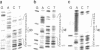Abstract
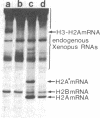
The control region of a sea urchin H2A histone gene may be functionally dissected into at least three DNA segments, which we have termed modulator, selector, and initiator elements. While the initiator and in particular the selector containing the T-A-T-A-A-A-T-A sequence are specificity elements that dictate the generation of faithful 5' ends to H2A mRNA, the modulators control the rate at which these specificity elements operate [Grosschedl, R. & Birnstiel, M. L. (1980) Proc. Natl. Acad. Sci. USA 77, 1432-1436]. By functional tests of in vitro mutated histone DNA in the Xenopus oocyte we have now discovered that the segment E of the A+T-rich spacer DNA lying at a considerable distance upstream of the conservative T-A-T-A-A-A-T-A sequence is a strong modulator element of H2A gene transcription. Deletion of this element creates a 15- to 20-fold H2A-specific down mutation. Segment E by itself cannot elicit initiation of transcription except in coordination with the prelude sequence of the H2A gene. The nucleotide sequence of the relevant spacer element showing modulator activity has been determined and found to contain a pattern of T and A runs as well as a series of inverted repeats. Additional pre-H2A spacer mutants, including a spacer inversion mutant, have been constructed in vitro, that, when injected into the oocyte nucleus, modulate the expression of the H2A gene by an overall factor as large as 100. Other factors controlling promoter activity are discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bendig M. M., Thomas T., Folk W. R. Regulatory mutants of polyoma virus defective in DNA replication and the synthesis of early proteins. Cell. 1980 Jun;20(2):401–409. doi: 10.1016/0092-8674(80)90626-1. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. Deletions covering the putative promoter region of early mRNAs of simian virus 40 do not abolish T-antigen expression. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3865–3869. doi: 10.1073/pnas.77.7.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Birnsteil M. L. A regulatory sequence near the 3' end of sea urchin histone genes. Nucleic Acids Res. 1979 Jul 11;6(9):2997–3008. doi: 10.1093/nar/6.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Irminger J. C., Birnstiel M. L. Ubiquitous and gene-specific regulatory 5' sequences in a sea urchin histone DNA clone coding for histone protein variants. Nucleic Acids Res. 1980 Mar 11;8(5):957–977. doi: 10.1093/nar/8.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon F., O'Hare K., Perrin F., LePennec J. P., Benoist C., Cochet M., Breathnach R., Royal A., Garapin A., Cami B. Organisation and sequences at the 5' end of a cloned complete ovalbumin gene. Nature. 1979 Mar 29;278(5703):428–434. doi: 10.1038/278428a0. [DOI] [PubMed] [Google Scholar]
- Gluzman Y., Sambrook J. F., Frisque R. J. Expression of early genes of origin-defective mutants of simian virus 40. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3898–3902. doi: 10.1073/pnas.77.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman G., Iserentant D., Gheysen D., Fiers W. Characterization of the major altered leader sequence of late mRNA induced by SV40 deletion mutant d1-1811. Nucleic Acids Res. 1979 Dec 11;7(7):1799–1814. doi: 10.1093/nar/7.7.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Probst E., Birnstiel M. L. Transcriptional fidelity of histone genes injected into Xenopus oocyte nuclei. Nature. 1980 Nov 6;288(5786):100–102. doi: 10.1038/288100a0. [DOI] [PubMed] [Google Scholar]
- Konkel D. A., Maizel J. V., Jr, Leder P. The evolution and sequence comparison of two recently diverged mouse chromosomal beta--globin genes. Cell. 1979 Nov;18(3):865–873. doi: 10.1016/0092-8674(79)90138-7. [DOI] [PubMed] [Google Scholar]
- Kressmann A., Clarkson S. G., Telford J. L., Birnstiel M. L. Transcription of xenopus tDNAmet1 and sea urchin histone DNA injected into the Xenopus oocyte nucleus. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1077–1082. doi: 10.1101/sqb.1978.042.01.108. [DOI] [PubMed] [Google Scholar]
- Kressmann A., Hofstetter H., Di Capua E., Grosschedl R., Birnstiel M. L. A tRNA gene of Xenopus laevis contains at least two sites promoting transcription. Nucleic Acids Res. 1979 Dec 11;7(7):1749–1763. doi: 10.1093/nar/7.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S., Childs G., Kedes L. Sea urchin nuclei use RNA polymerase II to transcribe discrete histone RNAs larger than messengers. Cell. 1978 Sep;15(1):151–162. doi: 10.1016/0092-8674(78)90091-0. [DOI] [PubMed] [Google Scholar]
- Luse D. S., Roeder R. G. Accurate transcription initiation on a purified mouse beta-globin DNA fragment in a cell-free system. Cell. 1980 Jul;20(3):691–699. doi: 10.1016/0092-8674(80)90315-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nishioka Y., Leder P. The complete sequence of a chromosomal mouse alpha--globin gene reveals elements conserved throughout vertebrate evolution. Cell. 1979 Nov;18(3):875–882. doi: 10.1016/0092-8674(79)90139-9. [DOI] [PubMed] [Google Scholar]
- Portmann R., Schaffner W., Birnstiel M. Partial denaturation mapping of cloned histone DNA from the sea urchin Psammechinus miliaris. Nature. 1976 Nov 4;264(5581):31–34. doi: 10.1038/264031a0. [DOI] [PubMed] [Google Scholar]
- Probst E., Kressmann A., Birnstiel M. L. Expression of sea urchin histone genes in the oocyte of Xenopus laevis. J Mol Biol. 1979 Dec 15;135(3):709–732. doi: 10.1016/0022-2836(79)90173-6. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Kunz G., Daetwyler H., Telford J., Smith H. O., Birnstiel M. L. Genes and spacers of cloned sea urchin histone DNA analyzed by sequencing. Cell. 1978 Jul;14(3):655–671. doi: 10.1016/0092-8674(78)90249-0. [DOI] [PubMed] [Google Scholar]
- Scherer G. E., Walkinshaw M. D., Arnott S. A computer aided oligonucleotide analysis provides a model sequence for RNA polymerase-promoter recognition in E.coli. Nucleic Acids Res. 1978 Oct;5(10):3759–3773. doi: 10.1093/nar/5.10.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford J. L., Kressmann A., Koski R. A., Grosschedl R., Müller F., Clarkson S. G., Birnstiel M. L. Delimitation of a promoter for RNA polymerase III by means of a functional test. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2590–2594. doi: 10.1073/pnas.76.6.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Kédinger C., Corden J., Brison O., Chambon P. Specific in vitro initiation of transcription on conalbumin and ovalbumin genes and comparison with adenovirus-2 early and late genes. Nature. 1980 Jun 5;285(5764):367–373. doi: 10.1038/285367a0. [DOI] [PubMed] [Google Scholar]
- Wittig B., Wittig S. A phase relationship associates tRNA structural gene sequences with nucleosome cores. Cell. 1979 Dec;18(4):1173–1183. doi: 10.1016/0092-8674(79)90230-7. [DOI] [PubMed] [Google Scholar]
- Wyllie A. H., Laskey R. A., Finch J., Gurdon J. B. Selective DNA conservation and chromatin assembly after injection of SV40 DNA into Xenopus oocytes. Dev Biol. 1978 May;64(1):178–188. doi: 10.1016/0012-1606(78)90069-6. [DOI] [PubMed] [Google Scholar]
- Ziff E. B., Evans R. M. Coincidence of the promoter and capped 5' terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978 Dec;15(4):1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]