Abstract
Protein misfolding is a common cellular event that can produce intrinsically harmful products. To reduce the risk, quality control mechanisms are deployed to detect and eliminate misfolded, aggregated, and unassembled proteins. In the secretory pathway, it is mainly the endoplasmic reticulum-associated degradation (ERAD) pathways that perform this role. Here, specialized factors are organized to monitor and process the folded states of nascent polypeptides. Despite the complex structures, topologies, and posttranslational modifications of client molecules, the ER mechanisms are the best understood among all protein quality-control systems. This is the result of convergent and sometimes serendipitous discoveries by researchers from diverse fields. Although major advances in ER quality control and ERAD came from all model organisms, this review will focus on the discoveries culminating from the simple budding yeast.
In budding yeast, two main ER membrane-embedded complexes (Hrd1 and Doa10) recognize, translocate, and ubiquitinate misfolded proteins for degradation. Many other factors support this process.
Secreted and membrane proteins insert into the endoplasmic reticulum (ER) through the Sec61 translocon complex as unfolded molecules (Rapoport 2007). In the lumen, protein folding is assisted by chaperones and modifying enzymes, some of which are members of ER quality control (ERQC) pathways. Chaperone interactions keep immature proteins soluble to prevent aggregation and facilitate folding. Once folding and assembly are complete, proteins are sorted for transport to their final destinations. Proteins failing to fold correctly are retained and targeted for degradation by the ER-associated protein degradation (ERAD) pathways of ERQC (McCracken et al. 1996). This review will focus on mechanisms that differentiate folding intermediates, native proteins, and misfolded proteins in the ER and keep them on the correct processing track.
The discovery of protein quality control mechanisms in the ER originated from mammalian virus studies. The hemagglutinin (HA) and G glycoproteins of influenza and vesicular stomatitis viruses, respectively, fold and oligomerize in the ER (Copeland et al. 1986; Gething et al. 1986; Doms et al. 1988). Because these viruses bud from the plasma membrane, researchers observed that unfolded and misfolded molecules were stringently retained in the ER (Gething et al. 1986; Copeland et al. 1988; Doms et al. 1988). Together, these pioneering studies conceptualized the existence of an active protein quality control mechanism. Parallel studies showing that unassembled subunits of the T-cell receptor (TCR) complex rapidly degrades, suggested an ER-based mechanism to dispose of potentially harmful molecules (Lippincott-Schwartz et al. 1988; Bonifacino et al. 1989). Despite these monumental advances, the molecular mechanisms of ERQC remained elusive for years.
Using yeast as a model system for cell biology, investigators taking divergent paths serendipitously converged onto the nascent field of ERQC. Enormous strides were made in these early studies. The Wolf laboratory, focusing on the biogenesis of the vacuole (yeast lysosome), discovered that defective variants of its resident enzymes carboxypeptidase Y and proteinase A (CPY* and PrA*, respectively) never make it to the organelle but instead turn over in a pre-Golgi compartment (Finger et al. 1993). What degrades the misfolded enzymes, however, was unclear. A key genetic interaction discovered by Sommer and Jentsch provided a hint. They found that a UBC6 loss-of-function mutation suppressed the temperature-sensitive phenotype of sec61-2. This suggested that the functionally disruptive mutation in the translocon subunit also causes its degradation (Sommer and Jentsch 1993). More intriguingly, the finding linked the ubiquitin-proteasome system (UPS) to ERQC for the first time. The genetic link was confirmed functionally in follow-up studies and by the discovery that degradation of misfolded cystic fibrosis transconductance regulator (CFTR) protein was blocked by proteasome inhibitors (Ward et al. 1995; Biederer et al. 1996). The Hampton group cemented the generality of the system in ER protein degradation when they showed that the UPS regulates levels of HMG-CoA reductase (Hmg2) according to physiological need (Hampton and Rine 1994; Hampton and Bhakta 1997). Taking the biochemical route, the Brodsky and McCracken groups fractionated yeast cells and established the first in vitro system to study ER protein degradation (McCracken and Brodsky 1996; Werner et al. 1996; Brodsky et al. 1999). It was they who coined the now ubiquitous term, ER-associated degradation or ERAD.
EARLY STEPS OF ERQC: ARREST AND RELEASE
The translocation and folding of secretory and membrane proteins are assisted by ER resident chaperones and their cofactors. These include Hsp70 family members Kar2 and Lhs1, cochaperones of the DnaJ class (Jem1, Scj1), the nucleotide exchange factor Sil1, the Rot1 membrane-bound chaperone, the lectin-like Cne1, and thiol oxidoreductases (Eps1, Eug1, Mpd1, Mpd2, and Pdi1) (Tachibana and Stevens 1992; Brodsky et al. 1995; Matlack et al. 1997; Plemper et al. 1997; Silberstein et al. 1998; Gillece et al. 1999; Tyson and Stirling 2000; Nishikawa et al. 2001; Zhang et al. 2001; Tsai and Rapoport 2002; Wang and Chang 2003; Heiligenstein et al. 2006; Takeuchi et al. 2008; Hosoda et al. 2009; Sakoh-Nakatogawa et al. 2009; Vembar et al. 2010; Grubb et al. 2012). The fidelity of the selection process is critical because accumulation of aberrant protein conformers is the basis of numerous human diseases. For example, a single point mutation in the gene encoding α1-antitrypsin causes a severe deficiency resulting in bronchiectasis and pulmonary emphysema. Instead of being properly secreted from cells, the mutant protein is retained in the ER of hepatocytes (Hercz et al. 1978). Familial alleles of the gene encoding CFTR cause their retention and turnover in the ER (Cheng et al. 1990).
How ERQC sorts unfolded from folded proteins is not entirely clear. It could achieve this by actively retaining proteins meant to stay in the ER (unfolded proteins, misfolded proteins, ER resident proteins). All other proteins are simply exported by default. This was the basis of the “bulk flow” paradigm originally proposed by Rothman and coworkers (Wieland et al. 1987). The discovery of ER retention signals and receptors for resident proteins provided the basis of the proposal. The paradigm can be extended to ERQC by putting the job of retention in the hands of ER chaperones, whose very nature it is to associate with unfolded polypeptides. This simple model is confounded by the discovery of a large family of cargo sorting factors, including Erv26 and Erv29, which recognize export signals embedded in folded proteins and concentrates them at ER export sites (Dancourt and Barlowe 2010). Thus, in principle, an ERQC sorting mechanism could be through the formation of conformational export signals that form only when proteins are properly folded. Molecules failing to form export signals would be retained by default. This mode of sorting, if in operation, is insufficiently comprehensive for ERQC because some misfolded proteins can be exported out of the ER through COPII vesicles (Vashist et al. 2001; Wang and Ng 2010) using export signals that remain functional in the misfolded proteins (Kawaguchi et al. 2010). One purpose of the apparent “leakiness” is to provide a safety valve for conditions of stress, thus allowing other degradative pathways to assist when ERAD is saturated (Spear and Ng 2003). Otherwise, the inability to degrade aberrant proteins can lead to the increase of reactive oxygen species and cell death (Haynes et al. 2004). Taken together, a sorting mechanism that combines both retention and export is likely used by ERQC to provide the needed stringency at steady state and also allow some flexibility to handle increased substrate loads as conditions warrant.
Any sorting mechanism for ERQC is complicated further by the need to distinguish folding intermediates from misfolded proteins. Folding intermediates must stay in the ER because their maturation depends on resident chaperones and modifying enzymes. Misfolded proteins, on the other hand, must be captured and degraded. Some misfolded proteins form insoluble aggregates that are cleared by autophagy (Kruse et al. 2006; Ishida and Nagata 2009; Ishida et al. 2009). How these aggregates are recognized is unclear. Other ER-retained misfolded proteins remain soluble through their chaperone interactions (Nishikawa et al. 2001; Kabani et al. 2003). In some cases, these proteins can traffic out of the ER before they are retrieved from the Golgi for ERAD (Vashist et al. 2001; Kincaid and Cooper 2007; Hirayama et al. 2010). How cells differentiate this class from folding intermediates has puzzled the field for years. Unlike misfolded aggregates, there was no obvious physical difference between the two forms. How does a cell decide that a protein is misfolded and should be degraded versus one that is just in the process of folding?
JUDGE AND JURY: E3 UBIQUITIN LIGASES ORGANIZE SITES OF ERAD SUBSTRATE PROCESSING
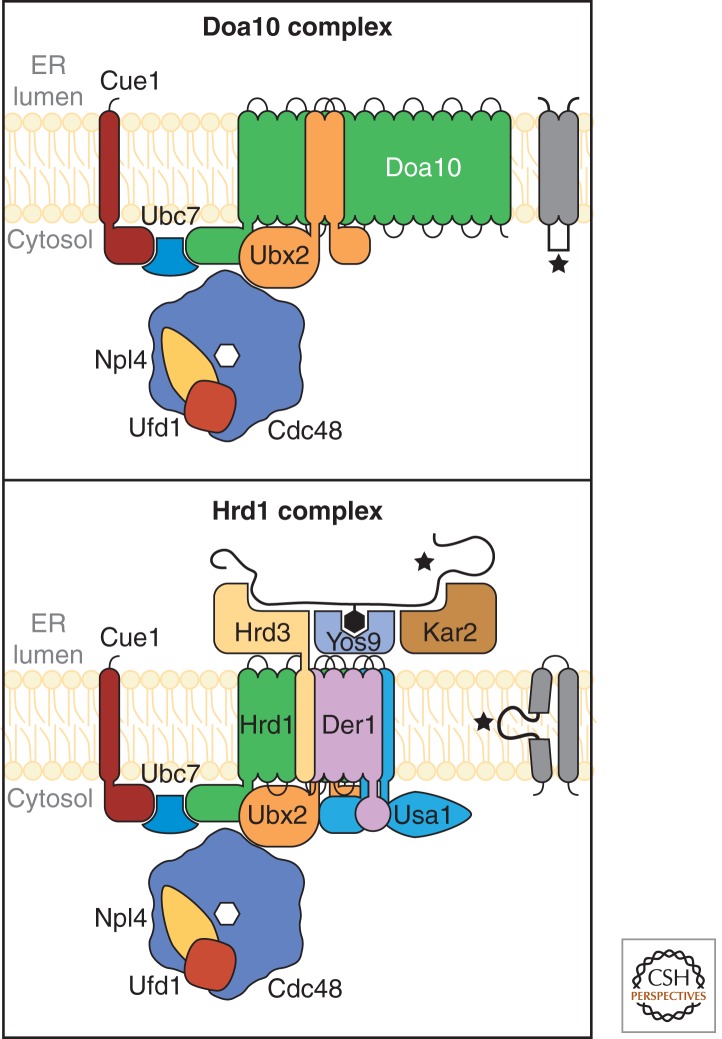
In budding yeast, the efforts of numerous groups contributed to the characterization of two ERAD complexes embedded in the ER membrane that function to recognize, translocate, and ubiquitinate substrates for degradation. These can be described as the Hrd1 and Doa10 complexes, named for the E3 ubiquitin ligases that differentiate them (Fig. 1). Hrd1 (also known as Der3) was identified from mutants unable to degrade Hmg2 and CPY* (Hampton et al. 1996; Hiller et al. 1996; Knop et al. 1996a; Hampton and Bhakta 1997; Bordallo et al. 1998; Cronin et al. 2000; Bays et al. 2001b). Doa10 was discovered from a genetic screen seeking genes required for the turnover of proteins bearing the Deg1 degradation signal (Swanson et al. 2001). Doa10 is dispensable for CPY* degradation, which is soluble, but required for the degradation of some normal and aberrant ER membrane proteins (Swanson et al. 2001; Huyer et al. 2004; Vashist and Ng 2004). A variety of other factors were identified for ERAD including Cue1, Ubc7, Ubc6, Der1, Hrd3, Cdc48, Npl4, Ufd1, Yos9, Htm1/Mnl1, and Ubx2 (Sommer and Jentsch 1993; Biederer et al. 1996, 1997; Hiller et al. 1996; Hampton and Bhakta 1997; Bordallo et al. 1998; Hill and Cooper 2000; Wilhovsky et al. 2000; Bays et al. 2001b; Hitchcock et al. 2001; Jakob et al. 2001; Nakatsukasa et al. 2001; Rape et al. 2001; Swanson et al. 2001; Walter et al. 2001; Ye et al. 2001; Jarosch et al. 2002; Rabinovich et al. 2002; Buschhorn et al. 2004; Bhamidipati et al. 2005; Kim et al. 2005; Neuber et al. 2005; Schuberth and Buchberger 2005). Biochemical analyses showed that Cdc48 forms a complex with Npl4 and Ufd1, Hrd3 with Hrd1, and Cue1 with Ubc7 (Biederer et al. 1997; Plemper et al. 1999; Gardner et al. 2000; Hoppe et al. 2000; Meyer et al. 2000). Genetic analysis showed that some genes including DER1, YOS9, HTM1/MNL1, HRD1, and DOA10 were needed for only subsets of substrates, whereas other genes like CUE1, UBC7, and CDC48 were more broadly required (Bordallo et al. 1998; Plemper et al. 1998; Swanson et al. 2001; Walter et al. 2001; Zhang et al. 2001; Taxis et al. 2003; Huyer et al. 2004; Ravid et al. 2006). A pattern emerged that some genes are specific for substrates containing luminal lesions (ERAD-L), for cytosolic lesions (ERAD-C), or required for all substrates (Taxis et al. 2003; Huyer et al. 2004; Vashist and Ng 2004; Willer et al. 2008). The significance of the pattern became clear through direct biochemical characterization of ERAD complexes.
Figure 1.
Schematic representation of Hrd1 and Doa10 complexes. Individual subunits of each complex are depicted with their known partners. (Top) The Doa10 complex monitors the folding state of cytosolic domains of membrane proteins (ERAD-C). (Bottom) The Hrd1 complex recognizes lesions of luminal domains of membrane and soluble proteins (ERAD-L) and of lesions within transmembrane domains (ERAD-M).
Using tandem affinity purification (TAP), Carvalho and Rapoport showed the existence of two distinct complexes (Carvalho et al. 2006). The surprise was not in the differences but in their similarities. Doa10 forms a complex with Cue1-Ubc7 and with Cdc48-Npl4-Ufd1-Ubx2 (Fig. 1, top). Hrd1 is associated with the same factors plus Hrd3, but also with Der1, Yos9, and a new factor Usa1 (Fig. 1, bottom). Hrd3 contains a large amino-terminal luminal domain and a carboxy-terminal transmembrane domain that interacts with Hrd1. The multispanning membrane protein Der1 might participate in the transfer of misfolded proteins from the ER to the cytosol (Knop et al. 1996a). The derlin-like protein Dfm1 is proposed to be involved in ERAD-C and -L degradation by interacting with Cdc48 (Schuberth and Buchberger 2005; Sato and Hampton 2006; Goder et al. 2008; Stolz et al. 2010). Usa1p links Der1 to Hrd1 (Carvalho et al. 2006). Usa1 contains two large amino- and carboxy-terminal cytosolic domains joined by two transmembrane segments. Both domains are important for ERAD activity and its interaction with the Hrd1p complex (Kim et al. 2009; Carroll and Hampton 2010). The N and C domains induce oligomerization of the Hrd1 complex and binds Der1, respectively (Horn et al. 2009). In this way, Usa1 functions as the major structural scaffold for the Hrd1 complex.
The organization of individual components of the Hrd1 and Doa10 ERAD complexes helped explain the genetic patterns observed previously for substrate specificity. An extended analysis showed that the Hrd1 complex is also required for membrane protein substrates bearing lesions within transmembrane segments (ERAD-M) (Carvalho et al. 2006). These advances provided a clear structural framework that was used to guide future studies.
THE VERDICT: SUBSTRATE SELECTION FOR DEGRADATION
Misfolded proteins can expose hydrophobic patches that would be buried in the native state. In the ER lumen, the molecular chaperones Kar2, Scj1, Jem1, and Pdi1 bind hydrophobic peptides and are required for ERAD (Silberstein et al. 1998; Gillece et al. 1999; Nishikawa et al. 2001; Thibault et al. 2011; Grubb et al. 2012). On the cytosolic side, Hsp26, Hsp42, Hsp70, Ydj1, and Hlj1 participate in ERAD depending on the substrate (Huyer et al. 2004; Youker et al. 2004; Ahner et al. 2007; Park et al. 2007; Vembar et al. 2009). Because most chaperones dissociate from proteins after folding, a prolonged interaction might seem to provide a simple mechanism for substrate selection. It is insufficient, however, because several misfolded proteins are known to accumulate stably in the ER (Knop et al. 1996b; Loayza et al. 1998; Kostova and Wolf 2005; Spear and Ng 2005; Kruse et al. 2006). For this reason, additional biochemical signatures or signals were proposed to be necessary for ERAD recognition.
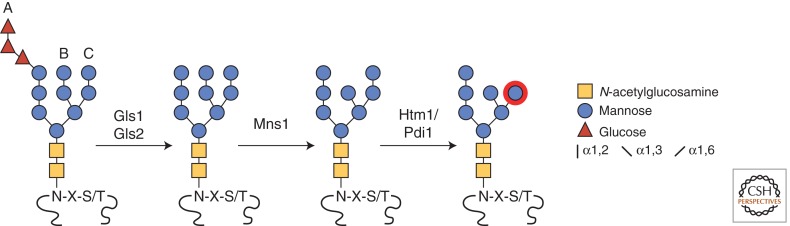
The best-characterized ERAD determinant exploits the structure of an N-linked glycan (Knop et al. 1996b). When the consensus sequence Asn-X-Ser/Thr (in which X is any amino acid other than proline) is encountered by oligosaccharyl transferase, the glycan is transferred en bloc from the dolichyl oligosaccharide Glc3-Man9-GlcNAc2-p-p-Dol substrate to the asparagine side chain (Fig. 2) (Burda and Aebi 1999). During folding cycle, glucosidase I and II (Gls1 and Gls2) sequentially removes three glucose residues leaving Man9-GlcNAc2. ER mannosidase I (Mns1) next trims the most distal mannose of branch B to generate Man8-GlcNAc2. Inhibition of any step impairs glycoprotein ERAD (Jakob et al. 1998; Hitt and Wolf 2004; Clerc et al. 2009). Together, these processing steps are not rapid so they provide a time window for the glycoprotein to fold. Should it remain unfolded, the Htm1 (also called Mnl1) mannosidase complexed with protein disulfide isomerase (PDI), mediates the next crucial step (Clerc et al. 2009; Sakoh-Nakatogawa et al. 2009). In vitro reconstitution experiments showed that the enzyme complex prefers unfolded polypeptides bearing the Man8-GlcNAc2 glycan. For these proteins, Htm1-PDI specifically cleaves the terminal mannose residue from the C branch (Fig. 2) (Gauss et al. 2011). This exposes a terminal α1,6-linked mannose, which is the ligand for the ERAD substrate receptor Yos9 (Buschhorn et al. 2004; Bhamidipati et al. 2005; Kim et al. 2005; Szathmary et al. 2005; Quan et al. 2008).
Figure 2.
Targeting glycoproteins for degradation. The core GlcNAc2-Man9-Glc3 glycan is rapidly added to the side-chain nitrogen of Asn (N) residues part of the consensus sequence Asn-X-Ser/Thr (N-X-S/T). This posttranslational modification occurs as soon as polypeptides enter the endoplasmic reticulum through the translocon. Subsequently, the three glucoses of branch A are trimmed consecutively by the glucosidase I (Gls1) and glucosidase II (Gls2) to generate GlcNAc2-Man9. Mannosidase I (Mns1) cleaves the α1,2-linked mannose of branch B to produce GlcNAc2-Man8. At this stage, folded and glycosylated proteins may leave the ER. However, glycosylated proteins failing to fold are recognized by Htm1/Pdi1, which cleaves the α1,2-linked mannose of branch C to yield the terminal α1,6-linked mannose residue as the Yos9 ligand (red circle).
The Man7-GlcNAc2 glycan structure alone is insufficient to target a substrate to ERAD (Xie et al. 2009). It was recognized that only specifically positioned substrate glycans can signal ERAD (Kostova and Wolf 2005; Spear and Ng 2005). It was later shown that the signal is bipartite, consisting of the Man7-GlcNAc2 glycan attached to a disordered segment (Xie et al. 2009). In this way, the positioned glycan functions as an intrinsic sensor for protein folding. If the protein folds by the time the signal glycan is processed to Man8-GlcNAc2, it escapes Htm1-PDI processing, leaving it free to exit the ER. Through this “glycan timer” mechanism, nascent polypeptides exceeding their set folding periods are degraded by ERAD.
A second type of ERAD glycan modification is more enigmatic. It was reported by several groups that some substrates are modified by O-mannosylation (Harty et al. 2001; Vashist et al. 2001; Nakatsukasa et al. 2004; Hirayama et al. 2008). During O-mannosylation, single mannose sugars are transferred covalently to serine and threonine residues, whose total numbers and distributions are currently unknown. The reaction is specifically performed by the Pmt1 and Pmt2 protein mannosyltransferases, which form a complex (Strahl-Bolsinger et al. 1999). The modification is required for efficient degradation of substrates, possibly by maintaining them in a more soluble form (Harty et al. 2001; Hirayama et al. 2008). Interestingly, the PMT1 and PMT2 genes are targets of the unfolded protein response, consistent with a stress-related function (Travers et al. 2000). How misfolded proteins are specifically targeted for O-mannosylation is unknown. However, a variety of ER factors including the Hrd1 complex, p24 family proteins, PDI, and Ero1 are associated with the Pmt1/Pmt2 complex suggesting possible mechanisms (Goder and Melero 2011). Furthermore, because ERAD is only nominally compromised when substrate O-mannosylation is reduced, a yet to be determined role in ERQC may be awaiting discovery.
The Hrd1 complex also mediates degradation of membrane proteins bearing defects in their transmembrane segments. Early insight into this mechanism was revealed by studies using Hmg2, a normal transmembrane protein. Hmg2 is the rate-limiting enzyme in sterol biosynthesis, whose levels are regulated by lipid signals within the membrane. When lipid levels in the pathway are high, Hmg2 is degraded by the Hrd1 pathway (Hampton et al. 1996; Gardner et al. 1998, 2001; Gardner and Hampton 1999a,b; Federovitch et al. 2008; Garza et al. 2009). Among misfolded proteins, the Hrd1 complex can also recognize aberrations within transmembrane domains (Carvalho et al. 2006). This mode has been termed ERAD-M. Unlike ERAD-L, whose substrates engage luminal components like Kar2 and Htm1-PDI first, it is the Hrd1 protein itself that recognizes transmembrane distortions (Sato et al. 2009). Some components like Htm1-PDI and Yos9 that are critical for ERAD-L are entirely dispensable for ERAD-M even as both pathways converge in the same Hrd1 complex (Kanehara et al. 2010). In the regulation of sterol biosynthesis, Hmg2 seems to have adapted principles of ERAD substrate recognition for its degradation. Hmg2 contains a conserved sterol-sensing domain (SSD) that detects pathway intermediates (Theesfeld et al. 2011). The SSD mediates a conformational change recognized by the Hrd1 complex that leads to Hmg2 ubiquitination and degradation (Shearer and Hampton 2004, 2005).
The Doa10 complex (Fig. 1, top) is mostly dedicated to substrates found on the cytosolic side of the ER membrane even though it can serve as a complement or backup to the Hrd1 complex (Gnann et al. 2004; Vashist and Ng 2004). Interestingly, not only is Doa10 required for the degradation of misfolded membrane proteins, it also serves as the E3 enzyme for degradation of some misfolded cytosolic proteins (Metzger et al. 2008; Lewis and Pelham 2009). For transmembrane proteins, Doa10 recognizes misfolded cytosolic domains (Huyer et al. 2004; Vashist and Ng 2004; Nakatsukasa et al. 2008). An in vitro ERAD system revealed that a substrate precomplex formed with Hsp70/Hsp40 mediates recognition and ubiquitination by Doa10 (Nakatsukasa et al. 2008). Examples of the requirement for cytosolic chaperones for transmembrane protein ERAD is growing and it is becoming clear that like luminal ERAD components, the requirements vary depending on the substrate (Ahner et al. 2007; Hrizo et al. 2007; Buck et al. 2010; Bell et al. 2011; Needham et al. 2011).
DEAD MAN WALKING: SUBSTRATE RETROTRANSLOCATION
Proteins enter the ER through the Sec61 translocon complex (Walter and Johnson 1994). Because the UPS is on the cytosolic/nucleoplasmic side of the ER membrane, misfolded proteins must retrotranslocate from the membrane (Fig. 3A). The mechanism for this event is unclear. In principle, there could exist multiple mechanisms to account for substrate diversity. For soluble substrates, a pore-like conduit seems to be a requirement. For these molecules, this process is likely more complex than import because substrates can be oligomeric, be modified by disulfide bonds, contain N-linked or O-linked glycans, or any combination of these. Given the small size of the Sec61 pore, it would be inadequate for many proteins to use it also as the retrotranslocon (Van den Berg et al. 2004). For transmembrane proteins containing cytosolic domains, a direct “pulling” mechanism could work in the absence of a pore, but the high energy cost might preclude the possibility. Alternatively, passage of transmembrane domains into a pore-like complex as an early step of retrotranslocation is possible. This would simply be the reverse of the forward membrane integration mechanism (Van den Berg et al. 2004; Ismail et al. 2006).
Figure 3.
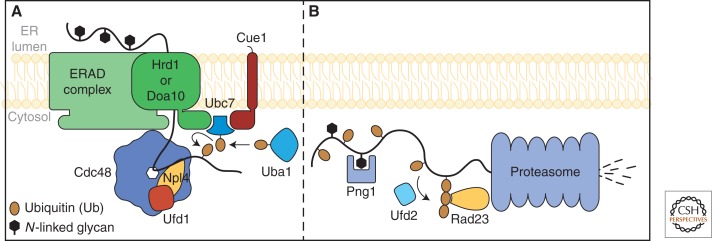
Retrotranslocation and degradation of ERAD substrates. (A) Proteins targeted for degradation by ERAD complex are retrotranslocated through a “retrotranlocon” (Hrd1, Doa10, and Sec61 are proposed to serve this role but only the E3s are shown for simplicity) with the mechanical force provided by the Cdc48-Npl4-Ufd1 AAA-ATPase complex. Polypeptides are subsequently ubiquitinated as depicted. (B) In the cytosol, Png1 removes N-linked glycans from substrates. Ufd2 can lengthen ubiquitin chains for some substrates. Polyubiquitinated species are recognized by Rad23p and passed to the proteasome for degradation.
Some steps of substrate retrotranslocation are resolved. Substrate recognition and targeting are usually coupled events of translocation mechanisms. This seems to be also true of ERAD. The ER chaperones Kar2, Jem1, and Scj1 are not just early engagers but also keep luminal ERAD substrates soluble, a state needed for retrotranslocation (Nishikawa et al. 2001). As discussed above, the Htm1-PDI complex is a key substrate recognition factor. The Htm1 mannosidase reaction generates the targeting signal recognized by Hrd3 and Yos9 proteins of the Hrd1 complex. Despite their seemingly glycan-specialized function, Hrd3 and Yos9 also bind nonglycosylated misfolded proteins and the core Hrd1 complex mediates the glycan-independent ERAD-L pathway (Bhamidipati et al. 2005; Gauss et al. 2006a,b; Kanehara et al. 2010). Thus, recent evidence reveals that Yos9 can improve the degradation efficiency of some nonglycosylated substrates (Benitez et al. 2011; Jaenicke et al. 2011). Furthermore, recent studies show that the Hrd1 and Yos9 play important roles in ER retention of misfolded proteins (Izawa et al. 2012). Along similar lines, PDI extends its functional versatility to ERAD. It is also required to target the nonglycosylated substrate Δgpαf to the export channel, to chaperone apolipoprotein B for ERAD, and provide redox activity required for CPY* turnover (Gillece et al. 1999; Grubb et al. 2012).
Polyubiquitination is not just a degradation signal for ERAD; it is also a critical step in retrotranslocation. Even in the earliest studies, it was observed that mutants defective in ubiquitination accumulate substrates in the ER (Hiller et al. 1996; Biederer et al. 1997). Soluble substrates can begin the export process but regress to the lumen should ubiquitination fail (Elkabetz et al. 2004). For transmembrane proteins, polyubiquitination is required for their extraction from ER membranes in vitro (Nakatsukasa et al. 2008). The polyubiquitin chain likely acts as a recognition signal for the p97 complex (homolog of yeast Cdc48p complex). The AAA+ ATPase protein Cdc48 forms a heterotrimeric complex with the cytosolic proteins Ufd1 and Npl4 and is required for ERAD (Meyer et al. 2000; Bays et al. 2001b; Braun et al. 2002; Jarosch et al. 2002; Rabinovich et al. 2002). Ubiquitinated substrates are retotranslocated to the cytosol by the action of the Cdc48 complex and targeted to the proteasome for degradation (Ye et al. 2001; Braun et al. 2002). The data suggest that p97 provides the mechanical force to move the polypeptide into the cytosol through ATP hydrolysis cycles (Ye et al. 2003). However, some substrates can retrotranslocate independently of the Cdc48 complex. Here, the mechanical force for extraction is likely provided by the proteasome 19S subunit (Russell et al. 1999; Ng et al. 2007).
The Cdc48 complex is anchored to the ER membrane through Ubx2. Ubx2 connects Cdc48 to the Doa10 complex by binding both (Neuber et al. 2005; Schuberth and Buchberger 2005; Wilson et al. 2006). The Ubx-domain protein Ubx4 is required for the ERAD activity of the Cdc48 complex (Alberts et al. 2009). UBX4 mutants accumulate substrates associated with Cdc48 suggesting that it is required for the final steps of extraction.
The identity of a translocation channel has been elusive. Although there are several candidates, there is no consensus to date. The most likely reason for the difficulty is the existence of multiple channels or multiple components required to assemble the channel(s). The first candidate proposed was the import channel Sec61 itself (Pilon et al. 1997; Plemper et al. 1998, 1999). Some SEC61 mutants are defective in ERAD at temperatures in which import is little affected (Plemper et al. 1998). So far, no direct evidence has been obtained for the export of soluble ERAD substrates through Sec61. However, a short-lived membrane substrate can be cross-linked to Sec61 through an in vivo intermolecular disulfide bond (Scott and Schekman 2008). Interestingly, a cryptic glycosylation site in the amino-terminal cytosolic domain becomes glycosylated, suggesting its transient localization there during the retrotranslocation process. However, a recent study shows that glycosylation could be owing to aberrant reengagement with the Sec61 translocon, which is eventually resolved through degradation via the Hrd1 pathway (Rubenstein et al. 2012). The ERAD factor Der1 was a proposed channel component because its mammalian homolog Derlin-1 can be cross-linked to MHC class I proteins in the process of retrotranslocation (Lilley and Ploegh 2004; Ye et al. 2004). However, some doubt to this hypothesis for the yeast system was raised by conditions that make Der1 entirely dispensable in ERAD (see below). In addition, Der1 is entirely dispensable for ERAD-M (Taxis et al. 2003; Carvalho et al. 2006; Kanehara et al. 2010). However, Der1’s essential function for ERAD-L and mostly transmembrane organization suggests a supporting, if not direct, role in forming the channel. Perhaps the most intriguing proposal is that misfolded glycoproteins are transported from the ER for degradation via lipid droplets (Ploegh 2007). In mammalian cells, the formation of lipid droplets and ERAD is linked via the protein AUP1. AUP1 is required for ERAD and localizes to both the Hrd1 complex and lipid droplets (Klemm et al. 2011). Despite this interesting link, a yeast strain defective in the formation of lipid droplets is competent for ERAD (Olzmann and Kopito 2011).
More recently, the Hrd1 protein itself was proposed to directly retrotranslocate ERAD-L substrates (Carvalho et al. 2010). Remarkably, overexpressing Hrd1p eliminates the requirement of the complex components Hrd3, Usa1, and Der1. Hrd1p oligomerization is required for this activity and it can be cross-linked with substrate at the early stages of retrotranslocation. Taken together, these data support the idea that Hrd1 itself forms all or parts of the retrotranslocation channel for ERAD-L. However, this role may not extend to ERAD-M because Hrd1 and Doa10 proteins are dispensable for the retrotranslocation of the fusion protein Hmg2-GFP in vitro (Garza et al. 2009).
LAST RITES: SUBSTRATE UBIQUITINATION AND PROTEASOME DEGRADATION
ERAD substrates engaged for degradation are polyubiquitinated and deglycosylated before being degraded into small peptides by the cytosolic proteasome (Fig. 3B). The essential cytosolic E1 ubiquitin-activating enzyme (McGrath et al. 1991) Uba1 activates ubiquitin by adenylation and attaches ubiquitin to one of its cysteine residues. Activated ubiquitin is subsequently transferred to the ERAD E2 ubiquitin-conjugating enzymes Ubc6 and Ubc7. Finally, ubiquitin is transferred to an ERAD substrate by the action of E3 ubiquitin ligase Hrd1 or Doa10. Both contain an amino-terminal cytosolic RING-type zinc finger domain required for its ligase activity (Bordallo and Wolf 1999). The Hrd1 RING domain binds and accepts ubiquitin from the membrane-associated protein E2 ubiquitin-conjugating ligase Ubc7 (Bays et al. 2001a). Hrd1 was proposed to support self-ubiquitination through its RING domain (Bazirgan et al. 2006). Lysine residues of ERAD substrates are the preferred ubiquitin acceptor but cysteine, threonine, serine, and even the amino terminus can be ubiquitin acceptors (Kerscher et al. 2006). ERAD substrates are polyubiquitinated by their E3s and through the action of the E4 chain-extension enzyme Ufd2 (Nakatsukasa et al. 2008).
Also in the cytosol, N-linked glycans are removed from ERAD substrates before degradation. The cytosolic enzyme Png1 catalyzes this reaction (Suzuki et al. 2000). Cells lacking PNG1 degrades CPY* nominally slower than wild type but other molecules like the ricin A chain are strongly impaired (Kim et al. 2006). Png1 generates free oligosaccharides (fOS) in the cytosol, which can be analyzed to determine the structures of endogenous ERAD substrate glycans. Analysis of fOS dependent on Png1 revealed the expected Man7-GlcNAc2 structure generated by the glycan timer cascade. More surprisingly, fOS structures consistent with Golgi processing were found abundantly, indicating that some ERAD substrates are retrieved from the Golgi before ERAD (Hirayama et al. 2010). Under stress when glucose is limiting, the levels of Png1-generated fOS increases dramatically suggesting elevated ERAD activity under those conditions (Chantret et al. 2011). The amino terminus of Png1 binds Rad23 (Biswas et al. 2004; Wang et al. 2009). Rad23 binds ubiquitinated proteins and transfers them to the proteasome for degradation (Schauber et al. 1998). Rad23 can also interact with Ufd2 (Kim et al. 2004). Interestingly, Ufd2 also binds Cdc48, potentially linking the ERAD machinery to the proteasome under some circumstances (Baek et al. 2011). Together, these findings suggest that ERAD substrates are rapidly polyubiquitinated, deglycosylated, and degraded in a localized environment near the Hrd1 and Doa10 complexes.
ACKNOWLEDGMENTS
The work in the authors’ laboratory is supported by funds from the Temasek Trust (G.T. and D.T.W.N.) and by the Singapore Millennium Foundation to G.T. (Postdoctoral fellowship).
Footnotes
Editors: Susan Ferro-Novick, Tom A. Rapoport, and Randy Schekman
Additional Perspectives on The Endoplasmic Reticulum available at www.cshperspectives.org
REFERENCES
- Ahner A, Nakatsukasa K, Zhang H, Frizzell RA, Brodsky JL 2007. Small heat-shock proteins select ΔF508-CFTR for endoplasmic reticulum-associated degradation. Mol Biol Cell 18: 806–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts SM, Sonntag C, Schafer A, Wolf DH 2009. Ubx4 modulates cdc48 activity and influences degradation of misfolded proteins of the endoplasmic reticulum. J Biol Chem 284: 16082–16089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek GH, Kim I, Rao H 2011. The Cdc48 ATPase modulates the interaction between two proteolytic factors Ufd2 and Rad23. Proc Natl Acad Sci 108: 13558–13563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY 2001a. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol 3: 24–29 [DOI] [PubMed] [Google Scholar]
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY 2001b. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell 12: 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazirgan OA, Garza RM, Hampton RY 2006. Determinants of RING-E2 fidelity for Hrd1p, a membrane-anchored ubiquitin ligase. J Biol Chem 281: 38989–39001 [DOI] [PubMed] [Google Scholar]
- Bell SL, Chiang AN, Brodsky JL 2011. Expression of a malarial Hsp70 improves defects in chaperone-dependent activities in ssa1 mutant yeast. PLoS ONE 6: e20047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez EM, Stolz A, Wolf DH 2011. Yos9, a control protein for misfolded glycosylated and non-glycosylated proteins in ERAD. FEBS Lett 585: 3015–3019 [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS 2005. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell 19: 741–751 [DOI] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T 1996. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J 15: 2069–2076 [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T 1997. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 278: 1806–1809 [DOI] [PubMed] [Google Scholar]
- Biswas S, Katiyar S, Li G, Zhou X, Lennarz WJ, Schindelin H 2004. The N-terminus of yeast peptide: N-glycanase interacts with the DNA repair protein Rad23. Biochem Biophys Res Commun 323: 149–155 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Suzuki CK, Lippincott-Schwartz J, Weissman AM, Klausner RD 1989. Pre-Golgi degradation of newly synthesized T-cell antigen receptor chains: Intrinsic sensitivity and the role of subunit assembly. J Cell Biol 109: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J, Wolf DH 1999. A RING-H2 finger motif is essential for the function of Der3/Hrd1 in endoplasmic reticulum associated protein degradation in the yeast Saccharomyces cerevisiae. FEBS Lett 448: 244–248 [DOI] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH 1998. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell 9: 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S 2002. Role of the ubiquitin-selective CDC48(UFD1/NPL4 )chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J 21: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Goeckeler J, Schekman R 1995. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci 92: 9643–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA 1999. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem 274: 3453–3460 [DOI] [PubMed] [Google Scholar]
- Buck TM, Kolb AR, Boyd CR, Kleyman TR, Brodsky JL 2010. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell 21: 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Aebi M 1999. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta 1426: 239–257 [DOI] [PubMed] [Google Scholar]
- Buschhorn BA, Kostova Z, Medicherla B, Wolf DH 2004. A genome-wide screen identifies Yos9p as essential for ER-associated degradation of glycoproteins. FEBS Lett 577: 422–426 [DOI] [PubMed] [Google Scholar]
- Carroll SM, Hampton RY 2010. Usa1p is required for optimal function and regulation of the Hrd1p ER-associated degradation (ERAD) ubiquitin ligase. J Biol Chem 285: 5146–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126: 361–373 [DOI] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA 2010. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantret I, Kodali VP, Lahmouich C, Harvey DJ, Moore SE 2011. Endoplasmic reticulum-associated degradation (ERAD) and free oligosaccharide generation in Saccharomyces cerevisiae. J Biol Chem 286: 41786–41800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O’Riordan CR, Smith AE 1990. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63: 827–834 [DOI] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M 2009. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol 184: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland CS, Doms RW, Bolzau EM, Webster RG, Helenius A 1986. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J Cell Biol 103: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland CS, Zimmer KP, Wagner KR, Healey GA, Mellman I, Helenius A 1988. Folding, trimerization, and transport are sequential events in the biogenesis of influenza virus hemagglutinin. Cell 53: 197–209 [DOI] [PubMed] [Google Scholar]
- Cronin SR, Khoury A, Ferry DK, Hampton RY 2000. Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spf1p. J Cell Biol 148: 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C 2010. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem 79: 777–802 [DOI] [PubMed] [Google Scholar]
- Doms RW, Ruusala A, Machamer C, Helenius J, Helenius A, Rose JK 1988. Differential effects of mutations in three domains on folding, quaternary structure, and intracellular transport of vesicular stomatitis virus G protein. J Cell Biol 107: 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabetz Y, Shapira I, Rabinovich E, Bar-Nun S 2004. Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates: Roles of endoplamic reticulum-bound p97/Cdc48p and proteasome. J Biol Chem 279: 3980–3989 [DOI] [PubMed] [Google Scholar]
- Federovitch CM, Jones YZ, Tong AH, Boone C, Prinz WA, Hampton RY 2008. Genetic and structural analysis of Hmg2p-induced endoplasmic reticulum remodeling in Saccharomyces cerevisiae. Mol Biol Cell 19: 4506–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH 1993. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem 218: 565–574 [DOI] [PubMed] [Google Scholar]
- Gardner RG, Hampton RY 1999a. A “distributed degron” allows regulated entry into the ER degradation pathway. EMBO J 18: 5994–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Hampton RY 1999b. A highly conserved signal controls degradation of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase in eukaryotes. J Biol Chem 274: 31671–31678 [DOI] [PubMed] [Google Scholar]
- Gardner R, Cronin S, Leader B, Rine J, Hampton R, Leder B 1998. Sequence determinants for regulated degradation of yeast 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell 9: 2611–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY 2000. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol 151: 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Shearer AG, Hampton RY 2001. In vivo action of the HRD ubiquitin ligase complex: Mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol Cell Biol 21: 4276–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza RM, Sato BK, Hampton RY 2009. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J Biol Chem 284: 14710–14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C 2006a. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol 8: 849–854 [DOI] [PubMed] [Google Scholar]
- Gauss R, Sommer T, Jarosch E 2006b. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J 25: 1827–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Kanehara K, Carvalho P, Ng DT, Aebi M 2011. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol Cell 42: 782–793 [DOI] [PubMed] [Google Scholar]
- Gething MJ, McCammon K, Sambrook J 1986. Expression of wild-type and mutant forms of influenza hemagglutinin: The role of folding in intracellular transport. Cell 46: 939–950 [DOI] [PubMed] [Google Scholar]
- Gillece P, Luz JM, Lennarz WJ, de La Cruz FJ, Romisch K 1999. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J Cell Biol 147: 1443–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnann A, Riordan JR, Wolf DH 2004. Cystic fibrosis transmembrane conductance regulator degradation depends on the lectins Htm1p/EDEM and the Cdc48 protein complex in yeast. Mol Biol Cell 15: 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V, Melero A 2011. Protein O-mannosyltransferases participate in ER protein quality control. J Cell Sci 124: 144–153 [DOI] [PubMed] [Google Scholar]
- Goder V, Carvalho P, Rapoport TA 2008. The ER-associated degradation component Der1p and its homolog Dfm1p are contained in complexes with distinct cofactors of the ATPase Cdc48p. FEBS Lett 582: 1575–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb S, Guo L, Fisher EA, Brodsky JL 2012. Protein disulfide isomerases contribute differentially to the endoplasmic reticulum-associated degradation of apolipoprotein B and other substrates. Mol Biol Cell 23: 520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Bhakta H 1997. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci 94: 12944–12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Rine J 1994. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol 125: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J 1996. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell 7: 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty C, Strahl S, Romisch K 2001. O-mannosylation protects mutant α-factor precursor from endoplasmic reticulum-associated degradation. Mol Biol Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Titus EA, Cooper AA 2004. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell 15: 767–776 [DOI] [PubMed] [Google Scholar]
- Heiligenstein S, Eisfeld K, Sendzik T, Jimenez-Becker N, Breinig F, Schmitt MJ 2006. Retrotranslocation of a viral A/B toxin from the yeast endoplasmic reticulum is independent of ubiquitination and ERAD. EMBO J 25: 4717–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercz A, Katona E, Cutz E, Wilson JR, Barton M 1978. α1-Antitrypsin: The presence of excess mannose in the Z variant isolated from liver. Science 201: 1229–1232 [DOI] [PubMed] [Google Scholar]
- Hill K, Cooper AA 2000. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J 19: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH 1996. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273: 1725–1728 [DOI] [PubMed] [Google Scholar]
- Hirayama H, Fujita M, Yoko-o T, Jigami Y 2008. O-mannosylation is required for degradation of the endoplasmic reticulum-associated degradation substrate Gas1*p via the ubiquitin/proteasome pathway in Saccharomyces cerevisiae. J Biochem 143: 555–567 [DOI] [PubMed] [Google Scholar]
- Hirayama H, Seino J, Kitajima T, Jigami Y, Suzuki T 2010. Free oligosaccharides to monitor glycoprotein endoplasmic reticulum-associated degradation in Saccharomyces cerevisiae. J Biol Chem 285: 12390–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock AL, Krebber H, Frietze S, Lin A, Latterich M, Silver PA 2001. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol Biol Cell 12: 3226–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitt R, Wolf DH 2004. DER7, encoding α-glucosidase I is essential for degradation of malfolded glycoproteins of the endoplasmic reticulum. FEMS Yeast Res 4: 815–820 [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102: 577–586 [DOI] [PubMed] [Google Scholar]
- Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, Sommer T, Jarosch E 2009. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell 36: 782–793 [DOI] [PubMed] [Google Scholar]
- Hosoda A, Tokuda M, Akai R, Kohno K, Iwawaki T 2009. Positive contribution of ERdj5/JPDI to endoplasmic reticulum protein quality control in the salivary gland. Biochem J 425: 117–125 [DOI] [PubMed] [Google Scholar]
- Hrizo SL, Gusarova V, Habiel DM, Goeckeler JL, Fisher EA, Brodsky JL 2007. The Hsp110 molecular chaperone stabilizes apolipoprotein B from endoplasmic reticulum-associated degradation (ERAD). J Biol Chem 282: 32665–32675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S 2004. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem 279: 38369–38378 [DOI] [PubMed] [Google Scholar]
- Ishida Y, Nagata K 2009. Autophagy eliminates a specific species of misfolded procollagen and plays a protective role in cell survival against ER stress. Autophagy 5: 1217–1219 [DOI] [PubMed] [Google Scholar]
- Ishida Y, Yamamoto A, Kitamura A, Lamande SR, Yoshimori T, Bateman JF, Kubota H, Nagata K 2009. Autophagic elimination of misfolded procollagen aggregates in the endoplasmic reticulum as a means of cell protection. Mol Biol Cell 20: 2744–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N, Crawshaw SG, High S 2006. Active and passive displacement of transmembrane domains both occur during opsin biogenesis at the Sec61 translocon. J Cell Sci 119: 2826–2836 [DOI] [PubMed] [Google Scholar]
- Izawa T, Nagai H, Endo T, Nishikawa S 2012. Yos9p and Hrd1p mediate ER retention of misfolded proteins for ER-associated degradation. Mol Biol Cell 23: 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke LA, Brendebach H, Selbach M, Hirsch C 2011. Yos9p assists in the degradation of certain nonglycosylated proteins from the endoplasmic reticulum. Mol Biol Cell 22: 2937–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CA, Burda P, Roth J, Aebi M 1998. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol 142: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CA, Bodmer D, Spirig U, Battig P, Marcil A, Dignard D, Bergeron JJ, Thomas DY, Aebi M 2001. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep 2: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T 2002. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol 4: 134–139 [DOI] [PubMed] [Google Scholar]
- Kabani M, Kelley SS, Morrow MW, Montgomery DL, Sivendran R, Rose MD, Gierasch LM, Brodsky JL 2003. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol Biol Cell 14: 3437–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Xie W, Ng DT 2010. Modularity of the Hrd1 ERAD complex underlies its diverse client range. J Cell Biol 188: 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi S, Hsu CL, Ng DT 2010. Interplay of substrate retention and export signals in endoplasmic reticulum quality control. PLoS ONE 5: e15532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kim I, Mi K, Rao H 2004. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell 15: 3357–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Spear ED, Ng DT 2005. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell 19: 753–764 [DOI] [PubMed] [Google Scholar]
- Kim I, Ahn J, Liu C, Tanabe K, Apodaca J, Suzuki T, Rao H 2006. The Png1-Rad23 complex regulates glycoprotein turnover. J Cell Biol 172: 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Li Y, Muniz P, Rao H 2009. Usa1 protein facilitates substrate ubiquitylation through two separate domains. PLoS ONE 4: e7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid MM, Cooper AA 2007. Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals. Mol Biol Cell 18: 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm EJ, Spooner E, Ploegh HL 2011. Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J Biol Chem 286: 37602–37614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH 1996a. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J 15: 753–763 [PMC free article] [PubMed] [Google Scholar]
- Knop M, Hauser N, Wolf DH 1996b. N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast 12: 1229–1238 [DOI] [PubMed] [Google Scholar]
- Kostova Z, Wolf DH 2005. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J Cell Sci 118: 1485–1492 [DOI] [PubMed] [Google Scholar]
- Kruse KB, Brodsky JL, McCracken AA 2006. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: One for soluble Z variant of human α-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell 17: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR 2009. Inefficient quality control of thermosensitive proteins on the plasma membrane. PLoS ONE 4: e5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429: 834–840 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Bonifacino JS, Yuan LC, Klausner RD 1988. Degradation from the endoplasmic reticulum: Disposing of newly synthesized proteins. Cell 54: 209–220 [DOI] [PubMed] [Google Scholar]
- Loayza D, Tam A, Schmidt WK, Michaelis S 1998. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol Biol Cell 9: 2767–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack KE, Plath K, Misselwitz B, Rapoport TA 1997. Protein transport by purified yeast Sec complex and Kar2p without membranes. Science 277: 938–941 [DOI] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL 1996. Assembly of ER-associated protein degradation in vitro: Dependence on cytosol, calnexin, and ATP. J Cell Biol 132: 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Karpichev IV, Ernaga JE, Werner ED, Dillin AG, Courchesne WE 1996. Yeast mutants deficient in ER-associated degradation of the Z variant of α-1-protease inhibitor. Genetics 144: 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JP, Jentsch S, Varshavsky A 1991. UBA 1: An essential yeast gene encoding ubiquitin-activating enzyme. EMBO J 10: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Maurer MJ, Dancy BM, Michaelis S 2008. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem 283: 32302–32316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G 2000. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J 19: 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Nishikawa S, Hosokawa N, Nagata K, Endo T 2001. Mnl1p, an α-mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J Biol Chem 276: 8635–8638 [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Okada S, Umebayashi K, Fukuda R, Nishikawa S, Endo T 2004. Roles of O-mannosylation of aberrant proteins in reduction of the load for endoplasmic reticulum chaperones in yeast. J Biol Chem 279: 49762–49772 [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132: 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham PG, Mikoluk K, Dhakarwal P, Khadem S, Snyder AC, Subramanya AR, Brodsky JL 2011. The thiazide-sensitive NaCl cotransporter is targeted for chaperone-dependent endoplasmic reticulum-associated degradation. J Biol Chem 286: 43611–43621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T 2005. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol 7: 993–998 [DOI] [PubMed] [Google Scholar]
- Ng W, Sergeyenko T, Zeng N, Brown JD, Romisch K 2007. Characterization of the proteasome interaction with the Sec61 channel in the endoplasmic reticulum. J Cell Sci 120: 682–691 [DOI] [PubMed] [Google Scholar]
- Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T 2001. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol 153: 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR 2011. Lipid droplet formation is dispensable for endoplasmic reticulum-associated degradation. J Biol Chem 286: 27872–27874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH 2007. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell 18: 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Romisch K 1997. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J 16: 4540–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH 1997. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388: 891–895 [DOI] [PubMed] [Google Scholar]
- Plemper RK, Egner R, Kuchler K, Wolf DH 1998. Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J Biol Chem 273: 32848–32856 [DOI] [PubMed] [Google Scholar]
- Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH 1999. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci 112: 4123–4134 [DOI] [PubMed] [Google Scholar]
- Ploegh HL 2007. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature 448: 435–438 [DOI] [PubMed] [Google Scholar]
- Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS 2008. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell 32: 870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol 22: 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S 2001. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107: 667–677 [DOI] [PubMed] [Google Scholar]
- Rapoport TA 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669 [DOI] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M 2006. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J 25: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein EM, Kreft SG, Greenblatt W, Swanson R, Hochstrasser M 2012. Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J Cell Biol 197: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Steger KA, Johnston SA 1999. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J Biol Chem 274: 21943–21952 [DOI] [PubMed] [Google Scholar]
- Sakoh-Nakatogawa M, Nishikawa S, Endo T 2009. Roles of protein-disulfide isomerase-mediated disulfide bond formation of yeast Mnl1p in endoplasmic reticulum-associated degradation. J Biol Chem 284: 11815–11825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato BK, Hampton RY 2006. Yeast Derlin Dfm1 interacts with Cdc48 and functions in ER homeostasis. Yeast 23: 1053–1064 [DOI] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY 2009. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell 34: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391: 715–718 [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A 2005. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol 7: 999–1006 [DOI] [PubMed] [Google Scholar]
- Scott DC, Schekman R 2008. Role of Sec61p in the ER-associated degradation of short-lived transmembrane proteins. J Cell Biol 181: 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AG, Hampton RY 2004. Structural control of endoplasmic reticulum-associated degradation: Effect of chemical chaperones on 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem 279: 188–196 [DOI] [PubMed] [Google Scholar]
- Shearer AG, Hampton RY 2005. Lipid-mediated, reversible misfolding of a sterol-sensing domain protein. EMBO J 24: 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein S, Schlenstedt G, Silver PA, Gilmore R 1998. A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J Cell Biol 143: 921–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Jentsch S 1993. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature 365: 176–179 [DOI] [PubMed] [Google Scholar]
- Spear ED, Ng DT 2003. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol Biol Cell 14: 2756–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear ED, Ng DT 2005. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol 169: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Schweizer RS, Schafer A, Wolf DH 2010. Dfm1 forms distinct complexes with Cdc48 and the ER ubiquitin ligases and is required for ERAD. Traffic 11: 1363–1369 [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Gentzsch M, Tanner W 1999. Protein O-mannosylation. Biochim Biophys Acta 1426: 297–307 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Park H, Hollingsworth NM, Sternglanz R, Lennarz WJ 2000. PNG1, a yeast gene encoding a highly conserved peptide:N-glycanase. J Cell Biol 149: 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev 15: 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA 2005. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell 19: 765–775 [DOI] [PubMed] [Google Scholar]
- Tachibana C, Stevens TH 1992. The yeast EUG1 gene encodes an endoplasmic reticulum protein that is functionally related to protein disulfide isomerase. Mol Cell Biol 12: 4601–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Kimata Y, Kohno K 2008. Saccharomyces cerevisiae Rot1 is an essential molecular chaperone in the endoplasmic reticulum. Mol Biol Cell 19: 3514–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Hitt R, Park SH, Deak PM, Kostova Z, Wolf DH 2003. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J Biol Chem 278: 35903–35913 [DOI] [PubMed] [Google Scholar]
- Theesfeld CL, Pourmand D, Davis T, Garza RM, Hampton RY 2011. The sterol-sensing domain (SSD) directly mediates signal-regulated endoplasmic reticulum-associated degradation (ERAD) of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase isozyme Hmg2. J Biol Chem 286: 26298–26307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault G, Ismail N, Ng DT 2011. The unfolded protein response supports cellular robustness as a broad-spectrum compensatory pathway. Proc Natl Acad Sci 108: 20597–20602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- Tsai B, Rapoport TA 2002. Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J Cell Biol 159: 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JR, Stirling CJ 2000. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J 19: 6440–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA 2004. X-ray structure of a protein-conducting channel. Nature 427: 36–44 [DOI] [PubMed] [Google Scholar]
- Vashist S, Ng DT 2004. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol 165: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DT 2001. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol 155: 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Jin Y, Brodsky JL, Hendershot LM 2009. The mammalian Hsp40 ERdj3 requires its Hsp70 interaction and substrate-binding properties to complement various yeast Hsp40-dependent functions. J Biol Chem 284: 32462–32471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Jonikas MC, Hendershot LM, Weissman JS, Brodsky JL 2010. J domain co-chaperone specificity defines the role of BiP during protein translocation. J Biol Chem 285: 22484–22494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Johnson AE 1994. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol 10: 87–119 [DOI] [PubMed] [Google Scholar]
- Walter J, Urban J, Volkwein C, Sommer T 2001. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J 20: 3124–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chang A 2003. Substrate recognition in ER-associated degradation mediated by Eps1, a member of the protein disulfide isomerase family. EMBO J 22: 3792–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ng DT 2010. Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control. Mol Biol Cell 21: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xin F, Liu X, Wang Y, An Z, Qi Q, Wang PG 2009. N-terminal deletion of peptide: N-glycanase results in enhanced deglycosylation activity. PLoS ONE 4: e8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83: 121–127 [DOI] [PubMed] [Google Scholar]
- Werner ED, Brodsky JL, McCracken AA 1996. Proteasome-dependent endoplasmic reticulum-associated protein degradation: An unconventional route to a familiar fate. Proc Natl Acad Sci 93: 13797–13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland FT, Gleason ML, Serafini TA, Rothman JE 1987. The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell 50: 289–300 [DOI] [PubMed] [Google Scholar]
- Wilhovsky S, Gardner R, Hampton R 2000. HRD gene dependence of endoplasmic reticulum-associated degradation. Mol Biol Cell 11: 1697–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer M, Forte GM, Stirling CJ 2008. Sec61p is required for ERAD-L: Genetic dissection of the translocation and ERAD-L functions of Sec61P using novel derivatives of CPY. J Biol Chem 283: 33883–33888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JD, Liu Y, Bentivoglio CM, Barlowe C 2006. Sel1p/Ubx2p participates in a distinct Cdc48p-dependent endoplasmic reticulum-associated degradation pathway. Traffic 7: 1213–1223 [DOI] [PubMed] [Google Scholar]
- Xie W, Kanehara K, Sayeed A, Ng DT 2009. Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol Biol Cell 20: 3317–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA 2003. Function of the p97–Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: Dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 162: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 42: 841–847 [DOI] [PubMed] [Google Scholar]
- Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL 2004. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell 15: 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL 2001. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell 12: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]