Abstract
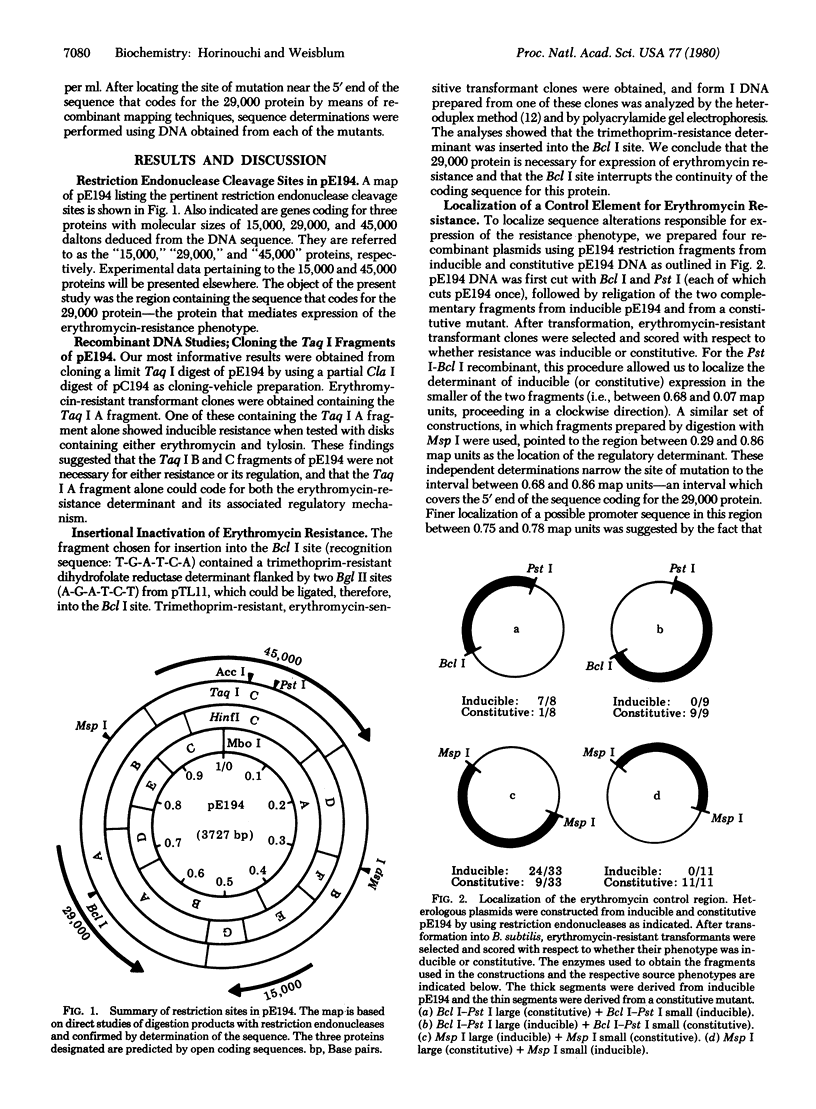
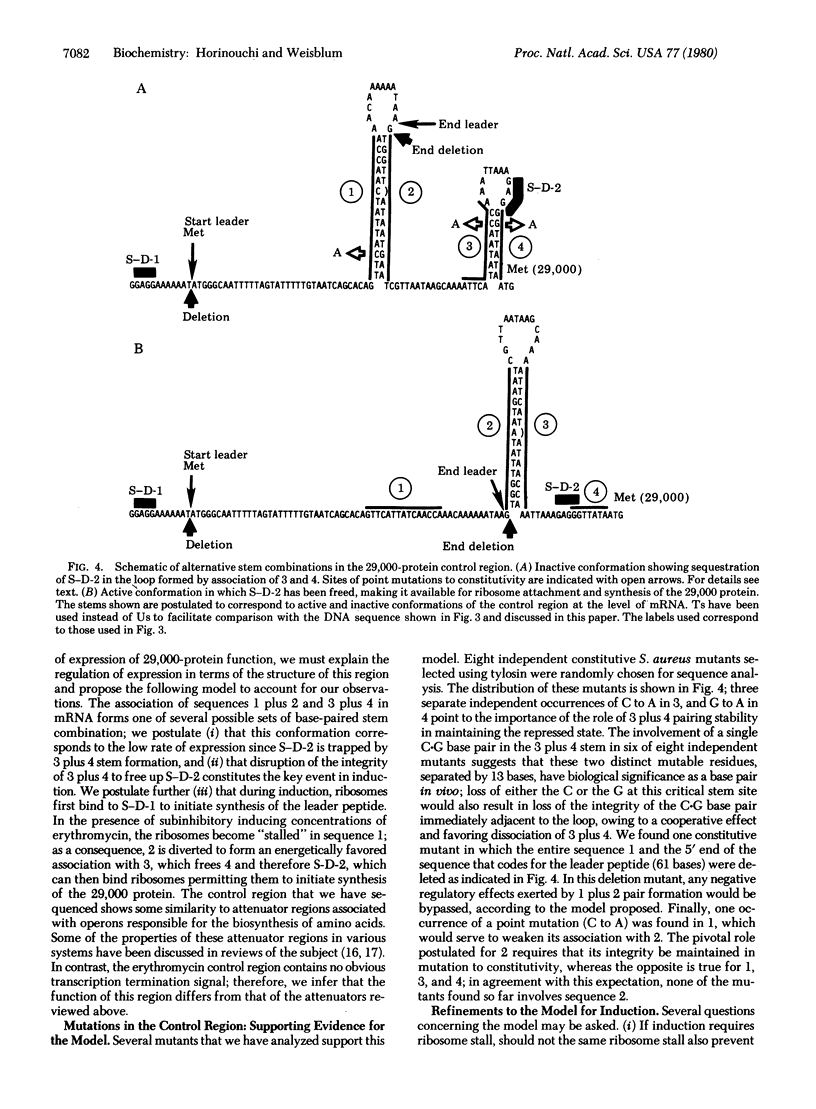
The nucleotide sequence of a gene in plasmid pE194 responsible for erythromycin-induced resistance, including regulation of the resistance phenotype, is reported. A DNA fragment from plasmid pE194, obtained by digestion with Taq I restriction endonuclease, was cloned in Bacillus subtilis by using pC194 as the plasmid cloning vector. Erythromycin-resistant, inducible transformant clones containing the Taq I fragment A were obtained in which the expression of resistance was similar to that found in the original pE194 background; an interpretative model of the regulation of the erythromycin-resistance determinant is proposed based on the sequence of the Taq I A fragment. The cloned Taq I A fragment consists of 1442 base pairs and has open reading frames capable of coding for a peptide and a protein containing 19 and 243 amino acids, respectively, referred to as the “leader peptide” and “29,000 protein.” Between the putative transcriptional start site and the ribosome binding site for 29,000-protein synthesis, the promoter region contains four complementary inverted repeat sequences named “1, 2, 3, and 4,” respectively, in which 1 is complementary to 2, 2 is complementary to 3, and 3 is complementary to 4. Sequence 1 encodes the COOH-terminal half of the leader peptide, whereas the ribosome binding site for synthesis of 29,000 protein is sequestered in a loop formed by the association of 3 and 4. The 29,000-protein promoter region does not appear to contain any transcription stop signal. We propose a model for regulation of erythromycin resistance according to which ribosomes engaged in leader peptide synthesis are partially inhibited by optimal inducing (i.e., subinhibitory) concentrations of erythromycin that, in turn, cause an accumulation of these partially inhibited (“stalled”) ribosomes in sequence 1. During induction, the translationally inactive states of association of the inverted repeats, postulated to be 1 plus 2 and 3 plus 4, respectively, are perturbed by a high level of stalled ribosome occupancy in sequence 1, and in the resultant redistribution, 2 associates with 3, freeing 4 and thereby freeing the ribosome binding site sequestered by the association of 3 and 4. Sequence alterations at the 5′ end of the 29,000-protein coding region associated with mutation to constitutive expression have been localized to the inverted complementary repeats, and determination of base changes in eight mutants are all capable of reducing the stability of the postulated stems in a manner consistent with predictions made by the model.
Keywords: antibiotics, ribosomes, gene expression, Staphylococcus aurcus, Bacillus subtilis
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cundliffe E. Antibiotics and polyribosomes. II. Some effects of lincomycin, spiramycin, and streptogramin A in vivo. Biochemistry. 1969 May;8(5):2063–2066. doi: 10.1021/bi00833a042. [DOI] [PubMed] [Google Scholar]
- Gryczan T., Shivakumar A. G., Dubnau D. Characterization of chimeric plasmid cloning vehicles in Bacillus subtilis. J Bacteriol. 1980 Jan;141(1):246–253. doi: 10.1128/jb.141.1.246-253.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordănescu S. Relationships between cotransducible plasmids in Staphylococcus aureus. J Bacteriol. 1977 Jan;129(1):71–75. doi: 10.1128/jb.129.1.71-75.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordănescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch Roum Pathol Exp Microbiol. 1976 Jan-Jun;35(1-2):111–118. [PubMed] [Google Scholar]
- Johnston H. M., Barnes W. M., Chumley F. G., Bossi L., Roth J. R. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci U S A. 1980 Jan;77(1):508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. B., Calvo J. M. Alternative secondary structures of leader RNAs and the regulation of the trp, phe, his, thr, and leu operons. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6186–6190. doi: 10.1073/pnas.76.12.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci U S A. 1971 Apr;68(4):856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. Antibiotics as probes of ribosome structure: binding of chloramphenicol and erythromycin to polyribosomes; effect of other antibiotics. Antimicrob Agents Chemother. 1974 Mar;5(3):255–267. doi: 10.1128/aac.5.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai P. C., Wallace B. J., Davis B. D. Selective action of erythromycin on initiating ribosomes. Biochemistry. 1974 Oct 22;13(22):4653–4659. doi: 10.1021/bi00719a029. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Teraoka H. Binding of erythromycin to Escherichia coli ribosomes. Biochim Biophys Acta. 1966 Jan 18;114(1):204–206. doi: 10.1016/0005-2787(66)90272-3. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kawano N. Cloning vehicles for the homologous Bacillus subtilis host-vector system. Gene. 1980 Jul;10(2):131–136. doi: 10.1016/0378-1119(80)90130-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T. recE4-Independent recombination between homologous deoxyribonucleic acid segments of Bacillus subtilis plasmids. J Bacteriol. 1979 Sep;139(3):775–782. doi: 10.1128/jb.139.3.775-782.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. E., Burgess R. R. Escherichia coli RNA polymerase binding and initiation of transcription on fragments of lambda rifd 18 DNA containing promoters for lambda genes and for rrnB, tufB, rplC,A, rplJ,L, and rpoB,C genes. Gene. 1979 Aug;6(4):331–365. doi: 10.1016/0378-1119(79)90073-8. [DOI] [PubMed] [Google Scholar]
- WEAVER J. R., PATTEE P. A. INDUCIBLE RESISTANCE TO ERYTHROMYCIN IN STAPHYLOCOCCUS AUREUS. J Bacteriol. 1964 Sep;88:574–580. doi: 10.1128/jb.88.3.574-580.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B., Siddhikol C., Lai C. J., Demohn V. Erythromycin-inducible resistance in Staphylococcus aureus: requirements for induction. J Bacteriol. 1971 Jun;106(3):835–847. doi: 10.1128/jb.106.3.835-847.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland B. C., Szybalski W., Ris H. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy. Science. 1969 Mar 21;163(3873):1343–1348. doi: 10.1126/science.163.3873.1343. [DOI] [PubMed] [Google Scholar]


