Abstract
Translational control plays an essential role in the regulation of gene expression. It is especially important in defining the proteome, maintaining homeostasis, and controlling cell proliferation, growth, and development. Numerous disease states result from aberrant regulation of protein synthesis, so understanding the molecular basis and mechanisms of translational control is critical. Here we outline the pathway of protein synthesis, with special emphasis on the initiation phase, and identify areas needing further clarification. Features of translational control are described together with numerous specific examples, and we discuss prospects for future conceptual advances.
The control of protein synthesis defines the proteome and maintains homeostasis in the cell and the organism. Regulation occurs at different steps of translation; the initiation step is the most common target.
Protein synthesis is an indispensable process in the pathway of gene expression, and is a key component in its control. Regulation of translation plays a prominent role in most processes in the cell and is critical for maintaining homeostasis in the cell and the organism. The synthesis rate of a protein in general is proportional to the concentration and translational efficiency of its mRNA. Translational control governs the efficiency of mRNAs and thus plays an important role in modulating the expression of many genes that respond to endogenous or exogenous signals such as nutrient supply, hormones, or stress. Because the vast majority of eukaryotic mRNAs have quite long half-lives (>2 h) (Raghavan et al. 2002), rapid regulation of the cellular levels of the proteins they encode must be achieved by controlling their mRNA translational efficiencies and protein degradation rates. During early stages of viral infection (Walsh et al. 2012) and in cells lacking active transcription such as oocytes and reticulocytes, translational control is often the only mechanism to regulate the synthesis of proteins. Furthermore, protein synthesis accounts for a large proportion of the energy budget of a cell, especially one that is rapidly growing or biosynthetically active, and therefore requires tight regulation. Because protein synthesis is intimately integrated with cell metabolism, aberrations in its regulation contribute to a number of disease states. It is therefore apparent that a detailed knowledge of the mechanisms that contribute to translational control is essential in understanding cell homeostasis and disease.
PROTEIN SYNTHESIS PATHWAY
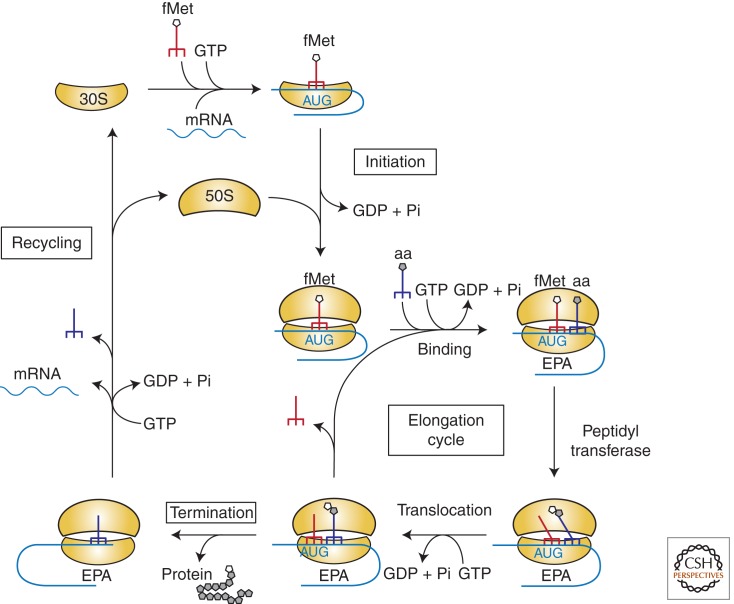
Protein synthesis is a highly conserved process that links amino acids together on ribosomes based on the sequence of an mRNA template. To appreciate the complex translation pathway in human cells, it is useful first to consider protein synthesis in bacteria. The bacterial pathway is coupled to transcription of the DNA into mRNA, made possible because no nuclear membrane separates these processes. It comprises four phases: initiation, elongation, termination, and recycling (Fig. 1). The initiation phase involves the binding of the small ribosomal subunit (30S) to an unstructured region in the mRNA (the Shine-Dalgarno region) that is complementary to a portion of the 16S rRNA, and is stabilized through an interaction between the ribosome-bound initiator formyl-methionyl-tRNA and the initiation codon, usually AUG. Although formation of the 30S initiation complex is promoted by three initiation factors, identification of the initiation site in the mRNA is based primarily on RNA–RNA interactions. The large ribosomal subunit (50S) then binds to form a 70S initiation complex, which contains the formyl-methionyl-tRNA in the ribosomal P-site and which is prepared to enter the elongation phase. Elongation involves three steps: the binding of an aminoacyl-tRNA whose anticodon is complementary to the mRNA codon in the ribosomal A-site; formation of a peptide bond by transfer of the amino acid or peptide attached to the tRNA in the P-site to the aminoacyl-tRNA in the A-site; and translocation of the newly formed peptidyl-tRNA from the A-site to the P-site, together with the mRNA. These reactions are promoted by a number of elongation factors and by the ribosome itself, with rRNA playing a particularly notable part. When a termination codon (UAA, UAG, UGA) enters the A-site, termination factors bind to the ribosome and promote the hydrolysis of the peptidyl-tRNA. Ribosomes are then recycled through interactions with a number of protein factors to generate ribosomal subunits capable of undergoing another round of protein synthesis. Detailed descriptions of the bacterial pathway are found in a number of reviews (Laursen et al. 2005; Noller 2007).
Figure 1.
Pathway of protein synthesis in bacteria. The simplified cartoon shows the four phases of protein synthesis and how the ribosomes, tRNAs, mRNA, and GTP interact. Not shown are the initiation, elongation, and termination factors that promote the reactions. Following initiation, each turn of the elongation cycle results in the addition of another amino acid (gray pentagon) to the growing peptide chain (not shown). Termination occurs when a termination codon (UAA, UAG, UGA) appears in the ribosomal A-site and involves hydrolysis of the peptidy-tRNA in the P-site. (Figure constructed by Nancy Villa, University of California, Davis.)
Protein synthesis in higher cells shares many similarities with that in bacteria. The genetic code is identical and the aminoacyl-tRNAs and their synthetases are very similar, but eukaryotic ribosomal subunits, named 40S and 60S, are larger and richer in protein, as illustrated by recent high-resolution structures (see Wilson and Cate 2012). Whereas the elongation phase is strongly conserved, the initiation and termination/recycling phases differ substantially. A conspicuous feature of eukaryotic protein synthesis is the fact that mRNAs are translated in the cytoplasm, making translation uncoupled from transcription. Mature eukaryotic mRNAs possess a m7G-cap at their 5′-terminus and, in most cases, a poly (A) tail at their 3′-terminus. They are generally monocistronic, unlike most bacterial mRNAs, and the pathway and mechanism for the formation of 40S and 80S initiation complexes differ substantially from those in bacteria. For example, a large number of initiation factors (at least 12) promote the binding of the mRNA and initiator methionyl-tRNAi (Met-tRNAi)—which is not formylated—to the 40S ribosomal subunit. Therefore 40S initiation complex formation involves numerous protein–RNA and protein–protein interactions, in contrast to what occurs in bacteria. Given the preeminence of the initiation phase in the regulation of protein synthesis, we develop the mechanism of eukaryotic initiation in considerable detail in the following section. Termination and recycling resemble the reactions in prokaryotes, except that different sets of proteins promote these phases. Eukaryotic initiation pathways are outlined in Figure 2; detailed descriptions of the molecular mechanisms are found in Hinnebusch and Lorsch (2012).
Figure 2.
Pathway of eukaryotic initiation. The simplified cartoon shows two types of initiation mechanisms (m7G-cap-dependent scanning and IRES-dependent internal), and how the ribosomes, methionyl-tRNAi, mRNA, and ATP/GTP interact. Not shown are the initiation factors or the possibility that scanning follows IRES-directed binding of the 40S ribosomal subunit during internal initiation. (Figure constructed by Nancy Villa, University of California, Davis.)
MECHANISM OF EUKARYOTIC INITIATION
To elucidate translational control mechanisms, it is essential to define the detailed molecular mechanism of protein synthesis. The major initiation pathway, scanning, involves binding of a 40S–Met-tRNAi complex to the 5′-terminus of an m7G-capped mRNA, followed by downstream scanning along the mRNA until an AUG (or near-cognate) initiation codon is recognized. The 60S ribosomal subunit then joins the 40S initiation complex to form an 80S initiation complex capable of entry into the elongation phase. These reactions are promoted by twelve or more initiation factors comprising over 25 proteins (see Hinnebusch and Lorsch 2012). Although much has been learned about how mammalian cells initiate protein synthesis, a number of gaps and uncertainties remain. For example, identification of initiation factors has been based on their stimulation of in vitro initiation assays constructed with purified components, and verified by genetic methods. However, the recent discoveries of new proteins apparently involved in the pathway (e.g., DHX29 [Parsyan et al. 2009] and DDX3 [Lai et al. 2008]) suggest that all relevant initiation factors may not have been identified. In addition, the relevance of some identified factors is uncertain. For example, eIF2A promotes the binding of Met-tRNAi to 40S ribosomal subunits, but its role in translation initiation is not well established (Komar et al. 2005; Ventoso et al. 2006). A newly identified factor, eIF2D, promotes tRNA binding into the ribosomal P-site in the absence of GTP, but its mechanism of action and role in initiation have not been defined (Dmitriev et al. 2010). eIF5A promotes protein synthesis, but whether it is involved in the initiation or elongation phase (Saini et al. 2009), or possibly just in formation of the first peptide bond, is controversial (reviewed in Henderson and Hershey 2011). eIF6 was first identified as an antiribosome association factor but its role in initiation was then questioned (Si and Maitra 1999); however, recent structural studies show clearly how binding to the nascent premature 60S subunit prevents junction with the 40S initiation complex (Klinge et al. 2011).
Despite its centrality, aspects of the scanning mechanism are not yet well elucidated. eIF4A is a well-established RNA helicase that functions while tethered to the cap-binding complex. However, does it continue to unwind RNA following 40S ribosomal subunit recruitment to the mRNA, or do other identified helicases such as DHX29 (Parsyan et al. 2009) and DDX3 (Lai et al. 2008) provide the helicase function during scanning? Are there mechanistic clues in the unusual bidirectionality of the helicase activity of eIF4A, and the departures from stoichiometry in the levels of some of the factors? Ribosome profiling methods detect initiation at numerous sites in a large number of mRNAs, some at non-AUG codons (Ingolia et al. 2011), but how such initiation events are regulated is unclear. Because rigorous kinetic analyses of many of the reactions in initiation have not been performed, we do not have a full description of their reaction rates, yet such information is essential for detecting and understanding regulation during initiation. In contrast, great progress has been made recently in elucidating the structure of eukaryotic ribosomes (see Ben-Shem et al. 2011; Klinge et al. 2011; Rabl et al. 2011; Wilson and Cate 2012), although atomic resolution structures of initiation complexes are still lacking.
Besides the scanning pathway, a sizable number of cellular mRNAs—generally estimated as 5%–10%—may use a different way to recruit the ribosome. Direct binding of the 40S ribosomal subunit to an internal region of the mRNA, called the internal ribosome entry site (IRES), bypasses the necessary recognition of the m7G cap (Fig. 2). IRES-mediated initiation is often used for the translation of viral mRNAs, and is reported for some cellular mRNAs as well (see Jackson 2012 for a critical analysis of cellular mRNAs containing IRESs). Still other initiation pathways have been described, involving shunting (Yueh and Schneider 2000; Pooggin et al. 2006), tethering (Martin et al. 2011), translation enhancers (Vivinus et al. 2001), the TISU element that frequently functions with mRNAs possessing very short 5′-UTRs (Elfakess et al. 2011), and a poly-adenylate leader in the 5′-UTR that appears to function in the absence of a number of the canonically required initiation factors (Shirokikh and Spirin 2008). Although we already possess much sophisticated knowledge of how these initiation pathways proceed, there remains much to be learned that is essential for a full understanding of translational control.
FEATURES OF TRANSLATIONAL CONTROL
Regulation of protein synthesis may occur at different steps of the pathway, with the initiation phase being the most common target. Which phase of protein synthesis is affected is often identified by determining polysome profiles (Merrick and Hensold 2000) and ribosome transit times (Fan and Penman 1970; Palmiter 1972). One of the salient features of translational control involves the number of mRNAs affected. A given mechanism might affect the translation of a single mRNA, a subset of mRNAs, or most mRNAs. Global regulation often is based on the activation or inhibition of one or more components of the translational machinery, whereas specific regulation frequently occurs through the action of trans-acting proteins (see Gebauer et al. 2012) or microRNAs (see Roux and Topisirovic 2012) binding to cis-elements in the mRNA. Some mRNAs are capable of escaping the effects of global activation or inhibition. Therefore, the response caused by a given mechanism may be complex in terms of the mRNAs affected. Methods that address this latter issue are ribosome profiling (Ingolia et al. 2009) involving high-throughput deep sequencing of ribosome-protected mRNA sequences, or DNA microarray technology, involving identification of mRNAs in size-fractionated polysomes (see Larsson et al. 2012). Such analyses of changes in ribosome profiles caused by a difference in physiological state enable identification of the mRNAs that are most affected. The polysome profiling method is particularly powerful, as it determines where ribosomes are positioned on essentially all cellular mRNAs at a specific point in time, thereby shedding light on the phase of protein synthesis that changes.
Another important feature of translational control is that a change in physiological state can activate multiple regulatory mechanisms that affect the rate of protein synthesis. Such redundancy complicates mechanistic studies, because interfering with one mechanism does not necessarily alter the overall extent of inhibition or activation. A further complication is that a given mechanism may itself cause only a minor change in protein synthesis rate. However, when multiple weak mechanisms act on the system together, significant translational control can result. Mechanisms that are modest in their action are especially difficult to elucidate, as their effects sometimes only slightly alter a specific reaction rate. To detect and assess the importance of such mechanisms, sophisticated and highly accurate kinetic analyses are required, and are increasingly being pursued.
REGULATORY MECHANISMS
Recruitment of the mRNA to the 40S ribosomal subunit is thought to be the rate-limiting step of initiation, and is often modulated. The binding of methionyl-tRNAi also is frequently regulated, and subsequent steps such as scanning and initiator codon recognition may be affected as well. How are these reactions regulated? The cellular levels of the canonical initiation factors differ in various cell or tissue types, thereby affecting initiation rates. Modulating the activities of the initiation factors by phosphorylation is often used to regulate global rates of protein synthesis. The best-studied examples are phosphorylation of eIF2α, which results in an inhibition of Met-tRNAi binding to ribosomes (see Hinnebusch and Lorsch 2012; Pavitt and Ron 2012), and phosphorylation of 4E-BPs (sequestors of the cap-binding protein, eIF4E), which relieves translational repression caused by decreased mRNA recognition and binding to ribosomes (see Hinnebusch and Lorsch 2012; Roux and Topisirovic 2012). Numerous other initiation factors are phosphorylated, often as targets of signal transduction pathways, as are ribosomes and the elongation factor eEF2, but how such events regulate protein synthesis is not yet well established. Besides phosphorylation, posttranslational modifications such as methylation, ubiquitination, and glycosylation, may affect protein synthesis, but these have not been studied extensively. One can anticipate that mass spectrometric methods will identify new modifications of importance in the near future.
mRNA levels appear not to be rate-limiting for global protein synthesis in many cells, as a substantial number of mRNAs are found as untranslated, free mRNPs rather than in active polysomes. However, mRNAs also can be sequestered in stress granules or P-bodies (see Decker and Parker 2012) or localized in specific regions of a cell’s cytoplasm (see Chao et al. 2012; Lasko 2012), indicating that mRNA accessibility can influence the efficiency of translation.
Regulation of translation through the action of microRNAs is an exciting new area of study. MicroRNAs can stimulate the degradation of mRNAs or affect protein synthesis directly (see Braun et al. 2012). The mode of regulation is mRNA-specific, although a single microRNA may affect a number of different mRNAs. Precise mechanisms whereby the microRNAs affect protein synthesis are yet to be elucidated. Recent in vitro experiments detecting early microRNA-based inhibition of protein synthesis prior to mRNA deadenylation may resolve the controversy of which effect is dominant (Fabian et al. 2009).
Trans-acting proteins affect initiation rates by binding to specific mRNAs and interfering with various steps of the pathway (see Gebauer et al. 2012). Such proteins frequently recognize a binding site in the 3′-UTR, yet affect events occurring near the 5′-m7G cap. These regulatory mechanisms often function during early development (see Gebauer et al 2012; Lasko 2012) via protein-mediated crosstalk between the two ends of the mRNAs. Indeed, active mRNAs are thought to be circularized through an interaction between the poly (A)-binding protein (PABP) and eIF4G, which is a component of the m7G-cap-binding complex (Wells et al. 1998). Some mRNAs, for example the histone mRNAs that lack a poly (A) tail, are circularized through specialized proteins that bind near the 3′-terminus of the mRNA and react with the cap-binding protein complex (Cakmakci et al. 2008). However, mutant yeast lacking the PABP-eIF4G interaction show normal translation rates (Park et al. 2011), which suggests that circularization does not invariably promote initiation, at least not in yeast. Another possibility, not yet established, is that circularization enhances the ability of a terminating ribosome to reinitiate on the same mRNA, perhaps by a mechanism that differs from the canonical scanning mechanism. During analyses of the generation of polysomes in vitro, the rate of addition of a new ribosome to a polysome was slower than the ribosome transit time, yet polysome size increased, suggesting that ribosomes already present on a polysome reinitiate more efficiently on the same mRNA than new ribosomes initiate (Nelson and Winkler 1987).
Although less commonly reported, the elongation and termination phases (see Dever and Green 2012) also are targets of translational control. The rate of elongation is thought to be maximal under most conditions (Ingolia et al. 2011), but can be inhibited by specific mechanisms. Whether such inhibition affects the elongation rates of all mRNAs equally is not well established. Only when elongation is slowed sufficiently, such that initiation is no longer rate-limiting, is the rate of protein synthesis affected. The occurrence of rare codons or strong secondary structure in the coding region of a mRNA is thought to slow the rate of elongation. Some mRNAs can thereby be induced to undergo a shift in reading frame at a specific region, generating a protein whose sequence and length differ from the unshifted one. For proteins that are to be inserted into the endoplasmic reticulum, their elongation is paused by the signal recognition particle, enabling the amino terminus of the nascent protein to dock onto the endoplasmic reticulum, after which elongation resumes (see Benham 2012). The rate of elongation affects the folding of proteins (see Cabrita et al. 2010); if elongation is too fast, e.g., when recombinant eukaryotic proteins are synthesized in bacteria, many proteins fail to fold properly unless the overall rate is reduced (Siller et al. 2010). Alternatively, slowing the elongation rate at specific regions of the mRNA may enhance proper folding (Zhang et al. 2009), further demonstrating the link between rates or elongation and protein folding. Furthermore, the folding of the nascent protein as it emerges from the large ribosomal subunit can affect the elongation rate, either positively or negatively (see Cabrita et al. 2010).
The termination phase also may be regulated (reviewed in Dinman and Berry 2007). Under most circumstances, termination is not rate-limiting for protein synthesis, because ribosomes are not found stacked up at the ends of mRNAs. However, termination can be suppressed, enabling either frame-shifting or read-through to occur, thereby extending the nascent protein at its carboxyl terminus. The UGA stop codon can be reprogrammed to enable the insertion of a seleno-cysteine residue rather than to terminate synthesis. Incorporated into proteins by the translational process itself, selenocysteine has been called the “21st amino acid,” and it is now followed by the 22nd—pyrrolysine, encoded by a UAG codon in some methanogenic archaea and bacteria (Atkins and Gesteland 2002). Such “amendments” to the elongation and termination steps are influenced by the sequence context of the codon, or by other features of the mRNA.
FUTURE PROSPECTS
New discoveries of the involvement of translational control in cell metabolism, proliferation and disease are being reported constantly. The ribosome profiling method has already identified unexpected changes in the translation of numerous specific mRNAs and can be expected to generate a vast amount of new data. Handling the plethora of information requires new and sophisticated bioinformatic methods that are rapidly being developed and refined (see Larsson et al. 2012). A challenge is presented by the proliferation of gene products, including alternatively processed mRNAs and protein isoforms, produced in higher cells. This diversification introduces additional levels of complexity that need to be accommodated in these analyses. Such high-throughput approaches do not generally elucidate details of the molecular mechanisms involved, however. To understand the observed changes in mRNA translation, many of them rather modest in extent, it is necessary to have a precise knowledge of the mechanism of protein synthesis.
What are the major challenges for understanding translational control mechanisms? We already have a fairly detailed description of the process of protein synthesis during the initiation, elongation, termination, and recycling phases. With the recent ability to obtain crystals of eukaryotic ribosomes (Ben-Shem et al. 2011; Klinge et al. 2011; Rabl et al. 2011), we can anticipate atomic level structures of ribosome complexes that are essential for describing how peptide bonds are formed and how the various factors interact on the surface of the ribosome to promote initiation, elongation, and termination. However, crystals of 40S and 80S initiation complexes have eluded researchers, and even cryo-electron microscopic approaches have not yet yielded sufficiently precise structures of these important intermediates. Another area lacking structural information at high resolution pertains to mRNAs. Although computer programs can predict structural motifs in RNA (Cruz and Westhof 2011), the actual 3-dimensional structures, especially those of the 5′-UTR, are only now beginning to be determined (Steen et al. 2011). Such detailed structures of mRNAs and their native mRNP complexes are eagerly awaited, as they surely are important for mRNA binding, scanning and initiator codon recognition during initiation. mRNA structures also affect the elongation and termination rates, thereby affecting protein folding and the regulation of frameshifting. So high-resolution mRNA structural information, especially pertaining to the initiation phase, is needed.
Another area in which our knowledge is limited pertains to the kinetics of the various reactions, interactions and conformational changes involved in protein synthesis. The elongation phase is relatively well characterized, especially for prokaryotes, but there are numerous initiation steps that are yet to be studied in detail. Insights into the kinetics of initiation complex formation have been gained from studies primarily performed with yeast components (reviewed in Lorsch and Dever 2010 and Hinnebusch and Lorsch 2012), yet much is yet to be learned. Do initiation factors form subcomplexes off the ribosome, or do they first bind to the 40S subunits, and if so, in what order? Which proteins mediate the binding of Met-tRNAi to 40S ribosomes, and how is that rate affected by other initiation factors? What is the rate of ribosome scanning of the 5′-UTR, and is this rate affected by changes in the activities of associated initiation factors, e.g., those involved in RNA helicase activity? Why do ribosomes appear to idle at the initiator AUG codon? That is, what limits their rapid progression into the elongation phase? Most kinetic analyses average the effects of many molecules over time. The application of single molecule studies to the kinetics of protein synthesis (see Petrov et al. 2012) likely will generate new views of how such reactions proceed. Additional work employing both single molecule and standard kinetic methods are needed to properly recognize and evaluate many of the translational control mechanisms that operate at the initiation phase, especially those mechanisms that only marginally affect reaction rates.
Complementing our constantly increasing understanding of the molecular mechanisms of translational control is the expectation that more and more examples of regulation at the level of protein synthesis will be discovered. A number of promising areas of research are featured in this volume. We anticipate that regulation by microRNAs will prove to be important for the translation for most mRNAs (Braun et al. 2012). How does the secretion or cotranslational folding of nascent proteins affect their synthesis (Cabrita et al. 2010; Benham 2012)? Other areas in which translational control already is firmly established are described in literature dealing with cell development (Lasko 2012), cancer (Ruggero 2012), synaptic plasticity and memory (Darnell and Richter 2012), and viruses (Walsh et al. 2012). As translational control mechanisms are better understood and as high throughput methods identify when such regulation occurs, we can confidently anticipate that we will learn of additional areas of cellular metabolism that are modulated through protein synthesis. Indeed, it is becoming clear that translational control and transcriptional control are comparably important in regulating gene expression.
The relevance and importance of protein synthesis in disease and medicine is increasingly recognized and appreciated. The dysregulation of protein synthesis in a specific disease provides a target for therapeutic intervention (see Malina et al. 2012). As our knowledge of the structures and detailed mechanisms of protein synthesis improve, this information can be applied to enable more rational drug design. Therefore, research in the area of translational control will contribute to a better understanding of many disease states and to the development of novel therapeutic agents.
Footnotes
Editors: John W.B. Hershey, Nahum Sonenberg, and Michael B. Mathews
Additional Perspectives on Protein Synthesis and Translational Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Atkins J, Gesteland R 2002. The 22nd amino acid. Science 296: 1409–1410 [DOI] [PubMed] [Google Scholar]
- *.Benham AM 2012. Protein secretion and the endoplasmic reticulum. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- *.Braun JE, Huntzinger E, Izaurralde E 2012. A molecular link between miRISCs and deadenylases provides new insight into the mechanism of gene silencing by microRNAs. Cold Spring Harb Perspect Biol 10.1101/cshperspecta012328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita DL, Dobson CM, Christodoulou J 2010. Protein folding on the ribosome. Curr Opin Struct Biol 20: 33–45 [DOI] [PubMed] [Google Scholar]
- Cakmakci N, Lerner R, Wagner E, Zheng L, Marzluff W 2008. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol Cell Biol 28: 1182–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chao JA, Yoon YJ, Singer RH 2012. Imaging translation in single cells using fluorescent microscopy. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J, Westhof E 2011. Sequence-based identification of 3D structural modules in RNA with RMDetect. Nat Methods 8: 513–521 [DOI] [PubMed] [Google Scholar]
- *.Darnell J, Richter J 2012. Cytoplasmic RNA binding proteins and the control of complex brain function. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Decker CJ, Parker R 2012. P bodies and stress granules and their possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dever TE, Green R 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J, Berry M 2007. Regulation of Termination and Recoding. In Translational control in biology and medicine (ed. Mathews MB, Sonenberg N, Hershey JWB), pp. 625–654 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Dmitriev S, Terenin I, Andreev D, Ivanov P, Dunaevsky J, Merrick W, Shatsky I 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R 2011. Unique translation initiation of mRNAs containing TISU element. Nucleic Acids Res 39: 7598–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M, Mathonnet G, Sundermeier T, Mathys H, Zipprich J, Svitkin Y, Rivas F, Jinek M, Wohlschlegel J, Doudna J, et al. 2009. Mammalian miRNA RISC recruits CAP1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Penman S 1970. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J Mol Biol 50: 655–670 [DOI] [PubMed] [Google Scholar]
- *.Gebauer F, Preiss T, Hentze MW 2012. From cis-regulatory elements to complex RNPs and back. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A, Hershey J 2011. The role of eIF5A in protein synthesis. Cell Cycle 10: 3617–3618 [DOI] [PubMed] [Google Scholar]
- *.Hinnebusch AG, Lorsch JR 2012. The mechanism of eukaryotic translation initiation: New insights and challengs. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N, Ghaemmaghami S, Newman J, Weissman J 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N, Lareau L, Weissman J 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Jackson R 2012. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948 [DOI] [PubMed] [Google Scholar]
- Komar A, Gross S, Barth-Baus D, Strachen R, Hensold J, Goss Kinzy T, Merrick W 2005. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J Biol Chem 280: 15601–15611 [DOI] [PubMed] [Google Scholar]
- Lai M, Lee Y, Tarn W 2008. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell 19: 3847–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Larsson O, Tian B, Sonenberg N 2012. The genome-wide landscape of translational control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lasko P 2012. mRNA localization and translational control in Drosophila. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen B, Sorensen H, Mortensen K, Sperling-Petersen H 2005. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69: 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch J, Dever T 2010. Molecular view of 43S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem 285: 21203–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Malina A, Mills JR, Pelletier J 2012. Emerging therapeutics targeting mRNA translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Barends S, Jaeger S, Schaeffer L, Prongidi-Fix L, Eriani G 2011. Cap-assisted internal initiation of translation of histone H4. Mol Cell 41: 197–209 [DOI] [PubMed] [Google Scholar]
- Merrick W, Hensold J 2000. The use of sucrose gradients in studies on eukaryotic translation. In Current protocols in cell biology, pp. 11.19.11–11.19.26 John Wiley & Sons, New York [Google Scholar]
- Nelson E, Winkler M 1987. Regulation of mRNA entry into polysomes. Parameters affecting polysome size and the fraction of mRNA in polysomes. J Biol Chem 262: 11501–11506 [PubMed] [Google Scholar]
- Noller H 2007. Structure of the Bacterial Ribosome and Some Implications for Translational Regulation. In Translational control in biology and medicine (ed. Mathews MB, Sonenberg N, Hershey JWB), pp. 41–58 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Palmiter R 1972. Regulation of protein synthesis in chick oviduct. II. Modulation of polypeptide elongation and initiation rates by estrogen and progesterone. J Biol Chem 247: 6770–6780 [PubMed] [Google Scholar]
- Park E, Walker S, Lee J, Rothenburg S, Lorsch J, Hinnebusch A 2011. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1-PABP mRNPs in vivo. EMBO J 30: 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A, Shahbazian D, Martineau Y, Petroulakis E, Alain T, Larsson O, Mathonnet G, Tettweiler G, Hellen C, Pestova T, et al. 2009. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc Natl Acad Sci 106: 22217–22222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pavitt GD, Ron D 2012. New insights into translational regulation in the endoplasmic reticulum unfolded protein response. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Petrov A, Chen J, O’Leary S, Tsai A, Puglisi JD 2012. Single-molecule analysis of translational dynamics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooggin M, Ryabova L, He X, Futterer J, Hohn T 2006. Mechanism of ribosome shunting in rice tungro bacilliform pararetrovirus. RNA 12: 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331: 730–736 [DOI] [PubMed] [Google Scholar]
- Raghavan A, Orgilvie R, Reilly C, Abelson M, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen P 2002. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res 30: 5529–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Roux PP, Topisirovic I 2012. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ruggero D 2012. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler D, Green R, Dever T 2009. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokikh N, Spirin A 2008. Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc Natl Acad Sci 105: 10738–10743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Maitra U 1999. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol Cell Biol 19: 1416–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller E, DeZwaan D, Anderson J, Freeman B, Barral J 2010. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J Mol Biol 396: 1310–1318 [DOI] [PubMed] [Google Scholar]
- Steen K, Siegfried N, Weeks K 2011. Selective 2′-hydroxyl acylation analyzed by protection from exoribonuclease (RNase-detected SHAPE) for direct analysis of covalent adducts and of nucleotide flexibility in RNA. Nat Protoc 6: 1683–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I, Sanz M, Molina S, Berlanga J, Carrasco L, Esteban M 2006. Translational resistance of late alphavirus mRNA to eIF2α phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 20: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivinus S, Baulande S, van Zanten M, Campbell F, Topley P, Ellis J, Dessen P, Coste H 2001. An element within the 5′ untranslated region of human Hsp70 mRNA, which acts as a general enhancer of mRNA translation. Eur J Biochem 268: 1908–1917 [DOI] [PubMed] [Google Scholar]
- *.Walsh D, Mathews MB, Mohr I 2012. Tinkering with translation: Protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S, Hillner P, Vale R, Sachs A 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2: 135–140 [DOI] [PubMed] [Google Scholar]
- *.Wilson DN, Cate JHD 2012. The structure and function of the eukaryotic ribosome. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueh A, Schneider R 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev 14: 414–421 [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z 2009. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16: 274–280 [DOI] [PubMed] [Google Scholar]