Abstract
Reactive oxygen species are increasingly perceived as players in plant development and plant hormone signalling pathways. One of these species, superoxide, is produced in the apoplast by respiratory burst oxidase homologues (rbohs), a family of proteins that is conserved throughout the plant kingdom. Because of the availability of mutants, the focus of research into plant rbohs has been on Arabidopsis thaliana, mainly on AtrbohD and AtrbohF. This study investigates: (i) a different member of the Atrboh family, AtrbohB, and (ii) several rbohs from the close relative of A. thaliana, Lepidium sativum (‘cress’). Five cress rbohs (Lesarbohs) were sequenced and it was found that their expression patterns were similar to their Arabidopsis orthologues throughout the life cycle. Cress plants in which LesarbohB expression was knocked down showed a strong seedling root phenotype that resembles phenotypes associated with defective auxin-related genes. These transgenic plants further displayed altered expression of auxin marker genes including those encoding the auxin responsive proteins 14 and 5 (IAA14 and IAA5), and LBD16 (LATERAL ORGAN BOUNDARIES DOMAIN16), an auxin-responsive protein implicated in lateral root initiation. It is speculated that ROS produced by rbohs play a role in root development via auxin signalling.
Key words: AtrbohB, auxin, Lepidium sativum, Rboh, reactive oxygen species, RNAi, root development, superoxide
Introduction
Over the last decade, widespread developmental roles for the reactive oxygen species (ROS) superoxide, hydrogen peroxide, and hydroxyl radicals throughout the plant life cycle have emerged (reviewed by Gapper and Dolan, 2006; Swanson and Gilroy, 2010). All three of these ROS are known to be produced in the apoplast of plant cells where they can participate in pathogen responses (Torres et al., 2006), biophysical changes in cell walls (i.e. weakening or strengthening) (Schopfer et al., 2002; Müller et al., 2009a ), and as mediators of systemic signalling (Miller et al., 2009).
Apoplastic superoxide can be produced by respiratory burst oxidase homologues (rbohs). These proteins are named after the homology of their catalytic domain to the mammalian respiratory burst oxidase subunit gp91phox (Keller et al., 1998). Electron transport and superoxide production occur when gp91phox accepts electrons intracellularly from NADH or NADPH and transfers them to extracellular O2. Rbohs consist of only one polypeptide with a gp91phox-homologous domain as a catalytic centre. They also contain six conserved transmembrane domains, a cytoplasmic FAD-binding domain and an N-terminal cytoplasmic domain with Ca2+-binding EF-hand motifs (Keller et al., 1998).
The first role to be characterized for rbohs was in ROS production during pathogen defence (reviewed by Torres et al., 2006). Since then, multiple roles of rboh activity have been identified in a variety of processes, including seedling elongation growth (Frahry and Schopfer, 2001), tip growth of root hairs (Foreman et al., 2003; Monshausen et al., 2007) and pollen tubes (Potocky et al., 2007), seed germination (Müller et al., 2009b ), and mechanosensing (Monshausen et al., 2009). Recently, the rbohs of Medicago truncatula have been linked to symbiotic nodule formation (Marino et al., 2011), and apoplastic superoxide as their reaction product has been linked to environmentally regulated allelopathic plant–plant interactions (Oracz et al., 2012). Rbohs thus seem to be involved in a large variety of developmental processes and responses to internal and external stimuli.
To date, ten rbohs (AtrbohA–AtrbohJ) have been identified in Arabidopsis. Of those, only three, namely AtrbohC, AtrbohD, and AtrbohF, have been formally characterized. AtrbohC was discovered as being allelic to Root Hair Deficient2 (RHD2). The rhd2 mutant exhibits impaired root hair growth, a phenotype associated with a reduced production of apoplastic superoxide (Foreman et al., 2003). The exact function of the apoplastic superoxide in root hair growth is unclear. However, a complex pattern of oscillating Ca2+, pH, and ROS pulses accompanies the growth process (Monshausen et al., 2007). AtrbohD and F have functions in plant defence responses to biotic stresses (Torres et al., 2002; Torres and Dangl, 2005). They also function in guard cells (Kwak et al., 2003); atrbohf/atrbohd double mutants display an inhibition of the ABA-induced increase in ROS and cytosolic Ca2+ and subsequent stomatal closure. The protein encoded by AtrbohD, which is ubiquitously expressed in Arabidopsis plants, is involved in systemic signalling by apoplastic ROS, which is triggered by wounding as well as abiotic stresses such as heat, cold, high-intensity light, and high salinity (Miller et al., 2009).
AtrbohC and AtrbohD, together with AtrbohA and AtrbohG, belong to the same phylogenetic group as determined by the construction of a phylogenetic tree based on rboh protein sequences from five species (Glycine max, Lotus japonicas, Oryza sativa, Arabidopsis thaliana, and Medicago sativa) (Marino et al., 2011). According to the tree, AtrbohF belongs to a second group together with AtrbohI, while AtrbohB is part of a separate group and is the only Arabidopsis rboh in its group, as is AtrbohE. Finally, AtrbohH and AtrbohJ are in the last group. It is of interest to characterize the rbohs from different phylogenetic groups in order to get a more complete picture of rboh functions, and to determine if the different groups have acquired different functions during evolution.
Müller et al. (2009b) elucidated a function of AtrbohB in seed after-ripening. AtrbohB is alternatively spliced in seeds depending on the after-ripening status of the seeds and the presence or absence of exogenous ABA. Our studies of AtrbohB are now extended to a characterization of its expression in other tissues and in other plant species. In order to explore the evolutionary conservation of rboh sequences and expression patterns, we looked at a close relative of Arabidopsis in our study: the Brassicaceae Lepidium sativum (garden cress, ‘cress’). Cress has several strategic advantages over Arabidopsis, such as seeds that are 40-50 times larger. Cress seedlings can easily be separated into their individual organs and tissues (Müller et al., 2009a ; Linkies et al., 2009). Five cress Rbohs (Lesarbohs) from separate phylogenetic groups were completely or partially cloned and sequenced and RNAi-lines were created for which LesarbohB expression is knocked down. Our phenotypic analysis of the transgenic lines points to a role for RbohB in cress seedling root development and in fertility.
Materials and methods
Plant materials
Arabidopsis plants (Columbia wild type, WT), enhancer-trap line AtrbohB-GUS (line CS24365 in Liu et al., 2005), and atrbohB, (all lines described in Müller et al., 2009b ) were grown in soil at 22 ºC under a 16-h photoperiod.
To generate the seedlings for stress treatments, mature seeds were plated on half-strength Murashige-Skoog (MS) media, pH 6.5, solidified with 1% agar and the plates were placed in germination conditions (22 ºC under a 16 h photoperiod). After 48 h, when all seeds had completed germination, the young seedlings were transferred to plates containing 2.5 mM H2O2 (oxidative stress), 150 mM NaCl (salt stress), or 6% mannitol (osmotic stress) for a duration of 48 h. As controls, seedlings were maintained for the 48 h on solid half-strength MS medium.
Lepidium sativum FR 14 and the lesarbohB-RNAi lines were grown in soil at 22 ºC under a 16-h photoperiod. Mature seeds were collected and plated on Petri dishes containing filter paper soaked with 6 ml of sterile water and the plates were placed in germination conditions (22 ºC under a 16-h photoperiod) to obtain seedlings.
GUS histochemical staining
AtrbohB-GUS and WT Arabidopsis materials at different stages in the life cycle were washed with deionized water, vacuum infiltrated with GUS staining solution (2 mM X-Gluc in 20 mM phosphate buffer, pH 6.2, 0.1% Triton X-100), and incubated overnight at 37 ºC with slow rotation. In some cases where low GUS activities occurred incubations were conducted for up to 3 d. The plant/seedling tissues were then transferred to 80% ethanol for several hours for destaining. The ethanol was exchanged as required. Destained plants were photographed on a lightbox with a Nikon Coolpix 4500 digital camera. WT plant and seedling tissues served as a negative control.
Cloning of Lesarbohs
Primers for the cloning of Lesarboh cDNAs were designed with the primer3 (Rozen and Skaletsky, 2000) tool in Geneious 4.8.5 based on Arabidopsis sequences as well as conserved domains of rbohs from different species. RT-PCRs were conducted on RNA extracted from cress seedlings and seeds. PCR fragments were cloned into pGEMTeasy-vectors (Promega, Karlsruhe, Germany), propagated in E. coli, extracted with a miniprep kit (Fermentas/ThermoScientific) and sequenced. Geneious software was used for alignments of sequences. 5’- and 3’-RACE was conducted with the corresponding Invitrogen kits (Invitrogen, Karlsruhe, Germany) following the manufacturer’s instructions. Sequences have been submitted to Genebank under the following accession numbers: LesarbohA JX312066, LesarbohB JX312067, LesarbohD JX312068, LesarbohF JX312069, and LesarbohH JX312070.
Generation of transgenic plants
Two fragments of the LesarbohB cDNA were amplified using the primer pairs RNAiB1for (5’-GGGG-attB1-GATGCAAAGCCACTGG TTCA-3’)/RNAiB1rev (5’-GGGG-attB1-CCAAAATTCTGATAATTC AGTTTT-3’), fragment length 437 bp, and RNAiB2for (5’-GGGG-attB1-CCCTAAATCCAAATTTGGACAT-3’)/RNAiB2rev (5’-GGGG-attB1-GGATCGAGTTCTTCCATGATT-3’), fragment length 330 bp. The fragments were cloned into Gateway vector pB7GWIWG2(I),0 via pDONR221 using the BP and LR cloning system (Invitrogen, Karlsruhe, Germany) as described in the manufacturer’s manual. The vectors were sequenced and cress plants were transformed by the floral dip method as described for Arabidopsis plants (Clough and Bent, 1998), but with only 0.01% (v/v) Silwet in the dipping solution. Transgenic T1 plants were selected by spraying BASTA on 7-d-old seedlings and repeating the spraying twice more after 4 d and 7 d.
Characterization of the fertility of transgenic plants
Reciprocal crosses between WT plants and transgenic lesarbohb-RNAiB1 and lesarbohB-RNAiB2 plants, respectively, were performed by transferring the pollen with a thin brush to the stigma of plants whose anthers had been removed to prevent self-fertilization. All flowers of eight WT-plants were pollinated with pollen from lesarbohb-RNAiB1 and lesarbohB-RNAiB2 plants, respectively. Reciprocally, all flowers on five lesarbohb-RNAiB1 and 5 lesarbohB-RNAiB2 plants were pollinated with WT pollen. The seed yield was counted after natural drying of the siliques on the plants.
RNA extraction and cDNA synthesis
Tissues of Arabidopsis or cress were ground in liquid nitrogen and total RNA was extracted as described in Chang et al. (1993) with the following modifications: After the addition of CTAB buffer (2% hexadecyl trimethyl-ammonium bromide [CTAB], 2% polyvinylpyrrolidone [MW=40 000/K30], 100 mM TRIS-HCl, pH 8.0, 25 mM EDTA, pH 8.0, 2 M NaCl, 2% β-mercaptoethanol), the extracts were kept at 65 °C for 10 min. All chloroform:isoamylalcohol extractions were repeated twice. RNA was treated with DNase-I (Fermentas, Burlington ON, Canada) to remove any remaining genomic DNA. RNA was run on an agarose gel to check for degradation, and the quantity and purity of the RNA samples were determined with a nanodrop spectrophotometer (ND-2000C, ThermoScientific, Mississauga ON, Canada). Two micrograms of RNA were reverse transcribed using the EasyScript Plus kit (abmgood, Richmond BC, Canada) with a mixture of random hexamers and oligo-dT primers. cDNA from four biological replicate RNA samples was used for qRT-PCR.
qRT-PCR
qRT-PCRs were run in 15 µl reactions on an ABI7900HT machine (Applied Biosystems, Carlsbad CA, USA) using the PerfeCTa Sybr Green Supermix with ROX (Qanta Biosciences, Gaithersburg MD, USA). Primers were designed with the primer3 (Rozen and Skaletsky, 2000) tool in Geneious 4.8.5. The reaction mixture consisted of 150 ng cDNA (RNA equivalent), 7.5 µl supermix and 140 nM of each primer and were subjected to a temperature regime of 3min at 95 °C and 40 cycles of 15 s at 95 °C and 1min at 60 °C. A dissociation curve was run after each qPCR to validate that only one product had been amplified in each well.
The efficiency E of the primer pairs was calculated as the average of the Es of the individual reactions by using raw fluorescence data with the publicly available PCR Miner tool (http://www.miner.ewindup.info/version2) (Zhao and Fernald, 2005). The efficiency was then used to calculate transcript abundance for the individual samples as (1+E)(–CT). No-template-controls were included for each primer pair to check for contamination of the reaction solutions, and no-RT-controls were used to check for genomic DNA contamination in the RNA. Actin 7 (ACT7; At5g09810) and elongation factor 1-α (EF1α; At5g60390) were used as reference genes. For both Arabidopsis thaliana and Lepidium sativum, species-specific gene primers were created and used as described in Graeber et al. (2011).
Determination of superoxide production in seedling roots
Superoxide production was measured by photometric determination of the reduction of XTT (3’-[1-phenylamino-carbonyl]-3,4-tetrazolium]-bis[4-methoxy-6-nitro] benzenesulphonic acid hydrate) (Polysciences Inc., Eppelheim, Germany) as described in Müller et al. (2009b) with the following modification: eight whole seedlings were collected for each sample in 10 mM phosphate buffer, pH 6.5. As the hypocotyls and cotyledons are covered by a cuticle, only the superoxide production from the root was measured. This was confirmed by visual inspection of the samples, which only showed red staining (indicative of XTT-reduction) in the roots.
Results
Cloning and expression analyses of Rbohs of cress
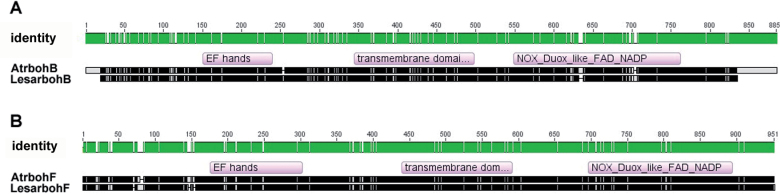
Using primers based on consensus sequences of conserved regions of Rboh genes from different species, or on the Arabidopsis gene sequences, two complete cress Rboh cDNAs were cloned. These were named LesarbohB (JX312067) and LesarbohF (JX312069) after their closest A. thaliana orthologues (Fig. 1). The predicted protein based on the LesarbohB cDNA shares 88% of its amino acid sequence with AtrbohB, which is also the highest hit in a pBLAST search. On the nucleotide level, there is 87% similarity between the two sequences. The closest pBLAST match for LesarbohF was a protein from Arabidopsis lyrata. By contrast, the highest BLAST hit on the nucleotide level was for rbohF of A. thaliana, in which there was 90% similarity. All of the major domains that rbohs are known to possess could be identified: EF-hands, transmembrane domains, and the catalytic site with its FAD and NADH/NADPH-binding pockets (Fig. 1; see Supplementary Figs. S1 and S2 at JXB online).
Fig. 1.
Alignment of the deduced amino acid sequences of the AtbohB and LesarbohB cDNAs. (A) Alignment of the AtrbohB amino acid sequence with that of its putative ortholog LesarbohB. (B) Alignment of the AtrbohF amino acid sequence with that of its putative orthologue LesarbohF. EF hand, transmembrane, and NOX/Duox like FAD/NADP binding domains of RbohB and RbohF of the two plant species were determined with the algorithm DELTA-BLAST (Domain Enhanced Lookup Time Accelerated BLAST) and are indicated in the alignment. Note that the amino acids are missing at the LesarbohB termini due to missing bases in the sequencing. Both the green and black sections in the sequences represent identical amino acids between the two proteins. The top panel with green filling also shows amino acid numbers. For an enlarged alignment detailing the specific amino acid residues, see Supplementary Fig. S1 for RbohB and Supplementary Fig. S2 for RbohF at JXB online.
Partial sequences for LesarbohA (JX312066), LesarbohD (JX312068), and LesarbohH (JX312070) were also obtained; these served as the basis for conducting qPCR analyses of gene expression. The five Lesarbohs (A, B, D, F, and H) belong to four of the five different groups on the phylogenetic tree of Marino et al. (2011) based on their sequence similarity to Atrbohs. Transcripts of these Lesarbohs were quantified by qRT-PCR analyses to examine their expression patterns and were compared with those exhibited by their Arabidopsis orthologues at different stages of the life cycle (Fig. 2). The orthologous genes of cress showed similar expression profiles: the RbohD and F orthologues were most strongly expressed in both species, followed by that corresponding to RbohB. In Arabidopsis, expression intensity of the transcripts differed by two orders of magnitude; differences were less pronounced in cress. RbohH showed weak or no expression in the various organs of both species. RbohA showed weak expression in cress; RbohA transcripts were undetectable in all tissues of Arabidopsis examined. This is in agreement with expression data published in the developmental map of the Arabidopsis eFP browser (Winter et al., 2007), which indicates that expression is exclusively in developing seeds.
Fig. 2.
Expression analysis of Lesarbohs and their Arabidopsis respiratory burst oxidase orthologues. Transcript abundance analysis for five members of the gene family determined by qRT-PCR in different plant tissues of L. sativum and A. thaliana. Note the similarities in intensity and expression profile between the two species. Averages of three biological replicates ± SE are presented. For the imbibed seed samples, seeds were harvested after 24 h on water in continuous light. Other samples included 5-d-old seedlings (5 d post-imbibition), and mature adult plants harvested 4 weeks post-germination. No AtrbohA transcript was found in any of the stages tested. Transcript abundance was corrected to the abundance of transcripts associated with the standard genes—the constitutive genes Actin 7 (ACT7; At5g09810) and Elongation Factor 1α (EF1α; At5g60390) for Arabidopsis and the corresponding genes (ACT7, HQ436350.1 and EF1α, HS981853.1) for L. sativum. n.d.=not detectable.
The expression patterns throughout the life cycle were also very similar between the two species (Fig. 2): RbohD showed strong constitutive expression, while RbohF was expressed strongly during all post-germinative stages and moderately in early-imbibed mature seeds. RbohH transcripts were found at low levels in roots and buds; maximum expression occurred in flowers. RbohB expression was low in mature seeds, but increased upon imbibition, and was most strongly expressed in seedlings and roots of adult plants in both species. Arabidopsis, but not cress, showed strong expression of RbohB in seedling cotyledons.
The expression patterns of AtrbohB throughout the life cycle of Arabidopsis was confirmed by histochemical analysis of an enhancer trap line, with a GUS coding sequence inserted 260bp upstream of the start codon for AtrbohB (Müller et al., 2009b ). GUS staining showed that expression was limited to mature embryos (Müller et al., 2009b), seedling hypocotyls and cotyledons, as well as to seedling root tips (Fig. 3). Within root tips, staining occurred mostly in the root cap and meristem areas, where cells do not undergo any major elongation (Fig. 3). GUS histochemical staining patterns corresponded well with the qRT-PCR data (Fig. 2). An exception was in relation to AtrbohB expression in adult roots (Fig. 2), which was not evident by histochemical staining (Fig. 3). Seedlings showed GUS-expression in the cotyledons even after several true leaves had been formed (Fig. 3).
Fig. 3.
GUS histochemical staining of seedlings and plants derived from an enhancer trap line with an insertion 300 bp upstream of the AtrbohB start codon. (A) Four-day-old seedling with staining in cotyledons, hypocotyl, and root tip. (B) Ten-day-old plantlet. Staining remains visible in cotyledons. (C) Enlarged image of root tip of 4-d-old seedling. Note that staining is confined to the cells which do not undergo elongation. (D) Plant at 24 d. No major areas of the adult plant show staining. The inset image shows that the cotyledons are still stained.
Intron retention in the AtrbohB gene is regulated by developmental and hormonal cues in Arabidopsis seeds (Müller et al., 2009b ). Therefore it was investigated if alternative splicing was evident in seedling tissues in which AtrbohB gene expression occurred (Figs 2,3). Alternative splicing of AtrbohB and LesarbohB transcripts was examined in response to ABA treatment of seedlings and seeds, and in response to stress treatments of seedlings (salt stress: 150 mM NaCl; osmotic stress: 6% mannitol, and oxidative stress: 2.5 mM H2O2) compared with the controls. Analyses conducted on Arabidopsis confirmed the phenomenon of alternative splicing of AtrbohB in the ABA-treated seeds; however no alternative splicing was observed in stress-exposed Arabidopsis seedlings. Further, no alternative splicing of the LesarbohB transcripts occurred in cress seeds and seedlings under any of the conditions.
Knock-down of Lesarboh B expression leads to reduced fertility and abnormal seedling root development in cress
In order to elucidate putative developmental roles of LesarbohB, the expression of the LesarbohB gene in cress was knocked down using two different RNAi-constructs, RNAiB1 and RNAiB2. Although different in length, both constructs target the sequences specifying a unique portion of the N-terminus of LesarbohB. Expression of both constructs had the same developmental effects. Following Agrobacterium-mediated floral dip transformation and BASTA selection, several independent seed-bearing lines were generated for each construct. BASTA-resistant T1-plants for both constructs produced less seeds than WT plants grown in parallel, with the transgenic lines producing an average of 19±4 seeds per plant as compared to 54±9 for the WT. Twenty-two per cent of the BASTA-resistant plants flowered, but produced no seeds; these were not included in the seed number calculation. In order to narrow down the possible cause of the reduced fertility, reciprocal crosses were made between the transgenic T1 plants, which are expected to be hemizygous, and WT plants. Pollinating a transgenic plant with WT pollen significantly increased the seed number compared with plants that were allowed to self-pollinate, and yielded an average of 35±7 seeds per plant. When WT plants were fertilized with pollen derived from transgenic plants, the seed numbers were fewer (24±8). It was therefore hypothesized that the pollen of the transgenic plants carrying the RNAi-construct may be impaired in its ability to effect pollination or fertilization. No other visible phenotype was identified in the adult plants and the dormancy and germination of seeds produced on plants displaying reduced fertility did not differ from the WT.
Strikingly, many of the T2 seedlings from transgenic lines of lesarbohb-RNAiB1 and lesarbohB-RNAiB2 showed severe developmental defects of their roots that appeared at about 5 d after germination: the lower part of the root was defective in tissue organization and turned into a thin thread-like structure with a thickened root tip (‘stage 1’; Fig. 4B). The thickened root tip fell off after a few days, and the root developed irregular lateral roots and callus-like tissues (‘stage 2’; Fig. 4C-E). Most of the affected seedlings dried out and died after transfer to soil.
Fig. 4.
Root phenotype of seedlings derived from Lesarboh-RNAi-knock-down lines. (A) Root of WT seedling. (B) Root of lesarbohB-RNAiB1 seedling 5 d after completion of germination: representative of phenotype stage 1. The lower part of the root is defective in tissue organization and turned into a thin thread-like structure with a thickened root tip. (C–E) Roots of lesarbohB-RNAiB1 seedlings, 10 d after completion of germination: representative of phenotype stage 2. The thickened root tips have fallen off, and the roots have developed irregular lateral roots and callus-like tissues. (F) Superoxide production as measured by XTT-reduction in WT seedlings and in the RNAi-seedlings at stages 1 and 2 of the root phenotype as indicated. (G) Expression of Lesarbohs in T2 seedlings that showed the root phenotype (“short roots”) versus seedlings that have WT-like roots (‘long roots’). Transcript abundance was corrected to the abundance of transcripts associated with the L. sativum standard genes Actin 7 (ACT7, HQ436350.1) and Elongation Factor 1α (EF1α, HS981853.1). Averages of 5 biological replicates ± SE are shown.
A comparison of the LesarbohB transcript level between WT-seedlings versus T2 seedlings with WT-like roots versus those with severely affected roots showed that expression was strongly reduced in the latter (Fig. 4G). This was associated with reduced superoxide production in the affected seedlings: superoxide production as measured by an XTT assay was significantly reduced in stage 1 seedlings. The generation of this ROS was higher once the unstructured lateral root growth set in and stage 2 was reached, but did not reach WT levels (Fig. 4F). There is no phenotype of comparable severity in any atrboh single mutant (Müller et al., 2009b ). The expression of the four other Lesarbohs, for which the respective sequences were cloned, was therefore tested. No significant changes in LesarbohA, LesarbohD, and LesarbohH transcript levels occurred; however, seedlings with defective root development showed a lower expression of LesarbohF than WT and unaffected seedlings (Fig. 4G). The effect on LesarbohF expression was much less pronounced than the effect on LesarbohB expression.
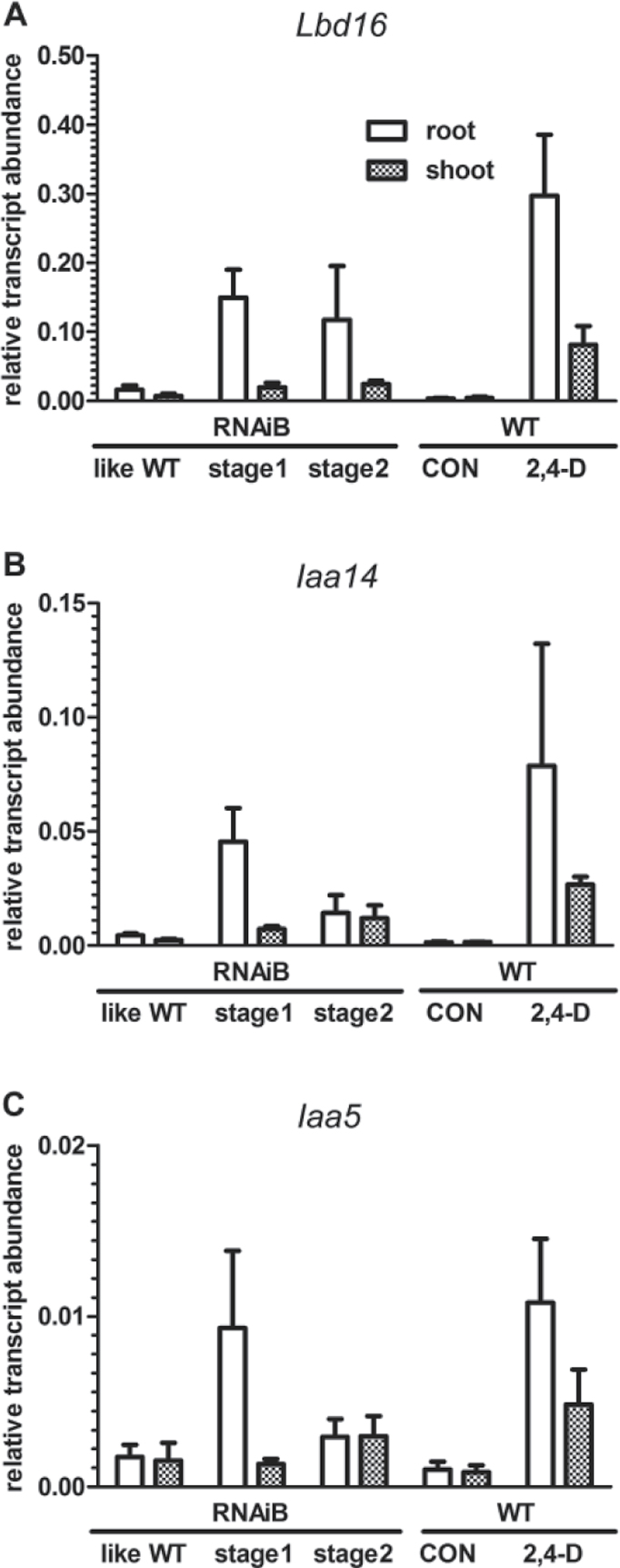
The root phenotype associated with the RNAi lines strongly resembled that connected with defective auxin-modulated processes (see Discussion). Therefore it was tested if Lesarboh-B and -F expression could be stimulated by auxin in WT seedlings. However, no statistically significant (P <0.05) change of expression was observed in seedling roots or shoots in response to 1h or 24 h exposure to the synthetic auxin 2,4-D (data not shown). LesarbohB and LesarbohF are therefore likely not directly regulated by auxin. It was also of interest to determine if the expression of auxin-marker genes (Paponov et al., 2008) was altered in the roots and shoots of seedlings of the RNAi lines that displayed the abnormal phenotypes. The expression of the marker genes was compared in the RNAi seedlings whose roots were in stages 1 and 2 of the phenotype (RNAiB stage1/2), with transgenic seedlings of equivalent age whose roots did not show a phenotype (RNAiB-likeWT), and with WT seedlings of equivalent age (Fig. 5). All transcripts were auxin-inducible as tested by the exposure of seedlings to 1 µM of the synthetic auxin 2,4-D for 24 h (Fig. 5, and data not shown). No differences in expression between WT and RNAi-lines were observed for the genes encoding the auxin transporter PIN1, the auxin-response factor ARF5, and the downstream effectors CKX6 (a cytokinin oxidase), and LBD29 (LATERAL ORGAN BOUNDARIES DOMAIN29). However, there were clear differences in the expression levels of genes encoding other auxin signalling components—auxin responsive proteins 14 and 5 (IAA-14 and -5), and LBD16, an auxin-responsive protein implicated in lateral root initiation (Okushima et al., 2007). IAA5 and IAA14 transcripts were induced in roots and shoots of the RNAi-seedlings in stages 1 and 2, with IAA5 showing strong induction particularly during stage 1 (Fig. 5C). LBD16 was induced strongly in roots of the lesarbohB-RNAi line (stages 1 and 2) compared with its control counterparts (WT- and RNAiB-like WT-seedlings); induction in shoots was less marked (Fig. 5A). The addition of exogenous auxin (0.5–2.5 µM 2,4-D or IAA) to the growth medium of WT Lepidium seedlings did not mimic the lesarbohB RNAi phenotype: exogenous auxin led to shortened roots, but did not produce the impaired root development that was observed in our RNAi-lines (data not shown). It was hypothesized that this is caused by the fact that the RNAi-effect is specific to the cells where RbohB is usually expressed, while exogenous auxin affects all cells in the root.
Fig. 5.
Expression of auxin-related genes in roots and shoots of L. sativum seedlings of wild-type and Lesarboh-RNAi knockdown lines. Transcript abundance for the genes encoding the LOB-domain containing protein (LBD) 16 (A), the auxin-responsive-protein, IAA14 (B) and the auxin-responsive protein IAA5 (C) as determined by qRT-PCR. Stage 1 corresponds to the phenotype shown in Fig. 4B, while stage 2 corresponds to the phenotype shown in Fig. 4C–E. Transcripts in wild-type seedlings were examined under 2,4-D treatment (24 h on 1 µM 2,4-D) and control conditions. Transcript abundance was corrected to the abundance of transcripts associated with the standard genes – the constitutive genes Actin 7 (ACT7; At5g09810) and Elongation Factor 1α (EF1α; At5g60390). Averages of three biological replicates ± SE are presented.
Discussion
Rbohs have been identified and described in a number of angiosperm species such as those belonging to the Brassicaceae (Arabidopsis thaliana) (Keller et al., 1998; Torres et al., 1998), the Solanaceae (Solanum tuberosum and S. lycopersicum; potato and tomato) (Kobayashi et al., 2006; Sagi et al., 2004), and in various other dicotyledonous and monocotyledonous plants such as Medicago truncatula (Marino et al., 2011), Citrullus colocynthis (Si et al., 2010), Nicotiana benthamiana (Yoshioka et al., 2003), Oryza sativa (Groom et al., 1996), Hordeum vulgare (Trijillo et al., 2006), and Zea mays (Lin et al., 2009). Sequence information retrieved from the NCBI databases shows that rbohs also exist in a number of other angiosperms, as well as in other evolutionary groups such as gymnosperms, the lycophyte Selaginella moellendorffii and the moss Physcomitrella patens.
While very similar expression patterns were observed between the orthologuous rbohs of cress and Arabidopsis, the expression patterns of rbohs from other more evolutionarily distant species (e.g. rice, alfalfa, and soybean) cannot be predicted based on their phylogenetic groups classified by Marino et al. (2010). For example, the rice Rboh genes, OsrbohD and OsrbohE, which are normally grouped with AtrbohH (an Rboh with an expression maximum in mature flowers of Arabidopsis), exhibit strong expression peaks in seeds and leaves, respectively, and only OsrbohE shows expression in flowers according to the rice eFP-browser (data from Jain et al., 2007).
It was of interest to elucidate the putative physiological function of LesarbohB by knocking-down the expression of the corresponding gene, a process which appeared to down-regulate at least one additional Lesarboh gene. The two major phenotypes that were observed in our RNAi-lines were reduced fertilization success and abnormal seedling root development. The flowers of the transgenic plants were anatomically normal, but their pollen was markedly less successful than WT pollen in terms of its ability to effect pollination or fertilization. This might be due to a reduced expression of rbohs in the pollen, as ROS play a role in pollen tube initiation (Speranza et al., 2012) and elongation (Potocky et al., 2007). In fact, an antisense construct against rbohs leads to decreased ROS production and pollen tube elongation in Nicotiana tabacum (Potocky et al., 2007), and reduced pollen tube initiation and elongation would indirectly affect fertilization success by preventing the pollen from reaching the egg cell. It is speculated that this is the case in our RNAi lines.
The root phenotype that we observed in the RNAi transgenic seedlings has not been described so far for a plant affected in Rboh-expression. Two stages in the defective root phenotype were noted: the first stage was characterized by the disintegration of the root leaving a thickened root tip that eventually died. This resembles the phenotype described for over-expression of genes encoding the serine/threonine kinase PINOID (Benjamins et al., 2001), so named in turn for its resemblance to the pin mutants, which are affected in polar auxin transport. PINOID is a positive regulator of polar auxin transport to the root tip.
The second stage of the root phenotype – the unstructured lateral root emergence – is somewhat reminiscent of the phenotype observed by Pasternak et al. (2005) in roots of seedlings that have been exposed to chemicals that cause oxidative stress, as well as in auxin-connected phenotypes. Over-expression of the auxin-response factor ARF5 in whole seedling roots or pericycle cells of a solitary-root (slr)-mutant background is associated with an inability of cells to initiate lateral roots because of a decreased auxin sensitivity; this leads to irregularly spaced and fused lateral roots (De Smet et al., 2010). Indeed these root characteristics were also observed in our RNAi transgenic lines. Irregular lateral root formation also occurs in the superroot (sur1)-mutants, mutants associated with increased endogenous auxin levels (Boerjan et al., 1995). Our transgenics thus exhibit a phenotype that resembles an enhanced response of the root to auxin.
The known functions of AtrbohD and AtrbohF include ABA-, methyl jasmonate-, and ethylene-regulated guard cell closure (Kwak et al., 2003; Suhita et al., 2004; Desikan et al., 2006). These studies were the first to suggest a role for rboh-mediated superoxide production in hormone signalling pathways (reviewed by Kwak et al., 2006). The notion that ROS play important roles in a variety of hormone signalling pathways as well as in the integration of hormone signals was confirmed by a number of subsequent studies (reviewed by Mori et al., 2009). Interestingly, apoplastic ROS down-regulates auxin signalling in Arabidopsis (Blomster et al., 2011). Given that the RNAi-transgenic plants of the present study have reduced apoplastic superoxide production, it is possible that the normal processes that down-regulate auxin signalling are affected, which might explain the similarity of the RNAi lines to phenotypes found in auxin over-producers. In conjunction with this an up-regulation of specific auxin-responsive genes occurred in the roots of the RNAi-lines, although this did not include the entire spectrum of auxin signalling components investigated.
While many Rboh genes are expressed in roots, the precise physiological functions of their encoded proteins in the roots are unclear. Root cells originate from the root meristem; these cells then go through a process of elongation and differentiation. Root tips are sites of active ROS production, particularly in the elongation zone of the root (Liszkay et al., 2004; Dunand et al., 2007; Tsukagoshi et al., 2010). Apoplastic ROS can participate in cell elongation (Liszkay et al., 2004; Müller et al., 2009a ), and in addition to aberrant root development, our RNAi transgenic seedlings also exhibited significantly shorter roots than those of WT seedlings. Thus, the transgenic lines might be affected in both elongation growth and in patterning. ROS have been connected to root cell differentiation in Arabidopsis: Vernoux et al. (2000) identified the gene ROOT MERISTEMLESS 1 that encodes an enzyme involved in glutathione biosynthesis, thus linking root meristem identity and activity to the redox state of the cells. The transition from cell division in the meristem to differentiation involves transcriptional up-regulation of a number of genes encoding peroxidases and is also accompanied by changes in ROS concentrations (Tsukagoshi et al., 2010). This fits with the failure of seedling root tips of the RNAi-lines to maintain a functional architecture. It was found that, in Arabidopsis, AtrbohB is expressed specifically in the root tip below the elongation zone. It is speculated that RbohB is responsible for the ROS production in this zone.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Alignment of the deduced amino acid sequences of the AtrbohB and LesarbohB cDNAs.
Supplementary Fig. S2. Alignment of the deduced amino acid sequences of the AtrbohF and LesarbohF cDNAs. sequences.
Acknowledgements
We gratefully acknowledge funding through a European Commission Marie Curie IOF grant for KM, and from a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant to ARK. The position and research of AL is funded by the Deutsche Forschungsgemeinschaft (DFG Le720/6), which is gratefully acknowledged.
References
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R. 2001. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport Development 128 4057–4067 [DOI] [PubMed] [Google Scholar]
- Blomster T, Salojärvi J, Sipari N, Brosche M, Ahlfors R, Keinänen M, Overmyer K, Kangsjärvi J. 2011. Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphological responses in Arabidopsis Plant Physiology 157 1866–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Onckelen HV, Montagu MV, Inzé D. 1995. superroot, a recessive mutation in Arabidopsis, confers auxin overproduction The Plant Cell 7 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees Plant Molecular Biology Reporter 11 113–116 [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana The Plant Journal 16 735–743 [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, et al. 2010. Bimodular auxin response controls organogenesis in Arabidopsis Proceedings of the National Academy of Sciences, USA 107 2705–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Last K, Harrett-Williams R, Tagliavia C, Harter K, Hooley R, Hancock JT, Neill S. 2006. Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis The Plant Journal 47 907–916 [DOI] [PubMed] [Google Scholar]
- Dunand C, Crevecoeur M, Penel C. 2007. Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases New Phytologist 174 332–341 [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchick V, Bothwell JHF, et al. 2003. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth Nature 422 442–446 [DOI] [PubMed] [Google Scholar]
- Frahry G, Schopfer P. 2001. NADH-stimulated, cyanide-restistant superoxide production in maize coleoptiles analysed with a tetrazolium-based assay Planta 212 175–183 [DOI] [PubMed] [Google Scholar]
- Gapper C, Dolan L. 2006. Control of plant development by reactive oxygen species Plant Physiology 141 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Wood AT, Leubner-Metzger G. 2011. A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study The Plant Cell 23 2045–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom QJ, Torres MA, Fordham-Skelton AP, Hammond-Kosack KE, Robinson NJ, Jones JDG. 1996. RbohA, a rice homologue of the mammalian gp91phox respiratory burst oxidase gene The Plant Journal 10 515–522 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. 2007. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress Plant Physiology 143 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Damude HG, Werner D, Doerner P, Dixon RA, Lamb C. 1998. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs The Plant Cell 10 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Kawakita K, Maeshima M, Doke N, Yoshioka H. 2006. Subcellular localization of Strboh proteins and NADPH-dependent O2-generating activity in potato tuber tissue Journal of Experimental Botany 57 1373–1379 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Mori CI, Pei Z, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JDG, Schroeder J. 2003. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis EMBO Journal 22 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Nguyen V, Schroeder JI. 2006. The role of reactive oxygen species in hormonal responses Plant Physiology 141 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Zhang Y, Jiang M-Y. 2009. Alternative splicing and differential expression of two transcripts of nicotine adenine dinucleotide phosphate oxidase B gene from Zea mays Journal of Integrative Plant Biology 51 287–298 [DOI] [PubMed] [Google Scholar]
- Linkies A, Müller K, Morris K, et al. 2009. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana The Plant Cell 21 3803–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Liszkay A, van der Zalm E, Schopfer P.
2004.
Production of reactive oxygen intermediates (
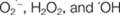 ) by maize roots and their role in wall loosening and elongation growth
Plant Physiology
136
3114–3123
[DOI] [PMC free article] [PubMed] [Google Scholar]
) by maize roots and their role in wall loosening and elongation growth
Plant Physiology
136
3114–3123
[DOI] [PMC free article] [PubMed] [Google Scholar] - Liu P-P, Koizuka N, Homrichhausen T, Hewitt J, Martin R, Nonogaki H. 2005. Large-scale screening of Arabidopsis enhancer-traps lines for seed germination-associated genes The Plant Journal 41 936–944 [DOI] [PubMed] [Google Scholar]
- Marino D, Andrio E, Danchin EGJ, Oger E, Gucciardo S, Lambert A, Puppo A, Pauly N. 2011. A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning New Phytologist 189 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Science Signaling. 2009;2:ra45. doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Messerli MA, Shi C, Gilroy S. 2007. Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs Proceedings of the National Academy of Sciences, USA 104 20996–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Weisenseel MH, Gilroy S. 2009. Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots The Plant Cell 21 2341–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori I, Murata Y, Uraji M. 2009. Integration of ROS and hormone signaling. In: delRio LA, A Puppo A.eds. Signaling and communication in plants: reactive oxygen species in plant signaling Heidelberg: Springer; 25–42 [Google Scholar]
- Müller K, Linkies A, Vreeburg RAM, Fry SC, Krieger-Liszkay A, Leubner-Metzger G. 2009. a In vivo cell wall loosening by hydroxyl radicals during cress seed germination and elongation growth Plant Physiology 150 1855–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Carstens AC, Linkies A, Torres MA, Leubner-Metzger G. 2009. b The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening New Phytologist 184 885–897 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct Activation of LBD/ASL genes in Arabidopsis The Plant Cell 19 1118–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oracz K, Voegele A, Tarkowská D, Jacquemoud D, Turecková V, Urbanová D, Strnad M, Sliwinska E, Leubner-Metzger G. 2012. Myrigalone A inhibits Lepidium sativum seed germination by interference with gibberellin metabolism and apoplastic superoxide production required for embryo extension growth and endosperm rupture Plant and Cell Physiology 53 81–95 [DOI] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JAH, Palme K. 2008. Comprehensive transcriptome analysis of auxin responses in Arabidopsis Molecular Plant 1 321–337 [DOI] [PubMed] [Google Scholar]
- Pasternak T, Potters G, Caubergs R, Jansen MAK. 2005. Complementary interactions between oxidative stress and auxins control plant growth responses at plant, organ, and cellular level Journal of Experimantal Botany 56 1991–2001 [DOI] [PubMed] [Google Scholar]
- Potocký M, Jones MA, Bezvoda R, Smirnoff N, Zárský V. 2007. Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth New Phytologist 174 742–751 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S.eds, Bioinformatics methods and protocols: methods in molecular biology Totowa, NJ: Humana Press; 365–386 [DOI] [PubMed] [Google Scholar]
- Sagi M, Davydof O, Orazova S, Yesbergenova Z, Ophir R, Stratmann JW, Fluhr R. 2004. Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum The Plant Cell 16 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P, Liszkay A, Bechtold M, Frahry G, Wagner A. 2002. Evidence that hydroxyl radicals mediate auxin-induced extension growth Planta 214 821–828 [DOI] [PubMed] [Google Scholar]
- Si Y, Dane F, Rashotte A, Kang K, Singh NK. 2010. Cloning and expression analysis of the Ccrboh gene encoding respiratory burst oxidase in Citrullus colocynthis and grafting onto Citrullus lanatus (watermelon) Journal of Experimental Botany 61 1635–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speranza A, Crinelli R, Scoccianti Geitmann A. 2011. Reactive oxygen species are involved in pollen tube initiation in kiwifruit Plant Biology 14 1438–8677 [DOI] [PubMed] [Google Scholar]
- Suhita D, Raghavendra AS, Kwak JM, Vavasseur A. 2004. Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure Plant Physiology 134 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson S, Gilroy S. 2010. ROS in plant development Physiologia Plantarum 138 384–392 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL. 2005. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development Current Opinion in Plant Biology 8 397–403 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG. 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygene intermediates in the plant defense response Proceedings of the National Academy of Sciences USA 99 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL. 2006. Reactive oxygen species signaling in response to pathogens Plant Physiology 141 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Onouchi H, Hamada S, Machida C, Hammond-Kosack KE, Jones JD. 1998. Six Arabidopsis homologues of the human respiratory burst oxidase (gp91phox) The Plant Journal 14 365–370 [DOI] [PubMed] [Google Scholar]
- Trijillo M, Altschmied L, Schweizer P, Kogel K-H, Hückelhoven R. 2006. Respiratory burst oxidase homologue A of barley contributes to penetration by the powdery mildew fungus Blumeria graminis f. sp. hordei Journal of Experimental Botany 57 3781–3791 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PB. 2010. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root Cell 143 606–616 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, et al. 2000. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development The Plant Cell 12 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, et al. An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, Jones JDG, Doke N. 2003. Nicotiana benthamiana gp91 phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans The Plant Cell 15 706–718 12615943 [Google Scholar]
- Zhao S, Fernald RD. 2005. Comprehensive algorithm for quantitative real-time polymerase chain reaction Journal of Computational Biology 12 1047–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.