade or so, congenital defects in certain DNA glycosylases have been identified, most notably in uracil DNA glycosylase (UNG), MutY Escherichia coli homolog glycosylase (MYH), and activation-induced cytidine deaminase (AICDA). Both AICDA and UNG are associated with the immunological phenotype of hyper-IgM syndrome (Revy et al. 2000; Imai et al. 2003). AICDA is a single-strand DNA (ssDNA) deaminase that is essential for class switch recombination (CSR) of Ig’s from IgM to other isotypes (IgG, IgA, etc.) and for somatic hypermutation (SHM) to refine antigen binding (Petersen et al. 2001; Petersen-Mahrt et al. 2002). UNG removes uracil from DNA, which can be generated by either cytosine deamination or replicative-incorporation of dUMP instead of dTMP. Therefore, UNG plays a vital role in repressing G/C-to-A/T transitions. Because AICDA-mediated uracil generation is essential for CSR, it is unsurprising that congenital defects in UNG would also result in hyper-IgM syndrome (Imai et al. 2003).
E. coli mutY is a component of the bacterial mismatch repair system that together with mutM reverts A/G and A/C mismatches. Oxidative damage converts guanine into 8-oxo-7, 8-dihydro-2′deoxyguanosine (8-oxo-G), which can mispair with adenine resulting in G/C-to-T/A transversions. MYH has nicking and glycosylase activity against A/G, A/C, and A/8-oxo-G mismatches, catalyzing the removal of the adenine base (Fromme et al. 2004). Such G/C-to-T/A transversions are frequently observed in the APC gene (adenomatous polyposis coli) associated with colorectal adenocarcinoma. Germline mutations in MYH cause MYH-associated juvenile polyposis, which presents with colorectal carcinoma and sometimes pilomatricomas (calcifying cutaneous tumors of hair matrix cells) (Jones et al. 2002; Baglioni et al. 2005).
Defective SSBR in Syndromal Ataxias
Congenital impairment of SSBR appears to be strongly associated with neurological deficits. Ataxia-oculomotor apraxia-1 (AOA-1) is caused by mutations in APTX, the gene encoding aprataxin. AOA1 is characterized by early-onset cerebellar ataxia, peripheral neuropathy, and oculomotor apraxia (Aicardi and Goutieres 1984). Aprataxin interacts with XRCC1 and APTX-mutated AOA1 cells are sensitive to agents that cause DNA single-strand breaks. Aprataxin is a member of the histidine triad family of nucleotide hydrolases and transferases. During SSBR, aprataxin resolves abortive ligation intermediates by catalyzing the nucleophilic release of adenylate groups from 5′-phosphate termini of single-strand breaks producing a 5′ phosphate that can be effectively ligated (Ahel et al. 2006). Therefore, it has been proposed that the neurological deficits in AOA-1 are likely the result of accumulating unrepaired single-strand breaks specifically in neurons (Ahel et al. 2006).
A defect in SSBR has also been documented in cells from patients with spinocerebellar ataxia with axonal neuropathy-1 (SCAN-1), a peripheral neuropathy characterized by moderate progressive ataxia, dysarthria, and cerebellar atrophy. All SCAN-1 patients identified to date carry the same neomorphic active site mutation (p.H493R) in TDP1 (tyrosyl-DNA phosphodiesterase 1) (Takashima et al. 2002). TDP1 removes Topo I-cleavable complexes (Topo I-CCs) from DNA. The neomorphic TDP1p.H493R allele has reduced enzymatic activity and accumulates with increased half-life on Topo I-CCs where it is thought to serve as a potent block to transcription and replication forks (El-Khamisy et al. 2005; Interthal et al. 2005).
Polynucleotide Kinase/Phosphatase and Microcephaly, Developmental Delay, and Seizure Syndrome
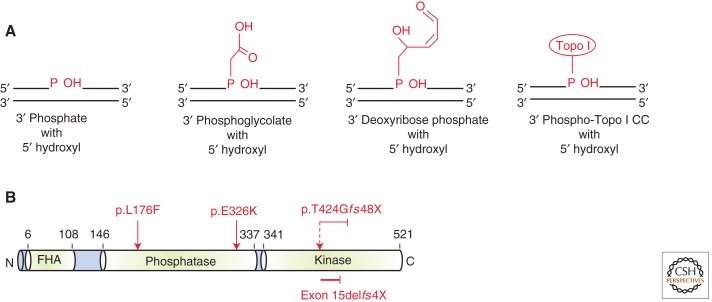
The most recent congenital defect identified in a component of the SSBR machinery is that of the dual kinase and phosphatase, polynucleotide kinase/phosphatase (PNKP) (Shen et al. 2010). Very often the termini of DNA strand breaks, whether induced by free radicals, Topo I, or the action of AP lyase and/or endonucleases, require processing to restore the 5′-phosphate and 3′-OH termini, essential for effective ligation (Fig. 2A). PNKP restores these termini as it possesses both 5′-kinase and 3′-phosphatase activity (Caldecott 2002; Weinfeld et al. 2011). PNKP is thought to play an active role in SSBR and DSBR by virtue of its FHA domain-mediated interaction between CK2 phosphorylation sites on both XRCC1 and XRCC4, respectively (Koch et al. 2004; Loizou et al. 2004). Multiple mutations in PNKP were identified by genome-wide linkage analysis in several consanguineous families with autosomal recessive severe primary microcephaly, marked developmental delay, hyperactivity, and intractable seizures (MCSZ) (Shen et al. 2010). These mutations were found in both the kinase and phosphatase domain of PNKP, usually also impacting on PNKP stability (Fig. 2B) (Shen et al. 2010). Subsequent analysis using recombinant versions of the MCSZ-associated mutant PNKPs have shown differential impacts of specific mutations on kinase and phosphatase activities (Reynolds et al. 2012). Collectively, the functional evidence indicates that PNKP activity is strongly impaired here, consistent with attenuated DNA breakage repair observed in MCSZ-patient-derived cells following H2O2 or camptothecin (CPT, a Topo I inhibitor) treatment (Shen et al. 2010; Reynolds et al. 2012).
Figure 2.
PNKP, its substrates and structure. (A) PNKP functions to clean up damaged termini at single-strand breaks to reconstitute the 3′-OH and 5′-phosphate (P) ends required for ligation. Some of the typical damaged termini requiring processing are shown here in red. The 3′-P, 5′-OH and phosphoglycolate termini are a consequence of ROS-induced DNA damage. The 3′-deoxyribose phosphates are produced by AP endonuclease action or by the AP lyase activity of certain DNA glycosylases. Topo I-cleavable complexes (CC) require the combined action of ubiquitin-dependent proteosomal degradation and TDP1 action to remove the covalently bound Topo I from DNA to repair the underlying strand nick. (B) Schematic representation of PNKP showing the juxtaposition of the phosphatase and kinase domains. The FHA domain is an important phosphoprotein-binding domain implicated in binding to CK2 phosphorylation sites on XRCC1 and XRCC4. MCSZ-patient mutations are shown in red.
Implications for Understanding Genotype–Phenotype Relationships
This fascinating defect further expands our understanding of the clinical consequences of impaired SSBR specifically with respect to its role in neurogenesis, as opposed to its presumed function in preventing ROS-induced neurodegeneration. The contrast here to AOA-1 and SCAN-1 is marked in this respect. MCSZ patients do not present with a neuropathy or ataxia but instead with a severe microcephaly without obvious postnatal progressive cerebellar degenerative or structural abnormalities suggestive of embryonic stem cell deficit typical of intrauterine programming (Fowden et al. 2006, 2008; Shen et al. 2010). The reasons for this are not clear but may reflect the role of PNKP in the repair of multiple types of damaged DNA break termini and perhaps also in DSBR. Interestingly, both LIG4 syndrome and Cernunnos/XLF-SCID patients show microcephaly (O’Driscoll et al. 2001; Buck et al. 2006).
The other marked clinical features characteristic of MCSZ are the intractable seizures coupled with developmental delay (Shen et al. 2010). The seizure phenotype, in particular, is not a general feature of known DNA repair or DNA damage response defective disorders, even those associated with profound microcephaly such as ATR-mutated Seckel syndrome (Goodship et al. 2000; O’Driscoll et al. 2003; O’Driscoll 2009b). The origins of this specific clinical feature are currently unclear. But, it is tempting to speculate that they may reflect some underlying deficit in mitochondrial function because of its strong association with seizures (Kudin et al. 2009; Waldbaum and Patel 2010; Folbergrová and Kunz 2012). The mitochondrial genome, by virtue of the fact that it lacks protective chromatin and resides in close proximity to the electron transport chain complexes, is subject to significant levels of ROS-mediated DNA damage. Consequently, the mitochondria contain several dedicated members of the BER-SSBR network to preserve the integrity of mitochondrial DNA (mtDNA) (de Souza-Pinto et al. 2008). For example, mitochondria contain several glycosylases (Nei and Nth family members), a truncated AP-endonuclease-1, a mitochondrial DNA ligase III, and mitochondrial-specific Topo I. See Alexeyev et al. (2013) for details of mitochondrial DNA repair mechanisms. Recently, PNKP has been found to possess a cryptic mitochondrial targeting motif that targets a proportion of nuclear PNKP to mitochondria where it associates with mitofilin, an inner-mitochondrial membrane component, and mediates repair of H2O2-induced mtDNA breaks (Tahbaz et al. 2012). It will be fascinating to ascertain whether impaired mtDNA repair and consequent mitochondrial dysfunction are features of MCSZ neurons.
NOVEL DISORDERS OF NER
NER is arguably one of the best characterized DNA repair pathways; essentially a “cut and paste” mechanism for the removal of helix distorting lesions from DNA, such as the cyclobutane pyrimidine dimers and 6-4 photoproducts formed following UV irradiation (Cleaver et al. 2009; see Scharer 2013 for details). NER subpathways include global genome NER (GGR) whereby helix distortion is recognized by XPC-HR23B and DDB and transcription-coupled NER (TCR), whereby RNA polymerase II blocking lesions are preferentially repaired from actively transcribing strands. TCR requires CSA/ERCC8 and CSB/ERCC6 (see Fousteri 2013 for details of TCR). Downstream from DNA damage recognition both GGR and TCR use the same machinery to locally unwind the helix around the lesion, site-specifically cleave the DNA either side of the lesion-containing strand, then filling in and ligating the resultant repair patch.
Congenital deficiency in GGR results in XP, which is caused by several distinct molecular defects including XPA, XPB(ERCC3), XPC, XPD (ERCC2), XPE (DDB2), XPF (ERCC4), XPG (ERCC5), and XPV (POLH), or trichothiodystrophy (TTD-A, but also specific defects in ERCC2 and ERCC3) (Cleaver et al. 2009). Defects in TCR cause Cockayne syndrome (CSA/ERCC8 and CSB/ERCC6). XP is characterized by severe photosensitivity, dramatically elevated skin cancer risk, and in severe instances, neurodegeneration. Cockayne syndrome (CS) is also characterized by photosensitivity but not elevated skin cancer risk. CS is a cachectic dwarfism associated with microcephaly, profound neurodegeneration, and progressive progeria (Nance and Berry 1992). TTD presents as an attenuated form of CS, again without elevated cancer risk but with ichthyosis and brittle hair and nails (Price et al. 1980; Yong et al. 1984; Stefanini et al. 1986). Several NER pathway components are also subunits of the multisubunit transcription factor TFIIH (e.g., TTDA, XPB, XPD), and it is thought that many of the developmental features observed in CS and TTD are attributed to reduced transcriptional capacity rather than defective DNA repair (Vermeulen et al. 2000; van der Pluijm et al. 2006; Gregg et al. 2011).
XFE (XPF-ERCC1) Progeria
The XPF-ERCC1 complex is the structure-specific endonuclease that makes the incision 5′ to the lesion during NER. XPF is the catalytic component, whereas ERCC1 is important for DNA binding. Cells defective in XPF-ERCC1 function are additionally hypersensitive to killing by ICL agents such as mitomycin C, thought to be a consequence of a role outside of core NER (Gregg et al. 2011). Mutations in XPF usually result in a mild form of XP including modest sun sensitivity (freckling) with a much delayed eventual appearance of skin cancer, generally from the second decade (Sijbers et al. 1996). This is somewhat at odds with a transgenic XPF-deficient mouse modeling human XPF mutations. The XPF patient XP23OS had a mild form of XP without evidence of skin cancer or neurodegeneration by the fourth decade of life (Zelle et al. 1980). In contrast, the transgenic Xpf-mutant mouse (Xpfm/m) of this patient showed overt postnatal growth delay and premature death by 3 wk of age associated with hepatocellular polyploidy typical of progeria (Tian et al. 2004). This extreme phenotype is also observed in mouse models of Ercc1 deficiency, which display evidence of accelerated aging coupled with progressive ataxia and dystonia suggestive of neurodegeneration (McWhir et al. 1993; Gregg et al. 2011).
Using whole genome-transcriptome analysis of Ercc1−/− mouse liver cells, Niedernhofer and colleagues identified a remarkable suppression of the somatotroph, lactotroph, and thyrotroph hormonal axes similar to what is observed in aged normal mice (Niedernhofer et al. 2006). These features potentially explain some of the phenotypes of the Xpf and Ercc1 mouse models and suggest a causal link between unrepaired DNA damage and aging. Furthermore, Niedernhofer and colleagues described an individual with marked growth retardation, microcephaly, photosensitivity, ataxia, and a profound rapidly progressive progeroid syndrome with a prepubescent onset eventually leading to premature death at the age of 16 yr (Niedernhofer et al. 2006). Cells from this patient were sensitive to killing by UV and showed severe defects in GGR (unscheduled DNA synthesis [UDS]) and TCR (impaired recovery of transcription following UV irradiation). Unexpectedly, this patient was found to harbor a homozygous missense mutation in XPF. Identification of this defect expanded the phenotype of XPF deficiency in humans from XP to a novel severe progeroid syndrome with overlapping features to CS.
ERCC1 and Cerebro-Oculo-Facial-Skeletal Syndrome
Subsequently, Jaspers and colleagues identified pathogenic defects in ERCC1 in a patient with a clinical diagnosis of cerebro-oculo-facio-skeletal (COFS) syndrome, a severe disorder characterized by growth retardation, microcephaly, congenital cataracts, facial dysmorphism, neurogenic artrogryposis (joint contractures), kyphoscoliosis, osteoporosis, and marked psychomotor disability (Jaspers et al. 2007). Interestingly, mutations in CSB/ERCC6, XPG/ERCC5, and XPD/ERCC2 had previously been identified in COFS patients (Hamel et al. 1996; Meira et al. 2000; Graham et al. 2001). Here, the ERCC1 defect was suggested following careful analysis of UDS, RNA synthesis, and the temporal recruitment of various NER components on UV irradiation of patient fibroblasts. Microinjection of recombinant XPF-ERCC1 reversed the severe UDS defect in the patient cells directly implicating this complex. Cells from this patient also showed marked hypersensitivity to killing by ICLs (Jaspers et al. 2007). A second patient presenting with progressive cortical atrophy, dementia, and premature death (37 yr) has also been briefly described (Imoto et al. 2007).
The severe clinical outcome of compromised XPF-ERCC1 function in humans appears distinct to core defects in NER components associated with XP. Because of the uniquely marked hypersensitivity to ICL agents of XPF-ERCC1-deficient cells compared with NER defects, it is tempting to speculate that these patients are particularly sensitive to some form of endogenously generated ICLs, as has been suggested for disorders such as Fanconi anemia (see section Endogenous DNA Damage and Its Implications for FA). Furthermore, ROS-induced ICL (e.g., Gua[8-5me]Thy) and cyclopurines such as the 8, 5′-cyclopurine-2′-deoxynucleosides have been shown to accumulate in Ercc1-defective mouse tissues, including brain, potentially representing an important endogenously generated DNA lesion in this context (Wang et al. 2012a,b).
UV-Sensitive Scaffold Protein A and UV-Sensitive Syndrome
No clear genotype-phenotype relationship between mutation site in CSA/ERCC8 and CSB/ERCC6 and clinical presentation has emerged (Cleaver et al. 2009). Mutations in each of these genes results in CS, but in stark contrast, they are also found in a few individuals with mild photosensitivity as their sole clinical feature (UV-sensitive syndrome [UVs]). Furthermore, other individuals presenting with photosensitivity alone were found not to harbor variants in either CSA/ERCC8 or CSB/ERCC6, along with defective TCR (Fujiwara et al. 1981; Itoh et al. 1994, 1995; Spivak 2005).
Recently, the causative genetic defect for this UVs syndrome was identified by several groups as a novel component of RNA polymerase II (RNA Pol II), termed UV-sensitive scaffold protein A (UVSSA, formerly known as KIAA1530) (Nakazawa et al. 2012; Schwertman et al. 2012; Zhang et al. 2012). Each group identified UVSSA using a different approach. Zhang and colleagues used microcell-mediated chromosome transfer into UVs syndrome patient fibroblasts to select for stable UV-resistant clones with normal TCR (Zhang et al. 2012). Comparative genomic hybridization array analysis refined the minimal complementing region, and candidate gene-containing BAC clones were used in a second set of complementation experiments, ultimately identifying KIAA1530. Schwertman and colleagues were using a SILAC-base proteomic approach to identify and characterize NER factors regulated by ubiquitination and identified KIAA1530 as a novel gene that impaired TCR on siRNA silencing, along with also implicating the deubiquitinating isopeptidase ubiquitin-specific protease 7 (USP7) in this process (Schwertman et al. 2012). Finally, Nakazawa and colleagues took the more direct route of applying exome sequencing to identify mutations in KIAA1530 in UVs syndrome individuals (Nakazawa et al. 2012).
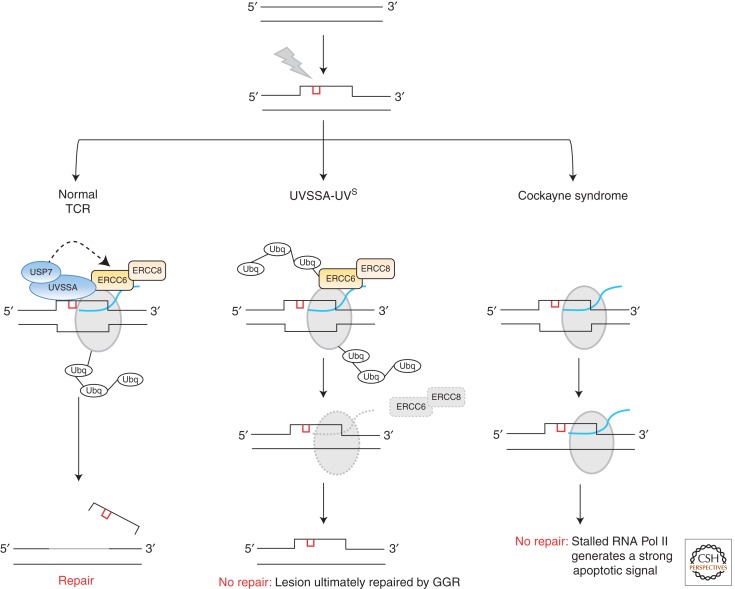
UV photoproducts are potent blocks to RNA Pol II-mediated transcription and TCR is dedicated to rapidly and efficiently removing these lesions enabling transcription resumption. UVSSA is thought to play some role in enabling stalled RNA Pol II to backtrack from the DNA lesion, thereby allowing access to the TCR machinery (Fig. 3). UVSSA interacts with TFIIH, CSB/ERCC6, and RNA Pol IIo (the elongating form of RNA Pol II) but also forms a complex with USP7; the latter interaction apparently being important for regulating the level of CSB/ERCC6. In UVSSA-defective cells, CSB/ERCC6 appears to be ubiquitinated and degraded after UV, likely potentiating RNA Pol II stalling and impairing recovery (Fig. 3) (Nakazawa et al. 2012; Schwertman et al. 2012; Zhang et al. 2012).
Figure 3.
TCR under specific contexts. UV photoproducts (red) create a localized distortion in the DNA helix prompting recognition and removal by nucleotide excision repair (NER). In actively transcribing regions of the genome, UV lesions create a block to RNA polymerase II (RNA Pol II [gray]), temporarily inhibiting RNA synthesis (blue) prompting engagement of transcription-coupled repair (TCR). In normal cells, stalled RNA Pol II is ubiquitinated in an ERCC6/ERCC8-dependent manner. The combined action of the ERCC6/ERCC8 and UVSSA/USP7 complexes somehow coordinate to enable stalled ubiquitinated RNA Pol II to be repositioned, thereby allowing access to the lesion for the NER machinery to remove the lesion. In the UVSSA-UVs situation, both ERCC6 and the stalled RNA Pol II remain ubiquitinated, likely prompting their degradation by the proteasome. Therefore, no repair occurs by rapid TCR and the lesion is left to be dealt with by global genome NER (GGR). In the context of Cockayne syndrome, ubiquitination of the stalled RNA Pol II does not occur and the polymerase remains stalled at the lesion, likely generating a very strong apoptotic signal owing to the failure to recover transcription.
Implications for Interpreting Genotype–Phenotype Relationships
So, if defective CSA/ERCC8, CSB/ERCC6, and UVSSA function all impair TCR and RNA Pol II recovery following UV, how can we rationalize the stark clinical differences between CS and UVs syndrome? Endogenously generated oxidative DNA damage may play a role here. CS patient-derived cells are sensitive to killing by ROS-generating agents such as H2O2, unlike UVSSA-defective UVs syndrome cells (Spivak and Hanawalt 2006; D’Errico et al. 2007; Nardo et al. 2009; Pascucci et al. 2012). Furthermore, CSA/ERCC8 and CSB/ERCC6 have been identified in mitochondria where they are thought to play a role in the repair of mtDNA, deficits of which may contribute to impaired neurogenesis and/or neurodegeneration, as discussed above for MCSZ (Kamenisch et al. 2010). It is not known yet whether UVSSA plays any role in mtDNA repair. An additional model to help explain the clinical differences between CS and UVs syndrome has been proposed by Nakazawa and colleagues (Nakazawa et al. 2012). Stalled RNA Pol II is stably ubiquitinated and backtracked in a CSA/ERCC8, CSB/ERCC6, and UVSSA-dependent process enabling TCR (Fig. 3). But, in UVSSA-UVs cells RNA Pol II can still be ubiquitinated in a CSA/ERCC8 and CSB/ERCC6-dependent manner, independent of UVSSA, leading to proteasomal degradation, thereby preventing transcription resumption (Fig. 3). In CS, both ubiquitin-dependent backtracking and degradation of RNA Pol II are impaired perhaps leading to a more deleterious prolonged arrest ultimately signaling to apoptosis (Fig. 3). Of course the possibility of additional as-yet-unknown roles of CSA/ERCC6 and CSB/ERCC8 cannot be ruled out.
DISORDERS OF HR AND ICL REPAIR: FANCONI ANAEMIA, FAMILIAL BREAST AND OVARIAN CANCER, AND KARYOMEGALIC INTERSTITAL NEPHRITIS
Fanconi anemia (FA) is the most frequent inherited cause of bone marrow failure (Shimamura and Alter 2010). This well-characterized devastating disorder follows a typical pattern of bone marrow failure in childhood–early teens before development of acute myeloid leukemia (AML) by late teens–early adulthood, with a median survival of 20 yr (Kutler et al. 2003; Rosenberg et al. 2003; Auerbach 2009; Shimamura and Alter 2010). The risk of developing solid tumors, particularly head and neck, is also elevated in FA adults. The hematopoetic system in FA is unstable showing frequent genetic reversion, mosaicism, and clonal expansion. Often, although not always, FA is associated with a combination of congenital abnormalities, including short stature, hyperpigmentation (café-au-lait spots), microphthalmia, and the archetypal radial-ray defects that range from hypoplasia to complete absence of the radius.
FA is multigenic and several new genetic defects have been described in FA patients since 2005, reinforcing the functional connection between the FA pathway and HR (Moldovan and D’Andrea 2009; Kim and D’Andrea 2012). Notable examples include defects in RAD51C (now FANCO), the BRCA2 interactor PALB2 (FANCN), and SLX4 (FANCP) (Reid et al. 2007; Xia et al. 2007; Meindl et al. 2010; Vaz et al. 2010; Crossan et al. 2011; Kim et al. 2011; Stoepker et al. 2011). SLX4-SLX1 endonuclease can resolve Holliday junctions in vitro (Andersen et al. 2009; Fekairi et al. 2009; Svendsen et al. 2009; Svendsen and Harper 2010). However, the physiological relevance of this has not been shown. More recently, a truncating nonsense mutation in RAD51 paralogue XRCC2 (p.Arg215*) was identified in a single Saudi Arabian patient of consanguineous parents showing typical FA phenotypes such as bilateral absent thumbs and cellular sensitivity to diepoxybutane (DEB), the standard diagnostic ICL sensitivity assay for FA (Shamseldin et al. 2012). The patient was 2.5 years old at the time of diagnosis without evidence of bone marrow failure or AML, yet.
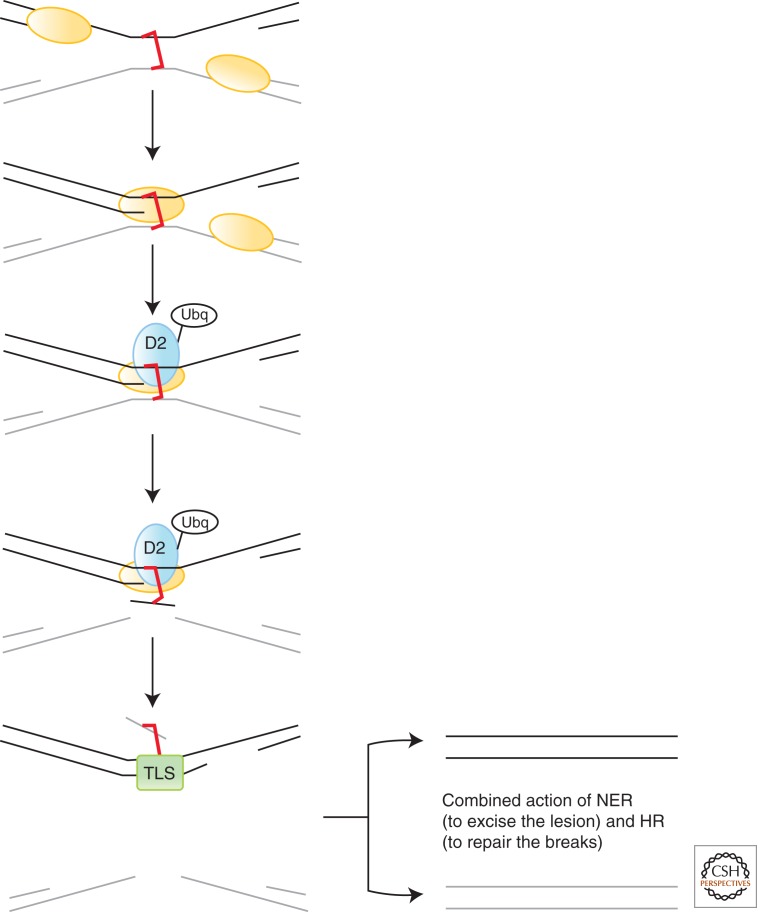
To date, 15 FA complementation groups/genes have been described; FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ/BRIP1, FANCL, FANCM, FANCN/PALB2, FANCO/RAD51C, and FANCP/SLX4. The FA pathway repairs ICLs in DNA, a highly toxic lesion (Fig. 4). The key molecular event in the FA pathway is the monoubiquitination of FANCD2 and FANCI by the FA core complex, an E3 ubiquitin ligase formed of FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM. Monoubiquitinated FANCD2 and FANCI then functionally interact with the remaining downstream FA proteins and factors such as BRCA1 and the recently described FAN-1 (FA-associated nuclease-1) nuclease (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). The mechanisms underlying the repair of ICLs are complex and only now starting to emerge (Kim and D’Andrea 2012). They involve functional interplay between the FA pathway, HR, and translesion synthesis (TLS) (Fig. 4). The reader is referred to Niedernhofer (2013) for a detailed review of DNA cross-link repair.
Figure 4.
ICL repair. An interstrand cross-link (ICL) poses a serious problem for replication and transcription. Here, two replication forks converge on an ICL (red). One of the forks is extended toward the ICL, whereas the other remains stalled and stabilized. The FA pathway is engaged and monoubiquitylated-FANC-D2 (D2-Ubq) is localized to the ICL. Excision of one strand occurs (gray), likely involving ERCC1-XPF and/or SLX4, depending on the context, generating a monoadducted lesion. Translesion synthesis (TLS) is engaged to allow bypass of the adducted base in the template strand (black). The resultant DNA double-strand break is thought to be repaired by homologous recombination, whereas the monoadduct is removed by NER.
Several structure-specific endonucleases, aside from FAN-1, are implicated in ICL repair. The full context-specific extent of their redundancy and/or functional hierarchy is as yet unclear. For example, epistasis analysis using a recently described chicken DT40 B-cell model for FAN1 deficiency suggested FAN1 operates independently of FANCC and FANCJ in response to ICL agents (Yoshikiyo et al. 2010). SLX4 is a scaffold protein that interacts with several endonucleases including MUS81-EME1 and XPF-ERCC1. It is thought that SLX4 is important for the recruitment of these alternate structure-specific endonucleases during ICL repair (Crossan and Patel 2012).
Endogenous DNA Damage and Its Implications for FA
Because of the stochastic nature of the developmental and hematological abnormalities in FA, it seems likely that these features are the consequence of impaired repair of some form of endogenous DNA damage (Crossan and Patel 2012). By-products and intermediates of normal oxidative metabolism, including reactive aldehydes (acetaldehyde, formaldehyde) and lipid peroxidation products, are capable of forming inter- and intrastrand DNA cross-links and DNA-protein cross-links, which may be relevant in this context. In support of this, cotargeting Aldh2 (aldehyde dehydrogenase 2, a reactive aldehyde catabolic enzyme) and Fancd2, potentiated leukemia development in mice, and knockout of ADH5 (alcohol dehydrogenase 5, a formaldehyde catabolic enzyme) is synthetic lethal with FANCL−/− in chicken DT40 cells (Langevin et al. 2011; Rosado et al. 2011).
Familial Breast and Ovarian Cancer
Other novel germline defects in HR-pathway components have also been described recently. Multiple mutations in RAD51C and RAD51D have been found in breast and ovarian cancer cohorts prompting some to call for screening for these genes in breast and ovarian cancer (Meindl et al. 2010; Loveday et al. 2011; Osorio et al. 2012). The difficulty in assigning pathogenicity for some missense variants, in the absence of functional evaluation has, however, led to some debate here (Loveday et al. 2012).
FAN1 and Karyomegalic Interstitial Nephritis
The identification of FAN1 as an ICL repair factor prompted suggestions that it likely represents a novel (as yet undescribed) FA causative defect, because a minority of FA patients exist that are not associated with defects in the known FANC genes (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). Recent evidence-based studies using patient-derived cells have challenged this, however. Trujillo and colleagues described four patients with 15q13.3 microdeletion involving seven genes but causing biallelic deletion of FAN1 associated with undetectable levels of FAN1 protein (Trujillo et al. 2012). These patients did not have FA or hematological problems but rather a complex developmental disorder; a likely consequence of the multigenic nature of the microdeletion typical of multiple genomic disorders (Colnaghi et al. 2011). Furthermore, despite these patient cells having a profound defect in FAN1 expression, they did not fall within the FA range in the standard DEB diagnostic assay or show pronounced G2 arrest typical of FA cells treated with ICL agents, only showing modest sensitivity to killing by ICL-forming agents compared with other FA cells (Trujillo et al. 2012). These findings are suggestive of a backup and/or alternate role for FAN1 in FANC pathway-mediated ICL repair in deference to other nucleases.
Using a combination of homozygosity mapping and exome sequencing in an attempt to identify novel nephronophthisis (NPHP)-related ciliopathy genes, multiple congenital defects in FAN1 were unexpectedly identified recently in several families showing karyomegalic interstitial nephritis (KIN) (Zhou et al. 2012). KIN is a rare NPHP-like chronic kidney disease caused by renal tubular degeneration and fibrosis, but specifically also associated with renal cell karyomegaly (enlarged nuclei) (Burry 1974; Mihatsch et al. 1979). Interestingly, polyploidy is not restricted to renal tissue in KIN patients but is often also observed in the lung, liver, and brain (Spoendlin et al. 1995; Monga et al. 2006). Lymphoblast and fibroblast cell lines from FAN1-mutated KIN patients showed hypersensitivity to killing and elevated chromosome aberration formation in response to ICL-forming agents, although quantitatively less so than cells from FANCA and FANCD2 patients (Zhou et al. 2012). Why impaired FAN1 function results in a chronic kidney disease in humans rather than FA is unclear but may have some origin in the relatively distinct/nonoverlapping tissue-specific expression of FA genes such as FANCD2, compared with FAN1 (Zhou et al. 2012). Of note, other defects in genes with known (MRE11, CEP164) or proposed roles in the DNA damage response (DDR) and/or cell cycle (ZNF423) have also recently been suggested to underlie NPHP (Chaki et al. 2012). The precise pathomechanism associating these defects specifically with chronic kidney disease in humans is currently unclear.
NOVEL DISORDERS OF IMPAIRED DDRs
The repair of DNA damage is intimately coordinated with complex interconnected signal transduction pathways enabling fundamental processes such as DNA damage detection and localized chromatin remodeling to facilitate cell-cycle checkpoint activation, DNA repair, and apoptosis induction. See the accompanying article on DNA repair in the context of chromatin by Genevieve Almouzni. Over the last decade, SUMOylation and ubiquitination have joined phosphorylation as essential posttranslational modifications of the DDR network (Cohn and D’Andrea 2008; Bekker-Jensen and Mailand 2010; Zlatanou and Stewart 2010; Xu and Price 2011). Subsequently, several disorders have emerged with congenital defects in key players in some of these events. The reader is referred to Marechal and Zou (2013) and Sirbu and Cortez (2013) for detailed descriptions of the signal transduction mechanisms involved in DNA damage detection and processing in mammals.
Microcephalic Primordial Dwarfisms and the DDR: Seckel Syndrome and Microcephalic Osteodysplastic Primordial Dwarfism Type II
Microcephalic primordial dwarfism (MPD) is the collective term for a family of clinically overlapping conditions typified by profound intrauterine and postnatal growth delay, severe microcephaly, and variable skeletal abnormalities from the subtle (clinodactyly, brachydactyly) to the overt (kyphosis, absent patellae). Two notable examples include Seckel syndrome (SS) and microcephalic osteodysplastic primordial dwarfism type II (MOPDii) (Seckel 1960; Majewski et al. 1982; Hall et al. 2004).
ATR-ATRIP SS
SS is an MPD usually presenting with symmetric dwarfism with disproportionate microcephaly. The first genetic defect identified in SS was a nonsynonymous missense mutation in ATR, the gene encoding aaxia telangiectasia and Rad3-related protein, the apical protein kinase of the DDR (O’Driscoll and Jeggo 2003; O’Driscoll et al. 2003; Cimprich and Cortez 2008). This mutation, identified by a homozygosity mapping approach, was found in five individuals from two related families (Goodship et al. 2000; O’Driscoll et al. 2003). The mutation caused variable missplicing of exon 9. This splicing mutation was modeled in the mouse germline creating an animal that recapitulated all of the SS clinical features observed in the index case, and more (e.g., pancytopenia) (Murga et al. 2009). This Atr-SS mouse model also provided clear evidence for the role of ATR during embryonic development, intrauterine programming, and the preservation of stem cell niches (O’Driscoll 2009b, 2009a). Recently, two more ATR-mutated SS individuals have been described, each with the same compound heterozygous mutations in ATR (Ogi et al. 2012). These defects were identified on a candidate-based approach following careful analysis of multiple ATR-dependent DDR end points. These additional patients further help define the clinical spectrum of ATR deficiency in humans, the most consistent feature being a severe disproportionate microcephaly, even relative to the markedly reduced body size.
ATR stably interacts with ATR-interacting protein (ATRIP) as part of the DDR, and gene knockdown approaches have shown that ATRIP deficiency phenocopies ATR deficiency (Cortez et al. 2001). The first example of congenital deficiency of ATRIP has also been described recently in a SS individual (Ogi et al. 2012). This defect was identified by a candidate-based approach. ATRIP-SS cells show defective ATR-dependent DNA damage signaling (e.g., γH2AX, pCHK1) and impaired G2-M cell-cycle checkpoint activation. In contrast to ATR-mutated individuals, the skeletal system was not disproportionately impacted here. Interestingly, MRI imaging did catalog an abnormal pituitary, which could be relevant to the severe growth delay.
PCNT and Microcephalic Osteodysplastic Primordial Dwarfism Type II
MOPDii as a clinical diagnosis is usually distinguished from other MPDs such as SS by virtue of its presentation as an asymmetric dwarfism and disproportionate short limbs with a more marked skeletal involvement (Hall et al. 2004). Nevertheless, this distinction is often not obvious as it can be very much age dependent. The first, and as yet to date, only genetic defect identified for MOPDii is that of PCNT, the gene encoding pericentrin, a large centrosomal protein, likely with a structural role therein (Griffith et al. 2008; Rauch et al. 2008). PCNT-mutated patient cells show altered microtubule spindles and supernumerary centrosomes. Homozygosity mapping of consanguineous families again played a vital role in gene identification here. Interestingly, these cells were also shown to be impaired in ATR and CHK1-dependent hydroxyurea (HU)-induced 53BP1 foci formation as well as defective ATR-dependent G2-M checkpoint activation, cellular features of ATR/ATRIP-SS (Griffith et al. 2008). It has been suggested that these defects have their origin at the level of CHK1 recruitment to and CDK1-Cyclin B activation at the centrosome (Griffith et al. 2008; Tibelius et al. 2009).
Congenital defects in multiple genes encoding proteins that localize to or function at the centrosome and microtubule spindles have been described in SS and primary microcephaly individuals (Bond et al. 2002; Trimborn et al. 2004; Bond et al. 2005; Al-Dosari et al. 2010; Barr et al. 2010; Bilguvar et al. 2010; Guernsey et al. 2010; Nicholas et al. 2010; Yu et al. 2010; Kalay et al. 2011; Sir et al. 2011; Hussain et al. 2012; Vulprecht et al. 2012). Interestingly, some of these defects have also been shown to be associated with impaired ATR-dependent DDR (Alderton et al. 2006; Smith et al. 2009).
Qvist and colleagues recently described novel defects in RBBP8, the gene encoding CtIP, in two families, one with SS and the other with a diagnosis of Jawad syndrome (microcephaly, mental retardation, digital abnormalities) (Kelly et al. 1993; Qvist et al. 2011). CtIP mediates resection of DNA DSBs to facilitate repair. See Rothstein (2013) and Jasin (in press) on the repair of strand breaks by HR. The dominant CtIP defects described here result in a failure to generate single-stranded DNA (ssDNA) following damage resulting in an acquired functional defect in ATR-signaling, because replication protein A (RPA)-coated ssDNA is the means through which ATR is recruited to DNA (Zou and Elledge 2003; Qvist et al. 2011). These findings are consistent with the previous description of an ATR-dependent checkpoint defect in cells from this SS family, further reinforcing the pathophysiological link between a compromised ATR-dependent DDR and MPD (Alderton et al. 2004).
Interestingly, PCNT-mutated MOPDii has been found to result in a severe insulin-resistant form of diabetes (Huang-Doran et al. 2011). This has implications for understanding the clinical presentation of this condition as well as patient management. It is unclear whether this is also a general feature of ATR/ATRIP-SS, although the Atr-SS mouse model did show a depressed somatotroph axis (Murga et al. 2009). Whether these features have an origin in impaired DDR or repair, similar to that described above for XFE progeria, ERCC1-COFS, and even CS, is certainly a possibility.
ATR and Autosomal Dominant Oropharyngeal Cancer Syndrome
Tanaka and colleagues recently described an unusual disorder comprising oropharyngeal cancer, pronounced dermal telangiectasias, and dental caries in 24 individuals from a large five-generation Caucasian pedigree originating from Indiana, United States (Tanaka et al. 2012). Homozygosity mapping identified the causal gene, unexpectedly, as ATR. Patients were heterozygous for a missense mutation in a highly conserved residue (p.Gln2144Arg) in the FAT domain of ATR. In contrast to ATR-SS, this mutation did not affect ATR expression, although patient fibroblasts showed mildly attenuated ATR-dependent phosphorylation of CHK1 and H2AX as well as reduced p53 accumulation following treatment with HU. Interestingly, loss of heterozygosity for the ATR locus was observed in the oropharyngeal tumor tissue. This syndrome represents the first example of germline mutation in ATR associated with a cancer syndrome representing a novel clinical outcome of impaired ATR function (Tanaka et al. 2012).
How ATR dysfunction in this context contributes to these clinical features is unclear. No malignancies have been reported in ATR/ATRIP-SS, although there are still too few cases to allow any conclusions to be drawn. But, the Atr-SS mouse model shows a conspicuous absence of tumors (Murga et al. 2009). In fact, crossing this strain into a p53−/− background revealed an unexpected synthetic lethality (Murga et al. 2009). This serendipitous finding, with its origin in modeling the human syndrome in mouse, is now being pursued from the perspective of ATR small molecule kinase inhibitors and their potential selective efficacy against p53-defective cancers (Toledo et al. 2011a,b).
RAD50 and Nimegen Breakage Syndromelike Disorder
The MRE11/RAD50/NBS1 (M/R/N) complex functions to tether DSBs and plays a role in optimal ATM activation at the site of the break. Defective ATM causes ataxia telangiectasia (A-T), a progressive neurodegenerative condition associated with immune dysfunction and elevated cancer incidence, specifically for lymphoma and leukemia (Lavin 2008). Pathogenic mutations in MRE11A, encoding MRE11, result in A-T-like disorder (A-T-LD), an attenuated form of A-T with mild ataxia and generally no evidence of malignancy (Taylor et al. 2004). Pathogenic mutations in NBN (previously termed NBS1), encoding NBS1, cause Nijmegen breakage syndrome (NBS). This disorder is characterized by growth retardation, microcephaly, combined immunodeficiency, and elevated lymphoma predisposition (Digweed and Sperling 2004). Cells from A-T, A-T-LD, and NBS all show sensitivity to killing by ionizing radiation, compromised DSB repair—specifically at heterochromatin, attenuated ATM-dependent phosphorylation of key substrates including p53, CHK2, SMC1, and KAP1 associated with impaired checkpoint activation following DSB formation. Recently, Waltes and colleagues described the first and as yet to date, only case of congenital deficiency in RAD50 in a single individual with a working clinical diagnosis of an NBS-like disorder (Waltes et al. 2009). The patient displayed growth retardation and microcephaly typical of NBS as well as the characteristic chromosome 7-14 translocation. But, no evidence of lymphoid malignancy or immune dysfunction was obvious up to 23 years of age. Cells from the RAD50-mutated patient were ionizing radiation sensitive, failed to form M/R/N foci following DSBs, showed impaired checkpoint activation, reduced ATM-dependent substrate phosphorylation (e.g., pSer15-p53, pSer957-SMC1, and pSer343-NBS1), and extremely low levels of RAD50 (Waltes et al. 2009).
RNF168-Deficiency Syndrome
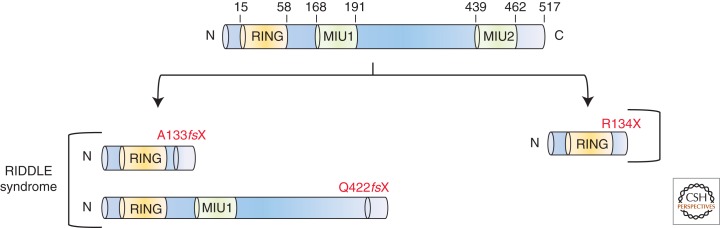
In recent years, ubiquitination and SUMOylation have emerged as fundamental posttranslation modifications orchestrating DSB repair. The RING finger E3-ubiquitin ligases RNF8 and RNF168, together with the HECT-domain-containing HERC2 ubiquitin ligase, sequentially ubiquitinate histones at DSBs, enabling the localized recruitment of factors such as 53BP1 and BRCA1 (Bekker-Jensen and Mailand 2010). Stewart and colleagues identified an individual with two truncating mutations in RNF168 associated with a disorder of hypogammaglobulinemia, short stature, mild motor impairment, and intellectual disability that they termed RIDDLE syndrome: radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties (Fig. 5) (Stewart et al. 2007, 2009). The inability of these patient cells to form IR-induced 53BP1foci led directly to the identification of the genetic defect in RNF168, based on candidates identified in a previously published siRNA screen (Kolas et al. 2007; Stewart 2009). A subsequent Rnf168−/− mouse model has provided evidence for a role of RNF168 in V(D)J and CSR (Bohgaki et al. 2011).
Figure 5.
Pathogenic defects in RNF168. Schematic representation of RNF168 showing the relative positions of the RING domain and the ubiquitin-binding domains (MIU1 and MIU2). The RIDDLE-syndrome-associated RNF168 defects are shown on the left-hand side, whereas the nonsense RNF168 defect described by Devgan and colleagues is shown on the right-hand side.
Devgan and colleagues recently identified the second known individual with a genetic defect in RNF168 (Devgan et al. 2011). In contrast to the RIDDLE syndrome case, this patient presented with complex condition associated with ataxia, ocular, and bronchial telangiectasia, but also with microcephaly, short stature, low IgA, and normal intelligence. This individual was homozygous for a primary truncating mutation downstream from the RING domain but upstream of the two MIU domains (motif interacting with ubiquitin) of RNF168 (Fig. 5). Because at least one of the MIU domains is preserved in the RIDDLE syndrome individual, Devgan and colleagues have proposed this as a potential mechanism to explain the clinical distinctions between these two RNF168-mutated individuals (Devgan et al. 2011). Collectively, these cases indicate that impaired RNF168 function impacts not only the immune system, where programmed DSB formation is a prerequisite for normal development, but also neurogenesis (microcephaly) and neuronal function (ataxia).
CONCLUDING REMARKS
The identification of congenital disorders of DNA repair and the DDR provides irrefutable evidence that the networks governing genomic stability are fundamentally important not only to prevent malignant transformation and neurodegeneration, but also, depending on context, for normal growth, development, neurogenesis, and immune system development.
In this overview, I have briefly reviewed only some of the key disorders described within the last decade, giving a flavor of the progress in this important area of the DNA repair field. But, there have been other exciting developments concerning congenital human disorders that I have not covered here. For example, growing evidence suggests that impaired genomic stability is associated with certain genomic disorders caused by gene copy number variation (CNV) (Colnaghi et al. 2011; Harvard et al. 2011; Outwin et al. 2011; Kerzendorfer et al. 2012). CNVs are a major cause of human congenital disorders (Lupski 2007; Hastings et al. 2009; Stankiewicz and Lupski 2010). The implications for DNA repair and DDR pathways in this context merits closer attention. There already exists tantalizing evidence to suggest that these pathways are sensitive to gene dosage (O’Driscoll 2008; Cabelof 2012; Depienne et al. 2012).
Congenital defects in the DNA replication licensing machinery have recently been identified in Meier-Gorlin syndrome (MGS), a MPD often associated with marked skeletal involvement (Gorlin et al. 1975; Ahmad and Teebi 1997; Bongers et al. 2001a,b, 2005; Bicknell et al. 2011a,b; Guernsey et al. 2011; de Munnik et al. 2012). Interestingly, mutations in MCM4 have been described in a clinically distinct syndrome of adrenal insufficiency growth retardation and selective natural killer cell deficiency (Casey et al. 2012; Gineau et al. 2012; Hughes et al. 2012). These human phenotypes are quite distinct to the mouse Mcm4Chaos3 allele (Shima et al. 2007). How can we explain these distinctions? What are the implications for these and MGS patients concerning cancer predisposition, or if cancer were to develop in this context, how would it be best treated? As our knowledge of the mechanics of these pathways grows, we will undoubtedly uncover more and more disorders. It is hoped that with the promising exciting potential of exome sequencing and genomic medicine we will be in a position to better diagnose and manage these conditions. Currently, congenital defects in DNA damage-induced ubiquitination and SUMOylation pathways appear to be underrepresented. It will be very interesting to observe how such conditions present clinically, and how this presentation will hopefully also inform on pathway function.
ACKNOWLEDGMENTS
The Human DNA Damage Response Disorders Group is funded by the UK Medical Research Council, Cancer Research UK (CR-UK), and Leukaemia Lymphoma Research. M.O’D. is a CR-UK Senior Cancer Research Fellow.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC 2006. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 443: 713–716 [DOI] [PubMed] [Google Scholar]
- Ahmad S, Teebi RJG 1997. Not a new Seckel-like syndrome but ear-patella-short stature syndrome. Am J Med Genet 70: 454. [PubMed] [Google Scholar]
- Ahnesorg P, Smith P, Jackson SP 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124: 301–313 [DOI] [PubMed] [Google Scholar]
- Aicardi J, Goutieres F 1984. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol 15: 49–54 [DOI] [PubMed] [Google Scholar]
- Alderton GK, Joenje H, Varon R, Borglum AD, Jeggo PA, O’Driscoll M 2004. Seckel syndrome exhibits cellular features demonstrating defects in the ATR signalling pathway. Hum Mol Genet 13: 3127–3138 [DOI] [PubMed] [Google Scholar]
- Alderton GK, Galbiati L, Griffith E, Surinya KH, Neitzel H, Jackson AP, Jeggo PA, O’Driscoll M 2006. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol 8: 725–733 [DOI] [PubMed] [Google Scholar]
- Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS 2010. Novel CENPJ mutation causes Seckel syndrome. J Med Genet 47: 411–414 [DOI] [PubMed] [Google Scholar]
- *.Alexeyev M, Shokolenko I, Wilson G, LeDoux S 2013. Mitochondrial DNA damage and repair—Critical analysis and update. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J 2009. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell 35: 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD 2009. Fanconi anemia and its diagnosis. Mutat Res 668: 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglioni S, Melean G, Gensini F, Santucci M, Scatizzi M, Papi L, Genuardi M 2005. A kindred with MYH-associated polyposis and pilomatricomas. Am J Med Genet A 134A: 212–214 [DOI] [PubMed] [Google Scholar]
- Baker K, Raymond FL, Bass N 2012. Genetic investigation for adults with intellectual disability: Opportunities and challenges. Curr Opin Neurol 25: 150–158 [DOI] [PubMed] [Google Scholar]
- Barr AR, Kilmartin JV, Gergely F 2010. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J Cell Biol 189: 23–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N 2010. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair 9: 1219–1228 [DOI] [PubMed] [Google Scholar]
- Bicknell LS, Bongers EMHF, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PAJ, et al. 2011a. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet 43: 356–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin C-A, Yeyati P, Al Sanna N, Bober M, et al. 2011b. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet 43: 350–355 [DOI] [PubMed] [Google Scholar]
- Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, et al. 2010. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohgaki T, Bohgaki M, Cardoso R, Panier S, Zeegers D, Li L, Stewart GS, Sanchez O, Hande MP, Durocher D, et al. 2011. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet 7: e1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond JRE, Mochida G, Hampshire D, Scott S, Askham J, Springell K, Mahadevan M, Crow Y, Markham A, Walsh CG, et al. 2002. ASPM is a major determinant of cerebral cortical size. Nat Genet 32: 316–320 [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet 37 353–355 [DOI] [PubMed] [Google Scholar]
- Bongers EMHF, Opitz JM, Fryer A, Sarda P, Hennekam RCM, Hall BD, Superneau DW, Harbison M, Poss A, van Bokhoven H, et al. 2001a. Meier-Gorlin syndrome: Report of eight additional cases and review. Am J Med Genet 102: 115–124 [DOI] [PubMed] [Google Scholar]
- Bongers EMHF, Van Bokhoven H, Van Thienen M-N, Kooyman MAP, Van Beersum SEC, Boetes C, Knoers NVAM, Hamel BCJ 2001b. The small patella syndrome: Description of five cases from three families and examination of possible allelism with familial patella aplasia-hypoplasia and nail-patella syndrome. J Med Genet 38: 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers E, Van Kampen A, Van Bokhoven H, Knoers N 2005. Human syndromes with congenital patellar anomalies and the underlying gene defects. Clin Genet 68: 302–319 [DOI] [PubMed] [Google Scholar]
- Bosma GC, Custer RP, Bosma MJ 1983. A severe combined immunodeficiency mutation in the mouse. Nature 301: 527–530 [DOI] [PubMed] [Google Scholar]
- Bryant HE, Helleday T 2004. Poly(ADP-ribose) polymerase inhibitors as potential chemotherapeutic agents. Biochem Soc Trans 32: 959–961 [DOI] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche M-C, Sanal O, Plebani A, Stephan J-L, Hufnagel M, le Deist F, et al. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299 [DOI] [PubMed] [Google Scholar]
- Burry AF 1974. Extreme dysplasia in renal epithelium of a young woman dying from hepatocarcinoma. J Pathol 113: 147–150 [DOI] [PubMed] [Google Scholar]
- Cabelof DC 2012. Haploinsufficiency in mouse models of DNA repair deficiency: Modifiers of penetrance. Cell Mol Life Sci 69: 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott K 2002. Polynucleotide kinase. A versatile molecule makes a clean break. Structure (Camb) 10: 1151. [DOI] [PubMed] [Google Scholar]
- Caldecott KW 2008. Single-strand break repair and genetic disease. Nat Rev Genet 9: 619–631 [DOI] [PubMed] [Google Scholar]
- Casey JP, Nobbs M, McGettigan P, Lynch S, Ennis S 2012. Recessive mutations in MCM4/PRKDC cause a novel syndrome involving a primary immunodeficiency and a disorder of DNA repair. J Med Genet 49: 242–245 [DOI] [PubMed] [Google Scholar]
- Chaki M, Airik R, Ghosh Amiya K, Giles Rachel H, Chen R, Slaats Gisela G, Wang H, Hurd Toby W, Zhou W, Cluckey A, et al. 2012. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150: 533–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev 16: 2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chiruvella KK, Liang Z, Wilson TE 2013. Repair of double-strand breaks by end-joining. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE 1968. Deficiency in repair replication of DNA in xeroderma pigmentosum. Nature 218: 652–656 [DOI] [PubMed] [Google Scholar]
- Cleaver JE 1969. Xeroderma pigmentosum: A human disease in which an initial stage of DNA repair is defective. Proc Nat Acad Sci 63: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE 1970. DNA repair and radiation sensitivity in human (xeroderma pigmentosum) cells. Int J Rad Biol 18: 557–565 [DOI] [PubMed] [Google Scholar]
- Cleaver JE, Lam ET, Revet I 2009. Disorders of nucleotide excision repair: The genetic and molecular basis of heterogeneity. Nat Rev Genet 10: 756–768 [DOI] [PubMed] [Google Scholar]
- Cohn MA, D’Andrea AD 2008. Chromatin recruitment of DNA repair proteins: Lessons from the Fanconi anemia and double-strand break repair pathways. Mol Cell 32: 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnaghi R, Carpenter G, Volker M, O’Driscoll M 2011. The consequences of structural genomic alterations in humans: Genomic disorders, genomic instability and cancer. Semin Cell Dev Biol 22: 875–885 [DOI] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ 2001. ATR and ATRIP: Partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Crossan GP, Patel KJ 2012. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J Pathol 226: 326–337 [DOI] [PubMed] [Google Scholar]
- Crossan GP, van der Weyden L, Rosado IV, Langevin F, Gaillard P-HL, McIntyre RE, Gallagher F, Kettunen MI, Lewis DY, Brindle K, et al. 2011. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet 43: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Yu Y, Gupta S, Cho Y-M, Lees-Miller SP, Meek K 2005. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol 25: 10842–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, West SC, Oettinger MA, Jeggo PA 2003. Non-homologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci 100: 2462–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Munnik SA, Bicknell LS, Aftimos S, Al-Aama JY, van Bever Y, Bober MB, Clayton-Smith J, Edrees AY, Feingold M, Fryer A, et al. 2012. Meier-Gorlin syndrome genotype-phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur J Hum Genet 20: 598–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Bouteiller D, Méneret A, Billot S, Groppa S, Klebe S, Charbonnier-Beaupel F, Corvol J-C, Saraiva J-P, Brueggemann N, et al. 2012. RAD51 haploinsufficiency causes congenital mirror movements in humans. Am J Hum Genet 90: 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM, et al. 2007. The role of CSA in the response to oxidative DNA damage in human cells. Oncogene 26: 4336–4343 [DOI] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Wilson DM III, Stevnsner TV, Bohr VA 2008. Mitochondrial DNA, base excision repair and neurodegeneration. DNA Repair 7: 1098–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devgan SS, Sanal O, Doil C, Nakamura K, Nahas SA, Pettijohn K, Bartek J, Lukas C, Lukas J, Gatti RA 2011. Homozygous deficiency of ubiquitin-ligase ring-finger protein RNF168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell Death Differ 18: 1500–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digweed M, Sperling K 2004. Nijmegen breakage syndrome: Clinical manifestation of defective response to DNA double-strand breaks. DNA Repair 3: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K 2003. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol 23: 5836–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW 2005. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434: 108–113 [DOI] [PubMed] [Google Scholar]
- Evans DG, Raymond FL, Barwell JG, Halliday D 2011. Genetic testing and screening of individuals at risk of NF2. Clin Genet 10.1111/j.1399-0004.2011.01816.x [DOI] [PubMed] [Google Scholar]
- Evers B, Helleday T, Jonkers J 2010. Targeting homologous recombination repair defects in cancer. Trends Pharmacol Sci 31: 372–380 [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong M-Q, Ruse C, Yates JR III, Russell P, et al. 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folbergrová J, Kunz WS 2012. Mitochondrial dysfunction in epilepsy. Mitochondrion 12: 35–40 [DOI] [PubMed] [Google Scholar]
- *.Fousteri M 2013. Transcription-coupled excision repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowden AL, Giussani DA, Forhead AJ 2006. Intrauterine programming of physiological systems: Causes and consequences. Physiology 21: 29–37 [DOI] [PubMed] [Google Scholar]
- Fowden AL, Forhead AJ, Coan PM, Burton GJ 2008. The placenta and intrauterine programming. J Neuroendocrinol 20: 439–450 [DOI] [PubMed] [Google Scholar]
- Fromme JC, Banerjee A, Huang SJ, Verdine GL 2004. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 427: 652–656 [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Ichihashi M, Kano Y, Goto K, Shimizu K 1981. A new human photosensitive subject with a defect in the recovery of DNA synthesis after ultraviolet-light irradiation. J Invest Dermatol 77: 256–263 [DOI] [PubMed] [Google Scholar]
- Gennery A, Slatter M, Bhattacharya A, Jeggo P, Abinun M, Flood T, Cant A 2005. Bone marrow transplantation for Nijmegan breakage syndrome. J Pediatr Hematol Oncol 27: 239. [DOI] [PubMed] [Google Scholar]
- Gineau L, Cognet C, Kara N, Lach FP, Dunne J, Veturi U, Picard C, Trouillet C, Eidenschenk C, Aoufouchi S, et al. 2012. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest 122: 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodship J, Gill H, Carter J, Jackson A, Splitt M, Wright M 2000. Autozygosity mapping of a seckel syndrome locus to chromosome 3q22.1-q24. Am J Hum Genet 67: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin R, Cervenka J, Moller K, Horrobin M, Witkop CJ 1975. Malformation syndromes. A selected miscellany. Birth Defects Orig Artic Ser 11: 39–50 [PubMed] [Google Scholar]
- Graham JM, Anyane-Yeboa K, Raams A, Appeldoorn E, Kleijer WJ, Garritsen VH, Busch D, Edersheim TG, Jaspers NG 2001. Cerebro-oculo-facio-skeletal syndrome with a nucleotide excision-repair defect and a mutated XPD gene, with prenatal diagnosis in a triplet pregnancy. Am J Hum Genet 69: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg SQ, Robinson AR, Niedernhofer LJ 2011. Physiological consequences of defects in ERCC1–XPF DNA repair endonuclease. DNA Repair 10: 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith E, Walker S, Martin C-A, Vagnarelli P, Stiff T, Vernay B, Sanna NA, Saggar A, Hamel B, Earnshaw WC, et al. 2008. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet 40: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, Babineau-Sturk T, Beis J, Dumas N, Evans SC, et al. 2010. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet 87: 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Matsuoka M, Jiang H, Evans S, Macgillivray C, Nightingale M, Perry S, Ferguson M, LeBlanc M, Paquette J, et al. 2011. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet 43: 360–364 [DOI] [PubMed] [Google Scholar]
- Hall JG, Flora C, Scott CI Jr, Pauli RM, Tanaka KI 2004. Majewski osteodysplastic primordial dwarfism type II (MOPD II): Natural history and clinical findings. Am J Med Genet A 130A: 55–72 [DOI] [PubMed] [Google Scholar]
- Hamel BC, Raams A, Schuitema-Dijkstra AR, Simons P, van der Burgt I, Jaspers NG, Kleijer WJ 1996. Xeroderma pigmentosum–Cockayne syndrome complex: A further case. J Med Genet 33: 607–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvard C, Strong E, Mercier E, Colnaghi R, Alcantara D, Chow E, Martell S, Tyson C, Hrynchak M, McGillivray B, et al. 2011. Understanding the impact of 1q21.1 copy number variant. Orphanet J Rare Dis 6: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G 2009. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T 2010. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis 31: 955–960 [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA 2008. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8: 193–204 [DOI] [PubMed] [Google Scholar]
- Huang-Doran I, Bicknell LS, Finucane FM, Rocha N, Porter KM, Tung YCL, Szekeres F, Krook A, Nolan JJ, O’Driscoll M, et al. 2011. Genetic defects in human pericentrin are associated with severe insulin resistance and diabetes. Diabetes 60: 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CR, Guasti L, Meimaridou E, Chuang C-H, Schimenti JC, King PJ, Costigan C, Clark AJL, Metherell LA 2012. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest 122: 814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain Muhammad S, Baig Shahid M, Neumann S, Nürnberg G, Farooq M, Ahmad I, Alef T, Hennies Hans C, Technau M, Altmüller J, et al. 2012. A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. Am J Hum Genet 90: 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee W-I, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol 4: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Imoto K, Boyle J, Oh S, Khan S, Ueda T, Nadem C, Slor H, Orgal S, Gadoth N, Busch D, et al. 2007. Patients with defects in the interacting nucleotide excision repair protein ERCC1 or XPF show xeroderma pigmentosum with later onset severe neurological degeneration. J Invest Dermatol 127 (Suppl): S92 [Google Scholar]
- Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ 2005. SCAN1 mutant Tdp1 accumulates the enzyme-DNA intermediate and causes camptothecin hypersensitivity. EMBO J 24: 2224–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Ono T, Yamaizumi M 1994. A new UV-sensitive syndrome not belonging to any complementation groups of xeroderma pigmentosum or Cockayne syndrome: Siblings showing biochemical characteristics of Cockayne syndrome without typical clinical manifestations. Mut Res 314: 233–248 [DOI] [PubMed] [Google Scholar]
- Itoh T, Fujiwara Y, Ono T, Yamaizumi M 1995. UVs syndrome, a new general category of photosensitive disorder with defective DNA repair, is distinct from xeroderma pigmentosum variant and rodent complementation group I. Am J Hum Genet 56: 1267–1276 [PMC free article] [PubMed] [Google Scholar]
- *.Jasin M. Cold Spring Harb Perspect Biol. doi: 10.1101/cshperspect.a012740. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers NGJ, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, Giglia-Mari G, Hoogstraten D, Kleijer WJ, Hoeijmakers JHJ, et al. 2007. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide rxcision repair and severe developmental failure. Am J Hum Genet 80: 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP 2002. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C → T:A mutations. Hum Mol Genet 11: 2961–2967 [DOI] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tuysuz B, Nurnberg G, et al. 2011. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet 43: 23–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenisch Y, Fousteri M, Knoch J, von Thaler A-K, Fehrenbacher B, Kato H, Becker T, Dollé MET, Kuiper R, Majora M, et al. 2010. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med 207: 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TE, Kirson L, Wyatt J 1993. Microcephaly and digital anomalies: A newly recognized syndrome of recessively inherited mental retardation. Am J Med Genet 45: 353–355 [DOI] [PubMed] [Google Scholar]
- Kerzendorfer C, Hannes F, Colnaghi R, Abramowicz I, Carpenter G, Vermeesch JR, O’Driscoll M 2012. Characterizing the functional consequences of haploinsufficiency of NELF-A (WHSC2) and SLBP identifies novel cellular phenotypes in Wolf–Hirschhorn syndrome. Hum Mol Genet 21: 2181–2193 [DOI] [PubMed] [Google Scholar]
- Kim H, D’Andrea AD 2012. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 26: 1393–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A 2011. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 43: 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D 2004. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J 23: 3874–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schöpf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavó E, Sartori AA, Hengartner MO, Jiricny J 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142: 77–88 [DOI] [PubMed] [Google Scholar]
- *.Krokan HE, Bjørås M 2013. Base excision repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudin AP, Zsurka G, Elger CE, Kunz WS 2009. Mitochondrial involvement in temporal lobe epilepsy. Exp Neurol 218: 326–332 [DOI] [PubMed] [Google Scholar]
- Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, Hanenberg H, Auerbach AD 2003. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101: 1249–1256 [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ 2011. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475: 53–58 [DOI] [PubMed] [Google Scholar]
- Lavin MF 2008. Ataxia-telangiectasia: From a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 9: 759–769 [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP 1996. The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochem Cell Biol 74: 503–512 [DOI] [PubMed] [Google Scholar]
- Li L, Moshous D, Zhou Y, Wang J, Xie G, Salido E, Hu D, de Villartay JP, Cowan MJ 2002. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J Immunol 168: 6323–6329 [DOI] [PubMed] [Google Scholar]
- Lieber MR 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Ann Rev Biochem 79: 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J 2010. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 329: 693–696 [DOI] [PubMed] [Google Scholar]
- Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, Sarno S, Meggio F, Pinna LA, Caldecott KW 2004. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 117: 17–28 [DOI] [PubMed] [Google Scholar]
- Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, Bowden G, Kalmyrzaev B, Warren-Perry M, Snape K, et al. 2011. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet 43: 879–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveday C, Turnbull C, Ruark E, Xicola RMM, Ramsay E, Hughes D, Warren-Perry M, Snape K, Eccles D, Evans DG, et al. 2012. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat Genet 44: 475–476 [DOI] [PubMed] [Google Scholar]
- Lupski JR 2007. Genomic rearrangements and sporadic disease. Nat Genet 39: S43–S47 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR 2002. Hairpin opening and overhang processing by an artemis/DNA-dependent protein kinase complex in nonhomologous End joining and V(D)J recombination. Cell 108: 781–794 [DOI] [PubMed] [Google Scholar]
- MacKay C, Déclais A-C, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. 2010. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski F, Ranke M, Schinzel A, Opitz JM 1982. Studies of microcephalic primordial dwarfism II: The osteodysplastic type II of primordial dwarfism. Am J Med Genet 12: 23–35 [DOI] [PubMed] [Google Scholar]
- *.Marechal A, Zou L 2013. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhir J, Selfridge J, Harrison DJ, Squires S, Melton DW 1993. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat Genet 5: 217–223 [DOI] [PubMed] [Google Scholar]
- Meek K, Kienker L, Dallas C, Wang W, Dark MJ, Venta PJ, Huie ML, Hirschhorn R, Bell T 2001. SCID in Jack Russell terriers: A new animal model of DNA-PKcs deficiency. J Immunol 167: 2142–2150 [DOI] [PubMed] [Google Scholar]
- Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP 2007. trans Autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol 27: 3881–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Dang V, Lees-Miller SP 2008. Chapter 2 DNA-PK: The means to justify the ends? In Advances in immunology (ed. Frederick WA), pp. 33–58 Academic, New York; [DOI] [PubMed] [Google Scholar]
- Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, Freund M, Lichtner P, Hartmann L, Schaal H, et al. 2010. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 42: 410–414 [DOI] [PubMed] [Google Scholar]
- Meira LB, Graham JM Jr, Greenberg CR, Busch DB, Doughty AT, Ziffer DW, Coleman DM, Savre-Train I, Friedberg EC 2000. Manitoba aboriginal kindred with original cerebro-oculo- facio-skeletal syndrome has a mutation in the Cockayne syndrome group B (CSB) gene. Am J Hum Genet 66: 1221–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihatsch M, Guda F, Zollinger HU, Heierli C, Thölen H, Reutter FW 1979. Systemic karyomegaly associated with chronic interstitial nephritis. A new disease entity? Clin Nephrol 12: 54–62 [PubMed] [Google Scholar]
- Moldovan G-L, D’Andrea AD 2009. How the Fanconi anemia pathway guards the genome. Ann Rev Genet 43: 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monga G, Banfi G, Salvadore M, Amatruda O, Bozzola C, Mazzucco G 2006. Karyomegalic interstitial nephritis: Report of 3 new cases and review of the literature. Clin Nephrol 65: 349–355 [DOI] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105: 177–186 [DOI] [PubMed] [Google Scholar]
- Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O 2009. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet 41: 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Sasaki K, Mitsutake N, Matsuse M, Shimada M, Nardo T, Takahashi Y, Ohyama K, Ito K, Mishima H, et al. 2012. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet 44: 586–592 [DOI] [PubMed] [Google Scholar]
- Nance MA, Berry SA 1992. Cockayne syndrome: Review of 140 cases. Am J Med Genet 42: 68–84 [DOI] [PubMed] [Google Scholar]
- Nardo T, Oneda R, Spivak G, Vaz B, Mortier L, Thomas P, Orioli D, Laugel V, Stary A, Hanawalt PC, et al. 2009. A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci 106: 6209–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AK, Khurshid M, Desir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A, et al. 2010. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet 42: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Niedernhofer LJ 2013. DNA crosslink repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, et al. 2006. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444: 1038–1043 [DOI] [PubMed] [Google Scholar]
- O’Driscoll M 2008. Haploinsufficiency of DNA damage response genes and their potential influence in human genomic disorders. Curr Genomics 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M 2009a. Life can be stressful without ATR. Nat Genet 41: 866–868 [DOI] [PubMed] [Google Scholar]
- O’Driscoll M 2009b. Mouse models for ATR deficiency. DNA Repair 8: 1333–1337 [DOI] [PubMed] [Google Scholar]
- O’Driscoll M, Jeggo PA 2003. Clinical impact of ATR checkpoint signalling failure in humans. Cell Cycle 2: 194–195 [PubMed] [Google Scholar]
- O’Driscoll M, Jeggo PA 2006. The role of double-strand break repair—Insights from human genetics. Nat Rev Genet 7: 45–54 [DOI] [PubMed] [Google Scholar]
- O’Driscoll M, Cerosaletti KM, Girard P-M, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer SE, Seidel J, et al. 2001. DNA ligase IV mutations identified in patients exhibiting development delay and immunodeficiency. Mol Cell 8: 1175–1185 [DOI] [PubMed] [Google Scholar]
- O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 33: 497–501 [DOI] [PubMed] [Google Scholar]
- O’Driscoll M, Gennery AR, Seidel J, Concannon P, Jeggo PA 2004. An overview of three disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair 3: 1227–1235 [DOI] [PubMed] [Google Scholar]
- Ogi T, Walker S, Stiff T, Hobson E, Limsirichaikul S, Prescott K, Suri M, Byrd PJ, Matsuse M, Mitsutake N, et al. 2012. Identification of the first ATRIP-deficient patient and novel mutations in ATR define a clinical spectrum for ATR-ATRIP Seckel syndrome. PLoS Genet 8: e1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio A, Endt D, Fernández F, Eirich K, de la Hoya M, Schmutzler R, Caldés T, Meindl A, Schindler D, Benitez J 2012. Predominance of pathogenic missense variants in the RAD51C gene occurring in breast and ovarian cancer families. Hum Mol Genet 21: 2889–2898 [DOI] [PubMed] [Google Scholar]
- Outwin E, Carpenter G, Bi W, Withers MA, Lupski JR, O’Driscoll M 2011. Increased RPA1 gene dosage affects genomic stability potentially contributing to 17p13.3 duplication syndrome. PLoS Genet 7: e1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascucci B, Lemma T, Iorio E, Giovannini S, Vaz B, Iavarone I, Calcagnile A, Narciso L, Degan P, Podo F, et al. 2012. An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell 11: 520–529 [DOI] [PubMed] [Google Scholar]
- Peterson SR, Kurimasa A, Oshimura M, Dynan WS, Bradbury EM, Chen DJ 1995. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc Nat Acad Sci 92: 3171–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. 2001. AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature 414: 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418: 99–104 [DOI] [PubMed] [Google Scholar]
- Price VH, Odom RB, Ward WH, Jones FT 1980. Trichothiodystrophy: Sulfur-deficient brittle hair as a marker for a neuroectodermal symptom complex. Arch Dermatol 116: 1375–1384 [DOI] [PubMed] [Google Scholar]
- Qvist P, Huertas P, Jimeno S, Nyegaard M, Hassan MJ, Jackson SP, Børglum AD 2011. CtIP mutations cause Seckel and Jawad syndromes. PLoS Genet 7: e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffan E, Hurst LA, Al Turki S, Carpenter G, Scott C, Daly A, Coffey A, Bhaskar S, Howard E, Khan N, et al. 2011. Early diagnosis of Werner’s syndrome using exome-wide sequencing in a single, atypical patient. Front Endocrinol 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, Ekici AB, van Essen AJ, Goecke TO, Al-Gazali L, Chrzanowska KH, et al. 2008. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science 319: 816–819 [DOI] [PubMed] [Google Scholar]
- Raymond FL, Whittaker J, Jenkins L, Lench N, Chitty LS 2010. Molecular prenatal diagnosis: The impact of modern technologies. Prenatal Diag 30: 674–681 [DOI] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al. 2007. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet 39: 162–164 [DOI] [PubMed] [Google Scholar]
- Resnick IB, Shapira MY, Slavin S 2005. Nonmyeloablative stem cell transplantation and cell therapy for malignant and non-malignant diseases. Transpl Immunol 14: 207–219 [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Lagelouse R, Gennery A, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102: 565–575 [DOI] [PubMed] [Google Scholar]
- Reynolds JJ, Walker AK, Gilmore EC, Walsh CA, Caldecott KW 2012. Impact of PNKP mutations associated with microcephaly, seizures and developmental delay on enzyme activity and DNA strand break repair. Nucleic Acids Res 40: 6608–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty AJ, Smith GCM, Recio M-J, Reis C, Dahm K, Fricke A, Krempler A, et al. 2004. A pathway of double strand break rejoining dependent upon ATM, Artemis, and proteins locating to g-H2AX foci. Mol Cell 16: 715–724 [DOI] [PubMed] [Google Scholar]
- Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA 2009. XLF-Cernunnos promotes DNA ligase IV–XRCC4 re-adenylation following ligation. Nucleic Acids Res 37: 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ 2011. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol 18: 1432–1434 [DOI] [PubMed] [Google Scholar]
- Rosenberg PS, Greene MH, Alter BP 2003. Cancer incidence in persons with Fanconi anemia. Blood 101: 822–826 [DOI] [PubMed] [Google Scholar]
- *.Rothstein R 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Scharer O 2013. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler W, Ruetsch NR, Amsler M, Bosma MJ 1991. Coding joint formation of endogenous T cell receptor genes in lymphoid cells from scid mice: Unusual P-nucleotide additions in VJ-coding joints. Eur J Immunol 21: 589–596 [DOI] [PubMed] [Google Scholar]
- Schwertman P, Lagarou A, Dekkers DHW, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JHJ, Demmers JAA, Fousteri M, Vermeulen W, et al. 2012. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet 44: 598–602 [DOI] [PubMed] [Google Scholar]
- Seckel HPG 1960. Bird-headed dwarfs: Studies in developmental anthropology including human proportions. In Bird-headed dwarfs: Studies in developmental anthropology including human proportions (ed. Thomas CT). S. Karger, Basel [Google Scholar]
- Shamseldin HE, Elfaki M, Alkuraya FS 2012. Exome sequencing reveals a novel Fanconi group defined by XRCC2 mutation. J Med Genet 49: 184–186 [DOI] [PubMed] [Google Scholar]
- Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, et al. 2010. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet 42: 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC 2007. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet 39: 93–98 [DOI] [PubMed] [Google Scholar]
- Shimamura A, Alter BP 2010. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev 24: 101–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei Y-F, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, et al. 1996. Xerderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86: 811–822 [DOI] [PubMed] [Google Scholar]
- Sir J-H, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D’Santos C, Woods CG, Gergely F 2011. A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet 43: 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sirbu BM, Cortez D 2013. DNA damage response: Three levels of DNA repair regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GC, Jackson SP 1999. The DNA-dependent protein kinase. Genes Dev 13: 916–934 [DOI] [PubMed] [Google Scholar]
- Smith E, Dejsuphong D, Balestrini A, Hampel M, Lenz C, Takeda S, Vindigni A, Costanzo V 2009. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat Cell Biol 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiácovo MP, et al. 2010. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 39: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak G 2005. UV-sensitive syndrome. Mutat Res-Fund Mol M 577: 162–169 [DOI] [PubMed] [Google Scholar]
- Spivak G, Hanawalt PC 2006. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair 5: 13–22 [DOI] [PubMed] [Google Scholar]
- Spoendlin M, Moch H, Brunner F, Brunner W, Burger H-R, Kiss D, Wegmann W, Dalquen P, Oberholzer M, Thiel G, et al. 1995. Karyomegalic interstitial nephritis: Further support for a distinct entity and evidence for a genetic defect. Am J Kidney Dis 25: 242–252 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR 2010. Structural variation in the human genome and its role in disease. Ann Rev Med 61: 437–455 [DOI] [PubMed] [Google Scholar]
- Stefanini M, Lagomarsini P, Arlett CF, Marinoni S, Borrone C, Crovato F, Trevisan G, Cordone G, Nuzzo F 1986. Xeroderma pigmentosum (complementation group D) mutation is present in patients affected by trichothiodystrophy with photosensitivity. Hum Genet 74: 107–112 [DOI] [PubMed] [Google Scholar]
- Stewart GS 2009. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle 8: 1532–1538 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Stankovic T, Byrd PJ, Wechsler T, Miller ES, Huissoon A, Drayson MT, West SC, Elledge SJ, Taylor AMR 2007. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci 104: 16910–16915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AWM, et al. 2011. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet 43: 138–141 [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Harper JW 2010. GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution. Genes Dev 24: 521–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahbaz N, Subedi S, Weinfeld M 2012. Role of polynucleotide kinase/phosphatase in mitochondrial DNA repair. Nucleic Acids Res 40: 3484–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H, Boerkoel CF, John J, Saifi GM, Salih MAM, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, et al. 2002. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet 32: 267–272 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Weinel S, Nagy N, O’Driscoll M, Lai-Cheong Joey E, Kulp-Shorten Carol L, Knable A, Carpenter G, Fisher Sheila A, Hiragun M, et al. 2012. Germline mutation in ATR in autosomal-dominant oropharyngeal cancer syndrome. Am J Hum Genet 90: 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AMR, Groom A, Byrd PJ 2004. Ataxia-telangiectasia-like disorder (ATLD)—Its clinical presentation and molecular basis. DNA Repair 3: 1219–1225 [DOI] [PubMed] [Google Scholar]
- Tian M, Shinkura R, Shinkura N, Alt FW 2004. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol Cell Biol 24: 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibelius A, Marhold J, Zentgraf H, Heilig CE, Neitzel H, Ducommun B, Rauch A, Ho AD, Bartek J, Krämer A 2009. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J Cell Biol 185: 1149–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Fernandez-Capetillo O 2011a. Targeting ATR and Chk1 kinases for cancer treatment: A new model for new (and old) drugs. Mol Oncol 5: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O 2011b. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol 18: 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn M, Bell SM, Felix C, Rashid Y, Jafri H, Griffiths PD, Neumann LM, Krebs A, Reis A, Sperling K, et al. 2004. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet 75: 261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo JP, Mina LB, Pujol R, Bogliolo M, Andrieux J, Holder M, Schuster B, Schindler D, Surrallés J 2012. On the role of FAN1 in Fanconi anemia. Blood 120: 86–89 [DOI] [PubMed] [Google Scholar]
- van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, Mari P-O, Tezcan I, Chen DJ, Zdzienicka MZ, et al. 2009a. A DNA-PKcs mutation in a radiosensitive T−B− SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest 119: 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg M, van Dongen JJ, van Gent DC 2009b. DNA-PKcs deficiency in human: Long predicted, finally found. Curr Opin Allergy Clin Immunol 9: 503–509 [DOI] [PubMed] [Google Scholar]
- van der Pluijm I, Garinis GA, Brandt RMC, Gorgels TGMF, Wijnhoven SW, Diderich KEM, de Wit J, Mitchell JR, van Oostrom C, Beems R, et al. 2006. Impaired genome maintenance suppresses the growth hormone-insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol 5: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, et al. 2010. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet 42: 406–409 [DOI] [PubMed] [Google Scholar]
- Vermeulen W, Bergmann E, Auriol J, Rademakers S, Frit P, Appeldoorn E, Hoeijmakers JH, Egly JM 2000. Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat Genet 26: 307–313 [DOI] [PubMed] [Google Scholar]
- Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, Liu F, Bestvater F, Raab MS, Zentgraf H, Izraeli S, et al. 2012. STIL is required for centriole duplication in human cells. J Cell Sci 125: 1353–1362 [DOI] [PubMed] [Google Scholar]
- Waldbaum S, Patel M 2010. Mitochondria, oxidative stress, and temporal lobe epilepsy. Epilepsy Res 88: 23–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, et al. 2009. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet 84: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao H, You C, Yuan B, Bahde R, Gupta S, Nishigori C, Niedernhofer LJ, Brooks PJ, Wang Y 2012a. Endogenous formation and repair of oxidatively induced G[8-5 m]T intrastrand cross-link lesion. Nucleic Acids Res 40: 7368–7374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Clauson CL, Robbins PD, Niedernhofer LJ, Wang Y 2012b. The oxidative DNA lesions 8,5′-cyclopurines accumulate with aging in a tissue-specific manner. Aging Cell 11: 714–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JNM 2011. Tidying up loose ends: The role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci 36: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiler R, Leber R, Moore BB, Vandyk LF, Perryman LE, Meek K 1995. Equine severe combined immunodeficiency—A defect in V(D)J recombination and DNA-dependent protein-kinase activity. Proc Natl Acad Sci 92: 11485–11489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. 2007. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 39: 159–161 [DOI] [PubMed] [Google Scholar]
- Xu Y, Price BD 2011. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle 10: 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong SL, Cleaver JE, Tullis GD, Johnston MM 1984. Is trichothiodystrophy part of the xeroderma pigmentosum spectrum? Clin Genet 36: 82S [Google Scholar]
- Yoshikiyo K, Kratz K, Hirota K, Nishihara K, Takata M, Kurumizaka H, Horimoto S, Takeda S, Jiricny J 2010. KIAA1018/FAN1 nuclease protects cells against genomic instability induced by interstrand cross-linking agents. Proc Natl Acad Sci 107: 21553–21557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, Sergi CM, Topcu M, McDonald MT, Barry BJ, Felie JM, et al. 2010. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet 42: 1015–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelle B, Berends F, Lohman PHM 1980. Repair of damage by ultraviolet radiation in xeroderma pigmentosum cell strains of complementation groups E and F. Mutation Res 73: 157–169 [DOI] [PubMed] [Google Scholar]
- Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S, Tahara H, Neilan EG, Honma M, Nohmi T, et al. 2012. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet 44: 593–597 [DOI] [PubMed] [Google Scholar]
- Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, et al. 2012. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet 44: 910–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanou A, Stewart GS 2010. A PIAS-ed view of DNA double strand break repair focuses on SUMO. DNA Repair 9: 588–592 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]