Abstract
The sirtuins are a family of proteins that act predominantly as nicotinamide adenine dinucleotide (NAD)-dependent deacetylases. In mammals seven sirtuin family members exist, including three members, Sirt3, Sirt4, and Sirt5, that localize exclusively within the mitochondria. Although originally linked to life-span regulation in simple organisms, this family of proteins appears to have various and diverse functions in higher organisms. One particular property that is reviewed here is the regulation of mitochondrial number, turnover, and activity by various mitochondrial and nonmitochondrial sirtuins. An emerging consensus from these recent studies is that sirtuins may act as metabolic sensors, using intracellular metabolites such as NAD and short-chain carbon fragments such as acetyl coenzyme A to modulate mitochondrial function to match nutrient supply.
In mammals there are seven sirtuin family members. They seem to act as metabolic sensors, modulating mitochondrial function to match nutrient supply. For example, Sirt1 senses NAD levels and influences mitochondrial biogenesis.
THE SIRTUIN FAMILY OF PROTEINS
Originally characterized in yeast as regulators of life span (Kaeberlein et al. 1999), the sirtuins are an evolutionarily conserved family of proteins that appear to exert a wide range of biological functions (Finkel et al. 2009). In mammals seven sirtuin family members exist. Sirt1 appears to be the closest mammalian homolog to yeast Sir2, the first member of the sirtuins linked to aging. Because of this homology, initial studies of mammalian sirtuins focused predominantly on the biology of Sirt1. Insights from these studies have implicated Sirt1 in the regulation of a variety of metabolic phenotypes including insulin secretion (Moynihan et al. 2005; Bordone et al. 2006), lipid mobilization from adipocytes (Picard et al. 2004), and regulation of glucose tolerance (Rodgers et al. 2005). With that said, the role of sirtuins in mammalian aging remains an open question, and even some of the earlier work in lower organisms involving Sir2 and life span has recently been questioned (Burnett et al. 2011).
Although there has been considerable attention directed toward Sirt1 biology, there is also a growing interest in understanding the function of the related family members. It is clear that each of the mammalian sirtuins has a distinct subcellular localization. Sirt1, Sirt6, and Sirt7 are nuclear proteins, although a fraction of Sirt1 can be found in the cytosol. Sirt2, on the other hand, is predominantly cytosolic, although again it can be found in the nucleus in certain situations (North and Verdin 2007). Finally, three sirtuins—Sirt3, Sirt4, and Sirt5—appear to be found exclusively in the mitochondria. From a human genetics point of view, the strongest association between aging and sirtuins is the association between polymorphisms in the mitochondrial Sirt3 and longevity (Rose et al. 2003).
The ability of sirtuins to influence metabolism and potentially life span is believed to revolve around the ability of sirtuin family members to function as protein deacetylases. In addition to this enzymatic function, Sirt4 can further act to ADP ribosylate target proteins. Unlike other protein deacetylases, sirtuins require nicotinamide adenine dinucleotide (NAD) as a cofactor in the deacetylation reaction (Imai et al. 2000). The link among NAD, NADH, and sirtuin activity has led many to believe that this family of proteins acts in some fashion as a sensor of energetic status. This may particularly be true in the mitochondria, where levels of NAD and NADH are high and where a disproportionate fraction of proteins appear to be acetylated (Kim et al. 2006).
In this review we will examine the link between sirtuin function and mitochondrial metabolism. While we highlight the role of those sirtuin family members that are uniquely mitochondrial in their localization, we will also discuss other sirtuins that can influence mitochondrial function, biogenesis, and turnover. Finally, although we discuss each sirtuin individually, there is a growing realization that one sirtuin family member can affect the function of other family members. A recent example is Sirt1-dependent regulation of Sirt6 expression in the control of hepatic metabolism (Kim et al. 2010b).
Sirt1 AND MITOCHONDRIAL FUNCTION
Evidence suggests that mitochondrial biogenesis is regulated at least in part by proliferator-activated receptor coactivator-1α (PGC-1α), a transcriptional coactivator of peroxisome proliferator-activated receptor-γ (PPARγ) as well as other transcription factors (Fernandez-Marcos and Auwerx 2011). It was therefore of considerable interest when it was shown that PGC-1α was in fact a deacetylation target of Sirt1 and that acetylation regulated PGC-1α activity (Nemoto et al. 2005; Rodgers et al. 2005). There are at least 13 lysine residues on PGC-1α that appear to be reversibly acetylated (Rodgers et al. 2005). Site-directed mutants that lack all 13 of these sites alter the ability of PGC-1α to regulate gene expression, although it remains unclear if all, or only a subset, of PGC-1α’s acetylation sites are truly regulatory in nature. Sirt1 appears to be the predominant in vitro and in vivo regulator of PGC-1α deacetylation. For instance, in vitro knockdown of Sirt1 in hepatic cells leads to increased PGC-1α acetylation with a corresponding reduction in a set of genes that are the rate-limiting enzymes responsible for hepatic gluconeogenesis (Rodgers et al. 2005). Similarly, both overexpression and knockdown studies support a role for Sirt1 in regulating PGC-1α activity through reversible deacetylation, which in turn has dramatic effects on in vivo hepatic glucose and lipid metabolism (Rodgers and Puigserver 2007; Erion et al. 2009). A similar relationship appears to exist in skeletal muscle. In particular, in skeletal muscle, fasting was shown to lead to a Sirt1-dependent deacetylation of PGC-1α, and this deacetylation appeared to be required for PGC-1α-dependent gene expression, including gene products required for effective mitochondrial biogenesis (Gerhart-Hines et al. 2007). Together these studies link Sirt1 and PGC-1α activities in metabolically active tissues such as the liver and skeletal muscle (Fig. 1).
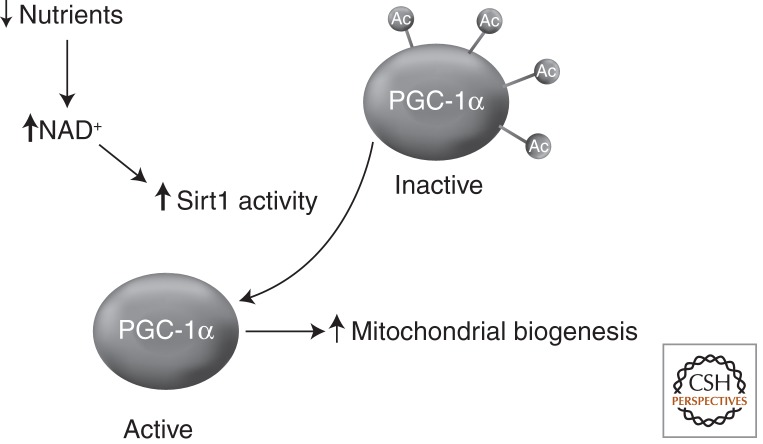
Figure 1.
Regulation of PGC-1α acetylation and activity by Sirt1. The transcriptional coactivator and regulator of mitochondrial biogenesis PGC-1α is, at least in part, regulated by lysine acetylation. In the setting of low nutrient availability, the intracellular and particularly the nuclear levels of NAD are believed to increase and lead to activation of Sirt1 enzymatic activity. This leads to PGC-1α deacetylation, resulting in increased PGC-1α activity and hence, ultimately, in an increase in mitochondrial number.
The above studies suggest that mitochondrial biogenesis might be regulated by tissue energetic status and that the sirtuins would represent important energy sensors in this homeostatic loop. Indeed, the notion that PGC-1α acetylation and function, and by extension mitochondrial activity, are regulated in a nutrient-dependent fashion by Sirt1 is appealing. Nonetheless, the concept that Sirt1 is in turn responding to nutrient-sensitive changes in basal NAD levels, although often invoked, has until recently had little experimental support. The difficulty in proving the supposition is that measurement and manipulation of NAD levels in various subcellular compartments is experimentally challenging. One recent approach is to carefully examine mice with a deletion of a major NAD-consuming enzyme, poly(ADP-ribose) polymerase-1 (PARP-1). These mice appear to have elevated NAD levels along with increased Sirt1 activity (Bai et al. 2011). Furthermore, consistent with increased NAD levels leading to increased PGC-1α activity, PARP-1−/− mice have increased mitochondrial content (Bai et al. 2011). Another recent report suggested that adiponectin, a secreted adipokine, could regulate intracellular NAD levels (Iwabu et al. 2010). Again, in these studies the addition of adiponectin to cells appeared to increase mitochondrial content through a Sirt1- and PGC-1α-dependent pathway. These results might be particularly important because metabolic diseases are often associated with low adiponectin levels (Hotta et al. 2000), as well as with mitochondrial dysfunction (Petersen et al. 2004), although the link between these two observations was previously unknown. Interestingly, these observations would also seem to support the growing link between genetic variants of Sirt1 and a person’s risk for developing obesity (Peeters et al. 2008; Zillikens et al. 2009).
The generation of new mitochondria through a Sirt1/PGC-1α-regulated pathway is complemented by another important connection between Sirt1 and the mitochondria. In particular, it would appear that Sirt1 is an important regulator of removing damaged mitochondria through the process of autophagy (Lee et al. 2008). The field of autophagy represents a rapidly expanding area of study, and a full review of the subject is not possible. Suffice it to say that autophagy is an evolutionarily conserved process present in organisms ranging from yeast to mammals. One of the major intracellular roles of autophagy is the removal of damaged organelles such as mitochondria. Evidence suggests that Sirt1 can stimulate autophagy and that Sirt1−/− tissues appear to accumulate abnormal-appearing mitochondria, consistent with what is seen in autophagy-deficient tissues (Lee et al. 2008). A cytoplasm-restricted mutant of Sirt1 can still stimulate autophagy, suggesting that this activity represents an extranuclear function of the protein (Morselli et al. 2011). The molecular basis for how Sirt1 stimulates autophagy is not clear. There is evidence that key molecules required for autophagy including Atg5 and Atg7 are direct targets of sirtuin-dependent deacetylation (Lee et al. 2008). In addition, the FoxO family of transcription factors, known targets of Sirt1 deacetylation as well as regulators of autophagy, has also been implicated (Hariharan et al. 2010; Kume et al. 2010). Nonetheless, although details remain to be elucidated, the notion that Sirt1 can regulate both the creation of new mitochondria as well as the removal of old mitochondria suggests a role for sirtuins in overall mitochondrial flux and in the maintenance of what may be viewed as “youthful” mitochondria in the cell.
A final area that we wish to discuss in which the biology of Sirt1 and the mitochondria intersect is the secretion of insulin by the pancreatic β cell. Previous studies have suggested that mitochondrial uncoupling protein-2 (UCP2) was an important negative regulator of insulin secretion by the β cell (Zhang et al. 2001). Indeed, UCP2 appears to be up-regulated in obese animals and UCP2-deficient mice have increased glucose-stimulated insulin release (Zhang et al. 2001). Subsequent studies identified Sirt1 as an important repressor of UCP2 expression in the pancreas (Moynihan et al. 2005; Bordone et al. 2006). It is believed that starvation may induce UCP2 expression in the β cell via a reduction in tissue NAD levels (Bordone et al. 2006). There is also evidence that a recently described extracellular, circulating form of the enzyme nicotinamide phosphoribosyltransferase (Nampt) may be an important in vivo mediator of insulin secretion (Revollo et al. 2007). The Nampt enzyme is a key enzyme in NAD biosynthesis and is responsible for the conversion of nicotinamide to nicotinamide mononucleotide, a metabolite that can in turn be converted directly to NAD. Again, these observations highlight the potential function of Sirt1 in linking metabolites such as NAD to the maintenance of overall metabolic homeostasis.
Sirt3 AND MITOCHONDRIAL FUNCTION
The studies we have reviewed indicating that the predominantly nuclear Sirt1 was an important regulator of mitochondrial function spurred interest in the biology of those sirtuin family members that directly localize to this organelle. Comparative analysis of total liver mitochondrial protein acetylation following distinct genetic knockout of each of the three mitochondrial-enriched sirtuins—Sirt3, Sirt4, and Sirt5—showed that Sirt3 is the major mitochondrial deacetylase (Lombard et al. 2007). At the same time, the dynamic flux in mitochondrial protein acetylation in response to changes in caloric load as illustrated by feeding and fasting (Kim et al. 2006), caloric restriction (Schwer et al. 2009), and caloric excess (Hirschey et al. 2011; Kendrick et al. 2011) suggest that, as is the case with Sirt1, Sirt3 may possess a nutrient-sensing regulatory role governing mitochondrial protein function.
Over the last few years, many laboratories have identified and functionally characterized mitochondrial targets of Sirt3 deacetylation and shown that Sirt3 does indeed modulate numerous mitochondrial pathways via protein lysine-residue deacetylation. As mitochondria are most readily identified as the “powerhouse” of the cell, we initially review the role of Sirt3 in mitochondrial bioenergetics. Sirt3 deacetylates and activates multiple steps in substrate catabolism, starting with the oxidation of fatty acids. As an example, Sirt3 has been shown under nutrient-restricted conditions to deacetylate and hence activate long-chain acyl coenzyme A (acyl-CoA) dehydrogenase to increase fatty acid β-oxidation (Hirschey et al. 2010; Hallows et al. 2011). Additional substrate catabolic pathway targets include glutamate dehydrogenase, which facilitates the oxidative deamination of glutamate to α-ketoglutarate, and the citric acid cycle enzyme isocitrate dehydrogenase 2 (IDH2) (Lombard et al. 2007; Schlicker et al. 2008). In keeping with its role as the final common denominator in mitochondrial energy production, numerous proteins in the electron transport chain have been shown to be directly deacetylated by Sirt3. Sirt3 activation has been shown to increase oxidative phosphorylation (Ahn et al. 2008; Bao et al. 2010a; Cimen et al. 2010) and to deacetylate and activate enzymes in complexes I and II of the electron transport chain (Ahn et al. 2008; Cimen et al. 2010; Finley et al. 2011; Kendrick et al. 2011), with additional deacetylation of proteins in complex V (Schlicker et al. 2008; Bao et al. 2010b).
An additional aspect of mitochondrial bioenergetics operational under caloric restriction or fasting conditions is the conversion of acetate to acetyl-CoA, a necessary step required for energy production in extrahepatic tissues and for the generation of ketones in the liver. Again, Sirt3 appears to have an important regulatory role in these pathways, as both acetyl-CoA synthetase 2 (Hallows et al. 2006; Schwer et al. 2006) and 3-hydroxy-3-methylglutaryl-CoA synthase 2 (Shimazu et al. 2010) are targets of Sirt3 deacetylation. A final metabolic pathway that is modulated by Sirt3 is the urea cycle, responsible for the detoxification of ammonia during amino acid metabolism. Here, too, Sirt3 deacetylates and activates a key enzyme, ornithine transcarbamoylase (Hallows et al. 2011).
Sirt3 is also emerging as a regulatory protein in the modulation of additional mitochondrial programs, including the deacetylation and inhibition of the mitochondrial ribosomal protein L10 (MRPL10) (Yang et al. 2009). This results in an NAD-dependent inhibition of mitochondrial protein synthesis, which might function as an energy-sparing response under nutrient-restricted conditions. Another mitochondrial protein that is inhibited by Sirt3 deacetylation is the mitochondrial matrix peptidyl–prolyl isomerase cyclophilin D (Ppif) (Hafner et al. 2010; Shulga and Pastorino 2010; Shulga et al. 2010). Our understanding of the function of cyclophilin D has expanded in recent years to include a role not only in increasing susceptibility to mitochondrial permeability transition (Baines et al. 2005; Nakagawa et al. 2005), but also in regulating mitochondrial calcium efflux with the concordant regulation of Ca2+-dependent mitochondrial enzyme activities (Elrod et al. 2010). An additional, albeit indirect, effect of Sirt3-dependent inactivation of cyclophilin D is the dissociation of hexokinase II from the mitochondria, which plays a role in the promotion of oxidative phosphorylation by Sirt3 (Shulga et al. 2010).
The mitochondrial sirtuins also appear to play an important role in the control of reactive oxygen species. This regulatory role may be particularly relevant to modulating the development of age-associated degenerative conditions. At the direct substrate level, the reactive oxygen species scavenging enzyme MnSOD is activated by Sirt3, and numerous lysine residues have been implicated in mediating this induction of enzyme activity (Qiu et al. 2010; Tao et al. 2010; Chen et al. 2011). Via a more indirect mechanism, the caloric restriction-associated activation of IDH2 has been shown to increase NADH, which is proposed to facilitate the increase in reduced glutathione levels found in association with Sirt3-mediated activation of IDH2 (Someya et al. 2010). The emerging role of Sirt3 in modulating various pathways in metabolism and stress modulatory programs is shown in Figure 2.
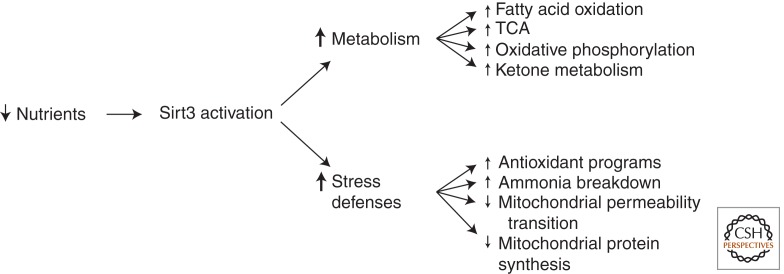
Figure 2.
A role for Sirt3 in metabolic adaptation and stress defense in the setting of low nutrients. The sirtuin family member Sirt3 is the predominant mitochondrial deacetylase. Multiple targets of Sirt3 have been described. These targets involve enzymes linked to substrate utilization as well as core components of the electron transport chain. In addition, Sirt3 has been linked to a wide array of other stress-related programs including the maintenance of redox homeostasis. See text for additional details.
Finally, a recently defined target of Sirt3, namely, mitochondrial aldehyde dehydrogenase 2 (ALDH2), has uncovered an additional biological role for the acetylation of lysine residues (Lu et al. 2011). In contrast to the prior targets of Sirt3 discussed above, the change in acetylation status of ALDH2 did not alter the activity of this dehydrogenase. However, because the reactive metabolite of the analgesic agent acetaminophen disrupts proteins by binding to lysine residues, the role of Sirt3 in acetaminophen metabolite binding to ALDH2 was explored (Lu et al. 2011). This analysis confirmed that lysine-residue acetylation can indeed function as an allosteric inhibitor of xenobiotic-reactive metabolite binding to mitochondrial proteins by exposing lysine residues to other posttranslational modifiers, and that in this instance Sirt3 activation has detrimental consequences (Silberman and Mostoslavsky 2011). These results might help explain the clinical observation that acetaminophen liver injury is exacerbated by fasting (Lu et al. 2011)
The investigations of the targets of Sirt3 collectively show that in response to caloric restriction or fasting, Sirt3 deacetylates a vast array of mitochondrial proteins, with a resulting panoply of effects including activation, inhibition, and allosteric modification of protein functioning. The physiological impact of these effects is beginning to be explored in a range of age-related or nutrient-sensitive diseases. On balance, recent observations suggest that Sirt3 activation is generally ameliorative with respect to the development of age-associated diseases such as cardiac dysfunction (Sundaresan et al. 2009; Hafner et al. 2010), hearing loss (Someya et al. 2010), metabolic syndromes and diabetes (Hirschey et al. 2011; Jing et al. 2011; Kendrick et al. 2011), and cancer (Kim et al. 2010a).
Sirt5 AND MITOCHONDRIAL FUNCTION
The identification of targets of Sirt5 in the mitochondria has been more limited, although the urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS-1) has been identified as a Sirt5 deacetylation target (Nakagawa et al. 2009). The activation of CPS-1 catalyzes ammonia to urea and would be expected to have ameliorative effects via the elimination of oxidative stress-promoting ammonium.
Recent work shows that in addition to acetyl groups, additional short-chain carbon fragments covalently attached to specific amino acids may be targets of Sirt5. These include the removal of succinyl and malonyl groups from lysine residues (Du et al. 2011; Peng et al. 2011). Succinyl-CoA and malonyl-CoA are metabolic intermediates of the tricarboxylic acid (TCA) cycle (Peng et al. 2011; Zhang et al. 2011). To date, very few specific protein targets of Sirt5-mediated desuccinylation or demalonylation have been identified. However, the levels of hepatic protein succinylation and malonylation are highly enriched in Sirt5 knockout mice (Du et al. 2011; Peng et al. 2011). Although these novel posttranslational modifications attributed to Sirt5 need to be fleshed out, they are compatible with the concept that the sirtuin proteins target lysine-residue modifications to alter metabolic functioning in response to changes in levels of various intracellular nutrients.
CONCLUSIONS
Although originally described in yeast, the mammalian sirtuins represent an intriguing family of proteins that appear to function as sensors and regulators of metabolic status. Included among this family’s diverse function is the coordinated control of mitochondrial activity. This regulation includes the creation and targeted destruction of mitochondria by Sirt1, as well as the regulation of substrate utilization and oxidative phosphorylation by Sirt3. Table 1 shows the metabolic substrates of Sirt1, −3, and −5 discussed in this review and summarizes the metabolic effects of these deacetylation reactions.
Table 1.
List of described sirtuin substrates, their subcellular localization, and the effect of deacetylation on protein function
| Sirtuin | Sirtuin substrate | Localization | Result of sirtuin action | References |
|---|---|---|---|---|
| Sirt1 | PGC-1α | Nucleus | Activates transcriptional coactivation | Nemoto et al. 2005; Rodgers et al. 2005 |
| FoxO | Nucleus | Activates transcription and autophagy | Hariharan et al. 2010; Kume et al. 2010 | |
| Atg5/7 | Cytosol | Deacetylation to promote autophagy | Lee et al. 2008 | |
| UCP2 | Nucleus | Repression of transcript | Zhang et al. 2001 | |
| Sirt3 | LCAD | Mitochondria | Activates enzyme, promotes lipid processing | Hirschey et al. 2010; Hallows et al. 2011 |
| GDH | Mitochondria | Activates enzyme, oxidative deamination of glutamate | Lombard et al. 2007 | |
| NDUFA9 | Mitochondria | Activates enzyme, up-regulates ETC activity | Ahn et al. 2008 | |
| SDHB | Mitochondria | Activates enzyme, up-regulates ETC activity | Cimen et al. 2010 | |
| ATP5A | Mitochondria | Activates enzyme, up-regulates ETC activity | Schlicker et al. 2008; Bao et al. 2010b | |
| OTC | Mitochondria | Activates enzyme, reduces ammonia toxicity and ROS | Hallows et al. 2011 | |
| ACS2, HMGCoAS2 | Mitochondria | Activate ketone metabolism | Hallows et al. 2006; Schwer et al. 2006; Shimazu et al. 2010 | |
| SOD2 | Mitochondria | Activates enzyme, reduces ROS | Qui et al. 2010; Tao et al. 2010; Chen et al. 2011 | |
| IDH2 | Mitochondria | Activates enzyme, reduces ROS | Schlicker et al. 2008; Someya et al. 2010 | |
| CypD (PPIF) | Mitochondria | Deactivates enzyme, prevents interaction with mPTP | Hafner et al. 2010; Shulga and Pastorino 2010; Shulga et al. 2010 | |
| ALDH2 | Mitochondria | Deacetylation by Sirt3 allows NAPQI binding to ALDH2, reducing its activity | Lu et al. 2011 | |
| MRPL10 | Mitochondria | Inhibition of mitochondrial ribosome | Yang et al. 2009 | |
| Sirt5 | CPS-1 | Mitochondria | Activates enzyme, reduces ammonia toxicity and ROS | Nakagawa et al. 2009 |
PGC-1α, proliferator-activated receptor coactivator-1α; UCP2, uncoupling protein-2; LCAD, long-chain acyl-CoA dehydrogenase; GDH, glutamate dehydrogenase; NDUFA9, NADH dehydrogenase subcomplex A9; SHDB, succinate dehydrogenase B subunit; ATP5A, ATP synthase subunit 5A; OTC, ornithine transcarbamoylase; ACS2, acyl-CoA synthetase 2; HMGCoAS2, 3-hydroxy-3-methylglutaryl-CoA synthase 2; SOD2, superoxide dismutase 2; IDH2, isocitrate dehydrogenase 2; CypD, cyclophilin D; ALDH2, aldehyde dehydrogenase 2; MRPL10, mitochondrial ribosomal protein L10; CPS-1, carbomoyl phosphate synthetase 1; ETC, electron transport chain; ROS, reactive oxygen species; mPTP, mitochondrial permeability transition pore; NAPQ1, N-acetyl-p-benzoquinoneimine.
In addition to these biochemical properties, there are tantalizing clues that sirtuins may play an important physiological role in overall metabolic homeostasis and perhaps in modulating age-related metabolic pathologies. Finally, although the predominant function of the sirtuin family revolves around NAD-dependent lysine deacetylation, other less-characterized enzymatic activities including ADP ribosylation, as well as other lysine modifications (e.g., demalonylation and desuccinylation), are just beginning to be explored. Although considerable gaps exist in our understanding, further dissection of sirtuin biology promises to provide important insight into how metabolic supply is coupled to mitochondrial activity.
Footnotes
Editors: Douglas C. Wallace and Richard J. Youle
Additional Perspectives on Mitochondria available at www.cshperspectives.org
REFERENCES
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T 2008. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci 105: 14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. 2011. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, et al. 2005. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature 434: 658–662 [DOI] [PubMed] [Google Scholar]
- Bao J, Lu Z, Joseph JJ, Carabenciov D, Dimond CC, Pang L, Samsel L, McCoy JP Jr, Leclerc J, Nguyen P, et al. 2010a. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J Cell Biochem 110: 238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Scott I, Lu Z, Pang L, Dimond CC, Gius D, Sack MN 2010b. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic Biol Med 49: 1230–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, et al. 2006. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic β cells. PLoS Biol 4: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, et al. 2011. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477: 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, Xiong Y 2011. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep 12: 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC 2010. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry 49: 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. 2011. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334: 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod JW, Wong R, Mishra S, Vagnozzi RJ, Sakthievel B, Goonasekera SA, Karch J, Gabel S, Farber J, Force T, et al. 2010. Cyclophilin D controls mitochondrial pore-dependent Ca2+ exchange, metabolic flexibility, and propensity for heart failure in mice. J Clin Invest 120: 3680–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, et al. 2009. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci 106: 11288–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Marcos PJ, Auwerx J 2011. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr 93: 884S–890S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R 2009. Recent progress in the biology and physiology of sirtuins. Nature 460: 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Haas W, Desquiret-Dumas V, Wallace DC, Procaccio V, Gygi SP, Haigis MC 2011. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS ONE 6: e23295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P 2007. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J 26: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, Sinclair DA 2010. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2: 914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci 103: 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devires MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, et al. 2011. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell 41: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J 2010. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res 107: 1470–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. 2010. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464: 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, et al. 2011. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell 44: 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Funahashi T, Arita Y, Takahashi M, Matsuda M, Okamoto Y, Iwahashi H, Kuriyama H, Ouchi N, Maeda K, et al. 2000. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol 20: 1595–1599 [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, et al. 2010. Adiponectin and AdipoR1 regulate PGC-1α and mitochondria by Ca2+ and AMPK/SIRT1. Nature 464: 1313–1319 [DOI] [PubMed] [Google Scholar]
- Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, Verdin EM, Kahn CR 2011. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci 108: 14608–14613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, et al. 2011. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J 433: 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23: 607–618 [DOI] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. 2010a. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, et al. 2010b. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab 12: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D 2010. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest 120: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T 2008. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci 105: 3374–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. 2007. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27: 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Bourdi M, Li JH, Aponte AM, Chen Y, Lombard DB, Gucek M, Pohl LR, Sack MN 2011. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep 12: 840–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Marino G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Benit P, Rustin P, Criollo A, et al. 2011. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol 192: 615–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S 2005. Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab 2: 105–117 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y 2005. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature 434: 652–658 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L 2009. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137: 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T 2005. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem 280: 16456–16460 [DOI] [PubMed] [Google Scholar]
- North BJ, Verdin E 2007. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS ONE 2: e784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters AV, Beckers S, Verrijken A, Mertens I, Roevens P, Peeters PJ, Van Hul W, Van Gaal LF 2008. Association of SIRT1 gene variation with visceral obesity. Hum Genet 124: 431–436 [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, et al. 2011. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics 110: M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI 2004. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429: 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D 2010. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 12: 662–667 [DOI] [PubMed] [Google Scholar]
- Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, et al. 2007. Nampt/PBEF/visfatin regulates insulin secretion in β cells as a systemic NAD biosynthetic enzyme. Cell Metab 6: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P 2007. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci 104: 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P 2005. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434: 113–118 [DOI] [PubMed] [Google Scholar]
- Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, et al. 2003. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol 38: 1065–1070 [DOI] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C 2008. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol 382: 790–801 [DOI] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E 2006. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci 103: 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB 2009. Calorie restriction alters mitochondrial protein acetylation. Aging Cell 8: 604–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, et al. 2010. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab 12: 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Pastorino JG 2010. Ethanol sensitizes mitochondria to the permeability transition by inhibiting deacetylation of cyclophilin-D mediated by sirtuin-3. J Cell Sci 123: 4117–4127 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shulga N, Wilson-Smith R, Pastorino JG 2010. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci 123: 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Silberman DM, Mostoslavsky R 2011. SIRT3 deacetylase: The Jekyll and Hyde sirtuin. EMBO Rep 12: 746–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA 2010. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143: 802–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP 2009. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest 119: 2758–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, et al. 2010. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell 40: 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cimen H, Han MJ, Shi T, Deng JH, Koc H, Palacios OM, Montier L, Bai Y, Tong Q, et al. 2009. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem 285: 7417–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CY, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, et al. 2001. Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, β cell dysfunction, and type 2 diabetes. Cell 105: 745–755 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y 2011. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol 7: 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillikens MC, van Meurs JB, Rivadeneira F, Amin N, Hofman A, Oostra BA, Sijbrands EJ, Witteman JC, Pols HA, van Duijn CM, et al. 2009. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes 58: 2828–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]