Abstract
Somatic mutation is a natural mechanism which allows plant growers to develop new cultivars. As a source of variation within a uniform genetic background, it also represents an ideal tool for studying the genetic make-up of important traits and for establishing gene functions. Layer-specific molecular characterization of the Pinot family of grape cultivars was conducted to provide an evolutionary explanation for the somatic mutations that have affected the locus of berry colour. Through the study of the structural dynamics along chromosome 2, a very large deletion present in a single Pinot gris cell layer was identified and characterized. This mutation reveals that Pinot gris and Pinot blanc arose independently from the ancestral Pinot noir, suggesting a novel parallel evolutionary model. This proposed ‘Pinot-model’ represents a breakthrough towards the full understanding of the mechanisms behind the formation of white, grey, red, and pink grape cultivars, and eventually of their specific enological aptitude.
Key words: Berry colour, grapevine, layer, molecular characterization, SSRs and SNPs, Vitis vinifera
Introduction
As a source of variation within a uniform genetic background, somatic mutation in plants is of theoretical and practical interest. A great deal of the genetic variation exploited in plant breeding comes from somatic variations that naturally arise in many plant groups and are used by growers in developing new cultivars that are agronomically and/or commercially superior to the parent stock. Indeed, many of these mutants have become a major source of horticultural staple crops (e.g. potato), fruit trees (Citrus spp., peach, banana, etc.), and ornamental plants (rose, dahlia, chrisanthemum, etc.) (Shamel and Pomeroy, 1936). It should be emphasized that the great majority of somatic mutations probably go unnoticed because they do not visibly affect easily detectable characteristics such as colour and growth habit of the plant (Hartmann and Kester, 1975).
Unlike higher animals, in higher plants somatic mutations can enter the germline and be transmitted to the progeny. Whether or not a somatic cell enters the germline depends on its localization in a defined cell layer (L) in the shoot apex (D’Amato, 1997). In dicots, gametes arise from the L2 lineage (Marcotigiano and Bernatzky, 1995). Indeed, most dicots have stratified apical meristems containing up to three layers of dividing cells, the combination of which gives rise to different plant tissues (Neilson-Jones, 1969). Mutations could be present in the entire meristem (bud sports) or only a portion (chimeras). Chimeras are composed of two or more genetically distinct tissue layers that grow adjacent to one another (Dermen, 1960). Even if a somatic mutation is not incorporated into a cell line that differentiates into gametes, it can still be perpetuated by asexual reproduction. This makes somatic mutation a valuable source of heritable variation especially in plants which are multiplied via vegetative propagation, including grapevine (Vitis vinifera L.). The growing scientific interest in the phenomenon of somatic mutation in grapevine is demonstrated by a list of recent publications. To date, in addition to anthocyanin production (Kobayashi et al., 2004; Walker et al., 2006), somatic genetic variation has been successfully used to identify genes involved in gibberellic acid signalling (Pinot meunier; Boss and Thomas, 2002), early berry morphogenesis (Fernandez et al., 2006), and flower development (Chatelet et al., 2007; Fernandez et al., 2010).
Among grapevine somatic variants, those affecting fruit colour are probably the most studied since they occur in several cultivars (Galet, 2000). Grape berry colour is due to the presence of a single pigment family, the anthocyanins, which vary greatly in concentration and composition depending on the grape cultivar (Mattivi et al., 2006). In many plants, anthocyanin biosynthesis is controlled by regulatory genes belonging to the Myb family of transcription factors (Koes et al., 2005). Two Myb-related transcription factor genes, VvMybA1 and VvMybA2, regulate anthocyanin biosynthesis in V. vinifera grapes. Inactivation of these two functional genes, through the insertion of the Gret1 retrotransposon in the VvMybA1 promoter and through a non-synonymous single nucleotide polymorphism (SNP) present in the VvMybA2 coding region, gives rise to a white berry phenotype (Kobayashi et al., 2004, 2005; Walker et al., 2007). Recently, several genetic and genomic studies revealed that the colour locus is a cluster of four Myb-like genes located on chromosome 2 (Walker et al., 2006; Matus et al., 2008; Azuma et al., 2009; Fournier-Level et al., 2009).
Since Pinot is a founder variety, it had several chances to undergo somatic mutations (Hocquigny et al., 2004; Lacombe et al., 2011). Most of these affected the ancestral black berry colour, and gave rise to cultivars such as in Pinot blanc and Pinot gris. The most established evolutionary model is that Pinot blanc arose from Pinot gris which arose from Pinot noir, even if the relationship between Pinot blanc and Pinot gris has not yet been fully explored (Viala and Vermorel, 1909; Pelsy, 2010). Pinot gris is reported to be a periclinal chimera of Pinot noir, but also in this case the exact nature of the genetic modification remains to be determined (Franks et al., 2002; Hocquigny et al., 2004). Chimerism in grapevine was first observed in the cell layers of shoot apical meristems (SAMs) as variability in the level of ploidy (cytochimeras). These studies demonstrated that the grapevine apical meristem is composed of only two distinct tissue layers, L1 and L2 (Einset and Pratt, 1954; Thompson and Olmo, 1963).
Investigation of somatic mutations affecting the fruit colour locus will improve knowledge of the genetics of grey- and white-skinned cultivars as well as promote understanding of the evolutionary events behind their origin. Here, a layer-specific structural analysis at both the genome-wide and berry colour locus level in several Pinot noir, Pinot gris, and Pinot blanc clones along with their naturally derived chimeras or sports is reported. The findings provide experimental evidence of a novel evolutionary model for the Pinot family of grape cultivars.
Materials and methods
Plant material and genomic DNA extraction
A set of 29 clones belonging to four V. vinifera cultivars were studied: (i) four clones of Pinot noir, including the sequenced Pinot noir clone ENTAV 115; (ii) 13 clones of Pinot gris, of which eight produced only grey-skinned berries, two bore wholly white-skinned berries, one produced berry sectorial chimeras and wholly white-skinned berry clusters, one produced bud sports (wholly mutated buds), and one displayed sectorial chimeras, wholly white-skinned berries, and wholly mutated buds; (iii) 10 clones of Pinot blanc; and (iv) two clones of Pinot meunier, as a somatic variant for an additional trait. The studied clones were registered by several European institutes and housed at Fondazione Edmund Mach (Italy), at the Laimburg Research Centre (Italy), and at the germplasm national repository of CRA-VIT (Italy) (Fig. 1; Supplementary Table S1 available at JXB online). For each clone, genomic DNA was isolated from 100–200mg of young leaf and/or berry skin (L1+L2) and 300–500mg of berry flesh and/or root (L2). To avoid contamination, berry flesh samples were isolated by dissecting the cells between the berry skin and the tissue surrounding seeds. After freeze-drying, leaf and root material was ground using an MM 300 Mixer Mill (Retsch Inc., Haan, Germany) and DNA extraction was performed using the DNeasy 96 Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. After liquid nitrogen grinding, DNA isolation from berry skin and flesh powders was carried out according to Lodhi et al. (1994). A total of 117 tissue-specific DNA samples were obtained and divided into a core set of 40 samples, representing 10 wild-type clones (four tissues) of the considered Pinot cultivars, and an extended set of 77 samples, containing derived chimeras and sports, and additional wild-type Pinot gris and Pinot blanc (Supplementary Table S1). It is important to emphasize that in each analysis the genetic make-up of L1 was derived from the difference between the L1+L2 (leaf, berry skin) and L2 (berry flesh, root) profiles.
Fig. 1.
Mosaic of berry colour variations in the studied Pinot somatic variants. (a) A berry cluster of Pinot noir; (b) berry clusters of Pinot blanc; (c) a berry cluster of wild-type Pinot gris and wholly white-skinned berry clusters from the same vine of Pinot gris; (d) wholly white-skinned berries of Pinot gris; and (e) a berry sectorial chimera of Pinot gris.
SSR analysis
For each simple sequence repeat (SSR) locus, primer sequence information, annealing temperature (Ta), and type of Taq polymerase are reported in Supplementary Table S2 at JXB online. Genomic DNA was amplified by PCR according to the following conditions: 20ng of DNA template, 1× PCR buffer, 1.5mM MgCl2, 0.2mM of each dNTP, 0.5mM fluorophore-labelled forward and reverse primer, 0.25U of Taq DNA polymerase, and milliQ water to 12.5ml PCR final volume. PCR thermocycling was performed with a 5/10min initial denaturation/activation step, followed by 35 cycles at 94 °C for 60 s, Ta for 45 s, and 72 °C for 90 s, with a final extension step of 7min at 72 °C. The presence of PCR products was assessed by electrophoresis with a 1.5% agarose gel and quantified by comparison with a Mass ruler DNA ladder mix (Thermo Fischer Scientific, Waltham, MA, USA). Capillary electrophoresis of PCR products was carried out on a 3730xl Genetic Analyzer (Life Tech, Foster City, CA, USA). Allele identification was performed by using GeneMapper v4.0 software (Life Tech).
SNPlex assay and data analysis
The SNPlex assay (Life Tech; Tobler et al., 2005) was carried out on ~40ng of genomic DNA fragmented by heat shock. Ten SNP sets, for a total of 430 electronic SNPs (eSNPs), were chosen based on the number of validated SNPs which were first identified in Pinot noir clone ENTAV 115 (Pindo et al., 2008; Vezzulli et al., 2008). SNPlex assay, capillary electrophoresis, and genotype analysis were carried out according to Pindo et al. (2008).
Sequencing analysis
The overall SNP regions, the VvMyb1, VvMyb2, and VvMyb3 promoter regions (encompassing the first exon), and the 3’ untranslated region (UTR), along with the VvMyb4 gene (exons and introns), were sequenced as described below. Flanking regions, between 300bp and 400bp long, were sequenced for each of the informative target SNPs in order to confirm the SNPlex results and to discover all possible polymorphic sites. PCR primers of VvMyb genes were derived from Fournier-Level et al. (2009), while PCR primers for SNP (non-coding) and gene (coding) regions were designed based on the Pinot noir and the PN40024 genomic sequence, respectively (Jaillon et al., 2007; Velasco et al., 2007), using the Primer3 software (Rozen and Skaletski, 2000) (Supplementary Table S3 at JXB online). PCRs were assembled using the following conditions: 10–20ng of genomic DNA, 1× PCR buffer (Life Tech), 1.5mM MgCl2, 0.2mM of each dNTP, 0.4 µM of each primer, 1U of Taq DNA polymerase (Life Tech), and water to a final volume of 12.5 µl. The DNA amplifications were performed using a 5min initial denaturation step, followed by 30 cycles at 94 °C for 30 s, Ta for 30 s, and 72 °C for 60 s, with a final extension step of 10min at 72 °C. The presence of PCR products was assessed by electrophoresis in a 1.5% agarose gel. In order to remove unincorporated dNTPs and primers, amplicons were enzymatically purified with exonuclease-phosphatase (ExoSAP-IT, GE Healthcare, Piscataway, NJ, USA). Sequencing reactions, post-sequencing purification, capillary electrophoresis, and polymorphism detection were carried out as reported by Vezzulli et al. (2008). The amplicon sequences were searched by BLAST-N (Altschul et al., 1990) against the genome assembly of both the highly heterozygous Pinot noir ENTAV 115 genotype (Velasco et al., 2007) and the near-homozygous PN40024 line (Jaillon et al., 2007).
Results
Layer-specific genome-wide analysis
Nine reference SSR markers (Supplementary Table S2 at JXB online) were used to confirm that the Pinot core set used in this study was true to type. According to this analysis, Pinot noir, Pinot gris, and Pinot blanc share the same genotype (Supplementary Table S4a), as reported by Regner et al. (2000). In contrast, being a grape chimera, Pinot meunier showed a triallelic profile at the reference locus VVS2 (Thomas and Scott, 1993) in agreement with Franks et al. (2002) (Supplementary Table S4a). In order to test for any additional chimeric genetic presence in the core set, these samples were also analysed with six SSR markers prone to triallelism (Supplementary Table S2). The triallelic SSR analysis did not reveal any chimeric state and confirmed that the Pinot family members share a uniform genetic background (Supplementary Table S4b).
Of the 430 eSNPs analysed on the Pinot core set in the SNPlex experiment, 284 satisfied the quality value, with a mean call rate of 98% per SNPset, and therefore were considered successful, while the remaining 146 were not included in the genetic analysis. Further testing of the 284 eSNPs showed that 220 were true positive (heterozygous) Pinot noir SNPs, while 64 were false positive. As anticipated by the SSR analysis, Pinot noir, Pinot blanc, and Pinot meunier share a highly uniform genetic background, since no differences were observed at 215 out of 220 SNP loci. Interestingly, unlike Pinot noir and Pinot blanc, Pinot gris clones had five SNPs (SNP4045, SNP7234, SNP4071, SNP7054, and SNP6166) that were not heterozygous in both L1+L2 and pure L2 tissues. Therefore, these SNPlex results did not allow discrimination of either clones or layers within each cultivar. Due to the already reported detection limits, such as preferential annealing, of this genotyping system (Pindo et al., 2008; Vezzulli et al., 2008), the derived SNP information was validated by sequencing.
The sequencing analysis of the Pinot core set at the five informative SNP loci (SNP4045, SNP7234, SNP4071, SNP7054, and SNP6166) mostly confirmed the genotype identified by SNPlex analysis. In fact, the SNPlex results were validated in all Pinot clones except for those of Pinot gris. Although appearing homozygous in each layer of the Pinot gris clones following SNPlex analysis, the five SNP regions had a layer-specific genetic make-up according to the sequencing analysis. In particular, while being heterozygous in the L1+L2-derived tissues, they were homozygous-like in the L2-derived tissues. However, in the heterozygous profiles, one allele was always under-represented, compared with the reference heterozygous profile of Pinot noir (Fig. 2). In addition to the five SNPs targeted by SNPlex assay, the resequencing analysis allowed for the detection of 15 point mutations along the five corresponding SNP regions in the L1+L2-derived tissues of the Pinot cultivars (Supplementary Table S5 at JXB online).
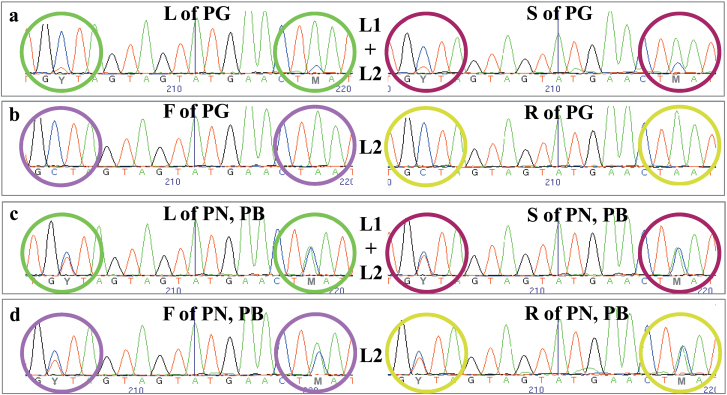
Fig. 2.
Comparison of the electropherograms obtained by resequencing the SNP6166 region among the studied Pinot colour somatic variants. Differently coloured circles highlight the point mutations Y (C/T) and M (A/C) in leaf (L), berry skin (S), berry flesh (F), and root (R) of Pinot noir (PN), Pinot gris (PG), and Pinot blanc (PB). Allelotyping and peak height are depicted. In particular, (a) shows the heterozygous state with an under-represented allele in L1+L2-derived tissues of PG, while (b) reveals the homozygous-like state in pure L2-derived tissues of PG. (c) and (d) show the fully heterozygous state in both L1+L2-derived and pure L2-derived tissues of PN and PB.
A BLAST-N search against the genome sequence of both the highly heterozygous Pinot noir ENTAV 115 genotype (Velasco et al., 2007) and the near-homozygous PN40024 line (Jaillon et al., 2007) located these five informative SNP regions along chromosome 2, near the Myb-related genes responsible for anthocyanin synthesis in grape.
Layer-specific structural analysis of the colour locus
To investigate further the genetic structure of this section of chromosome 2, the four Myb-related genes were amplified and sequenced in the Pinot core set (Supplementary Table S3 at JXB online). The main results (Fig. 3; Supplementary Table S5, S6) can be summarized as: (i) the VvMybA4 gene and the 3’UTR of VvMybA2 did not contain any polymorphisms in any Pinot genotype or layer; (ii) the VvMybA2 promoter as well as the VvMybA1 3’UTR and VvMybA1 first exon were heterozygous in both Pinot noir layers and in the L1 of Pinot gris, while showing a homozygous-like genetic make-up in the L2 layer of Pinot gris and in both Pinot blanc cell layers; (iii) the VvMybA1 promoter contained the Gret (non-functional) and non-Gret (functional) alleles in both Pinot noir cell layers and in the L1 cell layer of Pinot gris, while only the Gret (non-functional) allele was present in the L2 layer of Pinot gris and in both Pinot blanc cell layers; and (iv) primers designed within the VvMybA3 promoter and 3’UTR did not amplify a single target region, although different PCR conditions were applied on the Pinot core set.
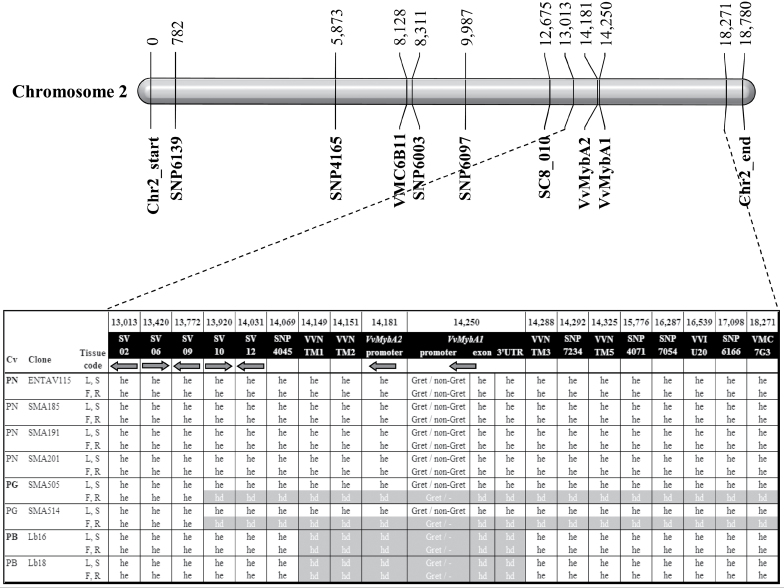
Fig. 3.
Chromosome 2 structure in the Pinot core set. A snapshot of the chromosome 2 region depicts different molecular markers showing either a heterozygous genetic state (he) or a heterozygous deletion (hd) in leaf (L), berry skin (S), berry flesh (F), and root (R) of Pinot noir (PN), Pinot gris (PG), and Pinot blanc (PB) registered clones.
In order to confirm the presence and to assess the extent of the homozygous-like region in the L2 layer of Pinot gris and in both cell layers of Pinot blanc, seven SSR markers and four SNP regions (Supplementary Table S2, S3 at JXB online) inserted among the Myb-related gene and the five informative SNP regions were genotyped in the Pinot core set. The VVNTM1 and VVNTM2 SSR markers were homozygous-like in the L2 layer of Pinot gris and in both Pinot blanc cell layers, but heterozygous in both Pinot noir cell layers and in the L1 layer of Pinot gris. Three SSR markers (VVNTM3, VVNTM5, and VVIU20) and four SNP regions (SNP7253, SNP0060, SNP8066, and SNP7002) were heterozygous in both layers of Pinot noir and Pinot blanc, and in the L1 layer of Pinot gris, but homozygous in the L2 cell layer of Pinot gris. VVNTM4 and VVNTM6 SSR markers did not display any polymorphisms in the Pinot genotypes and cell layers (Fig. 3; Supplementary Table S5, S6).
To extend the genetic and structural characterization upstream and downstream of the colour locus region, eight additional loci were analysed in the Pinot core set. In particular, four SSR markers were genotyped and four SNP regions were sequenced (Supplementary Table S2, S3 at JXB online). Out of the seven loci upstream of the SNP4045 region, the SC08_0146_026 SSR was not polymorphic, whereas the SC08_0146_010 SSR was heterozygous in each Pinot genotype and cell layer, as were the more upstream loci, namely SNP6097, SNP6003, VMC6B11, SNP4165, and SNP6139. The VMC7G3 SSR locus downstream of the SNP6166 region showed a heterozygous profile in both layers of Pinot noir and Pinot blanc, and in the L1 layer of Pinot gris, while this marker was homozygous-like in the L2 layer of Pinot gris (Fig. 3; Supplementary Table S5, S6). To map the 5’ border of the homozygous-like region in the L2 layer of Pinot gris more closely, six additional gene regions between the SC08_0146_010 and SNP4045 markers were analysed in the Pinot core set (Supplementary Table S3). Within this gap, the SV02, SV06, and SV09 regions were heterozygous, while SV08 was not polymorphic in any Pinot genotype or cell layer. SV10 and SV12 showed a heterozygous profile in both layers of Pinot noir and Pinot blanc, and in the L1 layer of Pinot gris, while both markers were homozygous-like in the L2 of Pinot gris (Fig. 3; Supplementary Table S5, S6).
In conclusion, this work, aimed at the confirmation and delimitation of the homozygous-like region in the L2 cell layer of Pinot gris and in both layers of Pinot blanc, led to the identification of 14 informative loci, in addition to the five SNP regions previously targeted by SNPlex analysis (Supplementary Table S5 at JXB online). In cultivars derived from asexual (vegetative) reproduction, such as the Pinot cultivars, this homozygous-like profile can only be ascribed to the presence of a deletion. Therefore, the ‘homozygous-like’ term corresponds to the genetic term ‘hemizygous’, which refers to the presence of a null allele. Thus, this study revealed that the Pinot gris deletion affects the L2 cell layer only and extends for a minimum of 4202kb and a maximum of 4350kb, while the Pinot blanc deletion occurs in both cell layers and ranges from 100kb to 179kb. These results are consistent with recent non-cell layer-specific findings, in which both Pinot blanc and Pinot gris were shown to be carrying a partially deleted allele at the berry colour locus (Hocquigny et al., 2004; Walker et al., 2006; Yakushiji et al., 2006).
In order to ascertain if this genetic structure exists in clones that originate from different viticultural areas, 13 of the 36 markers along chromosome 2 (Supplementary Table S6 at JXB online)-chosen as informative, well scattered, and easy to score-were screened on an extended set of plants consisting of mutants and additional wild-type Pinot blanc and Pinot gris. Of these, the eight Pinot blanc clones showed the same L1 and L2 cell layer genetic profiles as the core set Pinot blanc samples, and the six grey-berried clones of Pinot gris also confirmed the L1 and L2 layer genetic profiles of the core set Pinot gris samples. Moreover, additional Pinot gris clones bearing from wholly white-skinned berries to wholly mutated buds were shown to share the genetic make-up of the L2 cell layer of the wild-type Pinot gris clones. However, the grey skin sectors of sectorial chimeras possessed the L1 and L2 cell layer genetic profiles of the wild-type Pinot gris clones (Table 1).
Table 1.
Berry colour locus structure in the extended set of Pinot gris (PG) , along with its naturally derived mutants, and Pinot blanc (PB) clones, based on the analysis of 13 informative and well-scattered loci at the layer-specific level.
| Cv | Clone | State | Berry description | Tissue code | SV9 | SV10 | SNP4045 | VVNTM1 | VVNTM2 | VvMybA1promoter | VVNTM3 | SNP7234 | VVNTM5 | SNP7054 | VVIU20 | SNP6166 | VMC7G3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PG | SMA505 | MUT | L | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | |
| MUT | Wholly mutated buds | S | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | Wholly mutated buds | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | SMA514 | MUT | Wholly white-skinned berries | S | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd |
| MUT | Wholly white-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | 49/207FR | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | CL52 | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | CL53 | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | 457 | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | FEDIT13 | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | R6 | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | VCR5 | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | 10–05 | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | Wholly white-skinned berries | S | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | Wholly white-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | 10–10 | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | 513 | WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he |
| WT | Grey-skinned berries | F | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| MUT_wt | Grey sector of sectorial chimera | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| MUT_mut | White sector of sectorial chimera | S | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | Sectorial chimera | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | L | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | |||
| MUT | Wholly white-skinned berry clusters | S | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | Wholly white-skinned berry clusters | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PG | 516 | WT | L | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | |
| WT | Grey-skinned berries | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| WT | Grey-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT_wt | Grey sector of sectorial chimera | S | he | he | he | he | he | Gret/non-Gret | he | he | he | he | he | he | he | ||
| MUT_mut | White sector of sectorial chimera | S | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | Sectorial chimera | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | Wholly white-skinned berries | S | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | Wholly white-skinned berries | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| Cv | Clone | State | Berry description | Tissue code | SV | SV | SNP | VVN | VVN | VvMybA1 | VVN | SNP | VVN | SNP | VVI | SNP | VMC |
| MUT | L | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | |||
| MUT | Wholly mutated buds | S | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| MUT | Wholly mutated buds | F | he | hd | hd | hd | hd | Gret/– | hd | hd | hd | hd | hd | hd | hd | ||
| PB | 72FR | WT | L | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | |
| WT | White-skinned berries | S | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| WT | White-skinned berries | F | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| PB | CL55 | WT | L | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | |
| WT | White-skinned berries | S | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| WT | White-skinned berries | F | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| PB | CL54 | WT | L | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | |
| WT | White-skinned berries | S | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| WT | White-skinned berries | F | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| PB | 33W | WT | L | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | |
| WT | White-skinned berries | S | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| WT | White-skinned berries | F | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| PB | 209D | WT | L | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | |
| WT | White-skinned berries | S | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| WT | White-skinned berries | F | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| PB | 212D | WT | L | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | |
| WT | White-skinned berries | S | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| WT | White-skinned berries | F | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| PB | VCR5 | WT | L | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | |
| WT | White-skinned berries | S | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| WT | White-skinned berries | F | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| PB | VCR7 | WT | L | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | |
| WT | White-skinned berries | S | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he | ||
| WT | White-skinned berries | F | he | he | he | hd | hd | Gret/– | he | he | he | he | he | he | he |
L, leaf; S, berry skin; F, berry flesh; R, root; he, heterozygous genetic state; hd, heterozygous deletion.
Discussion
The layer-specific analysis of somatic variation
Somatic variation is important for genetic improvement in fruit trees, such as grape, where vegetative reproduction is used to propagate interesting new phenotypes, appearing as spontaneous sports. In spite of its relevance, little is known about the genetic and epigenetic mechanisms causing this variation, which might be in a chimeric state. For this reason, to study the Pinot somatic mutations, a layer-specific approach at a genomic scale was chosen, which turned out to be crucial.
In a first attempt to assess the impact of the chimerism on the intravarietal genetic diversity in the Pinot family of grape cultivars, a collection of clones belonging to the Pinot noir, Pinot gris, Pinot blanc, and Pinot meunier cultivars was analysed at a cell layer level with SSRs prone to triallelism. Given the intrinsic feature of SSRs in producing stutter bands, the layer-specific analysis prevented the identification of false-positive results. This study confirmed that the genetic background of the Pinot family members is highly uniform and showed the presence of the VVS2 microsatellite mutation in all tested Pinot meunier clones, in agreement with previous reports (Franks et al., 2002; Hocquigny et al., 2004). Unlike the present analysis, this mutation was also found in Pinot gris clones of different origin (Hocquigny et al., 2004). This suggests that the VVS2 locus is highly plastic and that this mutation most probably occurred independently after the Pinot gris and Pinot meunier divergence, namely after the respective mutations causing the partial loss of berry colour (Walker et al., 2007) and the hairy leaf phenotype (Boss and Thomas, 2002). This latter hypothesis is in contrast to the conclusion by Hocquigny et al. (2004) that Pinot meunier derives from Pinot gris, based on their non-layer-specific and non-genome-extensive approach. This discrepancy leads to a further hypothesis based on which these multiple mutations are recurrent. Therefore, it is not possible to speculate on which mutation occurred earlier than the other one.
Pinot cultivars show primitive morphological features analogous to those of the wild-type V. vinifera sbs. sylvestris, and are thus considered archaic cultivars (Levadoux, 1956). As one of the founder varieties and cultivated worldwide, Pinot has accumulated mutations leading to peculiar agronomical and enological aptitudes. The most relevant is the berry skin colour mutation leading to Pinot cultivars with different pigmentation. Berry skin colour mutants exist for a range of grape varieties (e.g. Grenache, Traminer, etc.) as well as for other fruit species (e.g. apple, pear, etc.) (Germplasm Resources Information Network, http://www.ars-grin.gov/). With regard to grapevine, much work has been carried out to investigate the genetic determinants of skin colour in V. vinifera germplasm collections (This et al., 2007; Walker et al., 2007; Fournier-Level et al., 2009, 2010), as well as in Vitis species and hybrids (Cadle-Davidson et al., 2008; Azuma et al., 2008, 2011; Shimazaki et al., 2011). However, the berry colour locus has been less explored in somatic variants of a grape variety (cépage, family of cultivars; Boursiquot and This, 1999), there being two reports, namely Giannetto et al. (2008) and Walker et al. (2006). In this latter study, somatic mutations of Cabernet Sauvignon, Malian, and Shalistin have been characterized at the molecular level, resulting in the development of the first evolutionary model (here reported as the ‘CabSau-model’) that explains the development of somatic colour mutants within a variety (Walker et al., 2006). Malian bearing bronze-skinned berries is a periclinal chimera of Cabernet Sauvignon, and Shalistin bearing white-skinned berries is a bud sport of Malian. The bronze berry phenotype of Malian is related to a large (>260kb) deletion in the colour locus, which includes the functional allele of VvMybA1 and VvMybA2 in the L2 cells, while the white berry phenotype of Shalistin is the result of a cellular rearrangement (or displacement) in Malian whereby the L2 cell layer (unpigmented phenotype) replaces the L1 cells (pigmented phenotype) (Walker et al., 2006).
The structural dynamics at the berry colour locus
In Pinot gris the findings revealed a very large heterozygous deletion of 4202–4350kb that represents a relevant structural change affecting about a quarter of chromosome 2 in the L2 cells. In addition to the VvMybA1 functional allele, this deletion encompasses one allele of 194 genes according to the current 12X grape reference genome annotation (http://www.genoscope.cns.fr). As the grape sex locus (Dalbò et al., 2000; Marguerit et al., 2009) was mapped ~10Mb upstream to the 5’ border of the deletion, it is relevant to emphasize that the occurrence of the mutation in the L2 cells, which gives rise to the gametes, did not prevent the current use of Pinot gris as a parental genotype for sexual propagation and thus in breeding programmes.
The present comprehensive study, in agreement with the first suggestion by Hocquigny et al. (2004), builds on the model suggested by Walker et al. (2006) that envisaged a large deletion-similar to the one in Malian-in the L2 of Pinot gris, based on a non-layer-specific cleaved amplified polymorphic sequences (CAPS) analysis. In addition to the layer localization of this deletion, the phenotype difference between Pinot gris and Pinot noir is probably the result of a displacement of L2 cells (unpigmented phenotype) into L1 cells (pigmented phenotype), as suggested by the different height of the peaks of the electropherograms in the L1+L2-derived tissues of Pinot gris (Fig. 2) and as proposed for Malian and Pinot gris by Walker et al. (2006).
Furthermore, the present results show that Pinot blanc harbours a short heterozygous deletion, in the 100–179kb range, encompassing both the VvMybA1 and VvMybA2 genes. As has occurred in Pinot gris, the deletion affects the VvMybA1 functional allele. However, unlike Pinot gris, this deletion is present in both L1 and L2 cells in Pinot blanc. A partially deleted functional VvMybA1 allele was suggested to exist in Pinot blanc by Yakushiji et al. (2006) based on a non-layer-specific Southern blot approach. Since the SAM is organized in separate layers, it is likely that the deletion affected an L2 cell, which subsequently multiplied, colonizing the L1 cells.
In conclusion, both Pinot gris and Pinot blanc bear a mutation causing loss of function.
The distinctiveness of Pinot gris bud sports and Pinot blanc
In this study, by scoring the plants of Pinot gris clones in the field, partially or wholly white-skinned berries (derived chimeras or sports) were observed with a frequency range of 1–19% for six of these clones (2010; data not shown). As expected, the chimeric instability was higher in the non-registered than in the registered clones. Considering that this phenomenon is affected by changes in the environment (Whitham et al., 1981), the occurrence of these Pinot gris-derived chimeras and sports is in agreement with what was previously described in the literature (Hocquigny et al., 2004; Furiya et al., 2008; Pelsy, 2010). In fact, their generation can occur as a consequence of a rearrangement (or displacement) of cell layers in an original chimera, being itself unstable (Dermen, 1960; Bergann, 1967). In the case of loss of a functional allele, as in Pinot gris, this mechanism is expected to generate dominant phenotypes in chimeric sectors that can be readily exposed to natural or anthropic selection (Fernandez et al., 2010). Such a mosaic of genetic variation, that more probably exists in plants that have multiple apical meristems and/or exhibit a propensity for clonal propagation (e.g. many grasses, shrubs, and fruit trees-such as grape) than in plants that exhibit a high degree of apical dominance and rarely clone (e.g. many conifers), implies that there is a potential ecological and adaptive significance (Whitham et al., 1981) and represents an untapped resource for investigations of cell interactions during plant development (e.g. Boss and Thomas, 2002).
In order to ascertain the complete cell displacement in the Pinot gris chimeras and bud sports and to exclude the hypothesis that Pinot blanc is a bud sport of Pinot gris, the layer-specific genetic analysis was extended to the Pinot gris-derived mutants and to additional Pinot blanc clones. The results revealed the presence of the 4202–4350kb deletion in all white skin sectors and wholly mutated skins of Pinot gris, confirming that these cells hold an L2 genetic origin, carrying the information for the unpigmented phenotype (Hocquigny et al., 2004). Colourless sectors of Pinot gris are analogous to the hairless sectors of Pinot meunier in which cells from the L2 layer displace cells of the L1 layer (Stenkamp et al., 2009). The finding are, indeed, in contrast to the study performed by Furiya et al. (2008), relying on one triallelic SSR profile based on a non-layer-specific survey in Pinot gris white-skinned berries. With regards to the additional Pinot blanc clones, they showed, at the cell layer level, the same genetic profile, which includes the 100–179kb deletion, as the Pinot blanc clones belonging to the core set. This result demonstrated that the Pinot gris clones and the Pinot blanc clones do not possess the same deletion pattern, suggesting that they have arisen from two independent ancestral mutations.
Besides deletions, structural changes can be caused by point mutations, duplications, inversions, aneuploidy, polyploidy, and extrachromosomal mechanisms of inheritance (Hartmann and Kester, 1975). Genomic loss can be retrotrasposition driven, and the rate at which transposon-induced mutations arise appears to be affected by the stress or shock experienced by the genome (McClintock, 1984). It seems probable that virus infection, very common in plants, may be among the most important natural stresses responsible for transposon activation. As recently observed in other Pinot noir clones (Carrier et al., 2012), it can be supposed that the structural changes that occurred in Pinot gris and in Pinot blanc were stress mediated, resulting in the activation of a mobile genetic element. It is known that retrotransposons (class I; Wicker et al., 2007) contribute to genome dynamics and to structural diversity in different plant species (Vitte and Bennetzen, 2006). In fact, unequal intrastrand (or illegitimate) recombination between the two long terminal repeats (LTRs), that terminate LTR retrotransposons, often generates solo LTRs, with the associated loss of one LTR and the internal sequences of the element (Roeder and Fink, 1980). Therefore, unequal recombination between homologous LTR retrotransposons at different genomic locations can cause large and net deletion (or duplication) of nuclear DNA between the elements (Garfinkel, 2005), similar to those detected here in Pinot gris and Pinot blanc.
The evolutionary model of Pinots
Based on the overall experimental data, in particular on the relevant difference in the size of the chromosome deletion, it is suggested that: (i) Pinot blanc is not a bud sport of Pinot gris; and (ii) Pinot blanc and Pinot gris arose as independent somatic mutations of Pinot noir.
Theoretically, the origin of a colourless berry mutant can be ascribed to two distinct models: (i) the sequential model, where the black-skinned berry ancestor gave rise to the grey-skinned which in turn gave rise to the white-skinned berry mutant; and (ii) the parallel model, where the black-skinned berry ancestor gave rise to the grey-skinned and the white-skinned berry mutants separately.
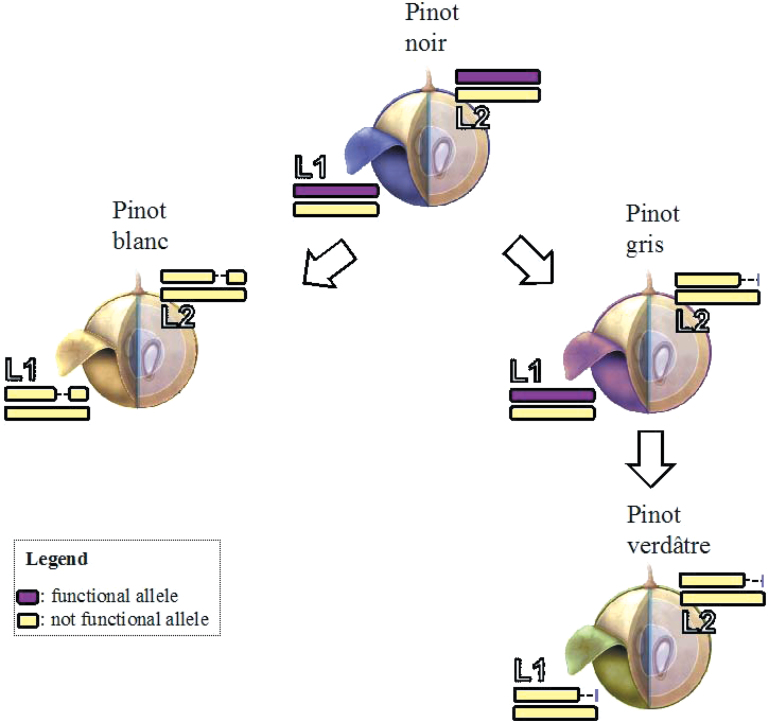
Here, the parallel model is proposed as the evolutionary model for the formation of Pinot berry colour somatic variants (Fig. 4). According to this novel model, the somatic mutants Pinot gris and Pinot blanc arose from the ancestral Pinot noir cultivar independently. This parallel model is named the ‘Pinot-model’, distinct from the previously reported sequential ‘CabSau-model’ (Walker et al., 2006). Moreover, these results elucidated the relationship between Pinot blanc and Pinot gris. Finally, the name Pinot verdâtre is suggested for the unpigmented bud sport of Pinot gris, holding a peculiar genetic make-up and a green-like phenotype.
Fig. 4.
Model of the formation of Pinot blanc, Pinot gris, and Pinot verdâtre from Pinot noir. A scheme represents the structural dynamics at the berry colour locus in the L1 and in the L2 of Pinot noir, Pinot blanc, Pinot gris, and Pinot verdâtre.
These findings represent a breakthrough towards the full understanding of the mechanisms behind the formation of white, grey, red, pink, and tintoreous grape cultivars, the overall phenotypes of which determine a specific enological aptitude.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of the studied Pinots.
Table S2. Primer information of the microsatellite markers used.
Table S3. Primer information of the studied regions distributed along chromosome 2.
Table S4. Genetic profile of the Pinot core set at the genome level.
Table S5. Genetic make-up of the Pinot core set at the berry colour locus.
Table S6. Genotypic state of the Pinot core set at all the analysed loci along chromosome 2.
Supplementary Material
Acknowledgements
The authors are grateful to L. Bianchedi (FEM, IT) for support in sample preparation, to A. Ciccotti (FEM, IT) for the in vitro maintenance of the IASMA grape clone collection, and to J. Terleth (Laimburg, IT) and L. Bavaresco (CRA-VIT, IT) for providing additional grape clone materials. The authors also wish to thank the Sequencing and Genotyping Platform team (FEM, IT) for helpful suggestions on SNPlex assay. We gratefully acknowledge E. De Paoli for helpful discussion. Finally, we wish to thank P.K. Boss for proofreading and critically reading the manuscript, and the anonymous reviewers for their useful suggestions. This work was supported by the S.A.M.P.LE. Vitis post-doc project funded by the Autonomous Province of Trento.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool Journal of Molecular Biology 215 403–410 [DOI] [PubMed] [Google Scholar]
- Azuma A, Kobayashi S, Goto-Yamamoto N, Shiraishi M, Mitani N, Yakushiji H, Koshita Y. 2009. Color recovery in berries of grape (Vitis vinifera L.) ‘Benitaka’, a bud sport of ‘Italia’, is caused by a novel allele at the VvmybA1 locus Plant Science 176 470–478 [DOI] [PubMed] [Google Scholar]
- Azuma A, Kobayashi S, Mitani N, Shiraishi M, Yamada M, Ueno T, Kono A, Yakushiji H, Koshita Y. 2008. Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin Theoretical and Applied Genetics 117 1009–1019 [DOI] [PubMed] [Google Scholar]
- Azuma A, Udo Y, Sato A, Mitani N, Kono A, Ban Y, Yakushiji H, Koshita Y, Kobayashi S. 2011. Haplotype composition at the color locus is a major genetic determinant of skin color variation in Vitis×labruscana grapes Theoretical and Applied Genetics 122 1427–1438 [DOI] [PubMed] [Google Scholar]
- Bergann F. 1967. The relative instability of chimerical clones: the basis for further breeding. In: Stubbe H, ed. Induced mutations and their utilization Berlin: Akademia Verlag; 287–300 [Google Scholar]
- Boss PK, Thomas MR. 2002. Association of dwarfism and floral induction with a grape ‘green revolution’ mutation Nature 416 847–850 [DOI] [PubMed] [Google Scholar]
- Boursiquot JM, This P. 1999. Essai de definition du cépage Progrès Agricole et Viticole 116 359–361 [Google Scholar]
- Cadle-Davidson MM, Owens CL. 2008. Genomic amplification of the Gret1 retroelement in white-fruited accessions of wild Vitis and interspecific hybrids Theoretical and Applied Genetics 116 1079–1094 [DOI] [PubMed] [Google Scholar]
- Carrier G, Le Cunff L, Dereeper A, Legrand D, Sabot F, Bouchez O, Audeguin L, Boursiquot J-M, This P. Transposable elements are a major cause of somatic polymorphism in Vitis vinifera L. PLoS One. 2012;7:e32973. doi: 10.1371/journal.pone.0032973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelet P, Laucou V, Fernandez L, Sreekantan L, Lacombe T, Martinez-Zapater JM, Thomas MR, Torregrosa L. 2007. Characterization of Vitis vinifera L. somatic variants exhibiting abnormal flower development patterns Journal of Experimental Botany 58 4107–4118 [DOI] [PubMed] [Google Scholar]
- Dalbò MA, Ye GN, Weeden NF, Steinkellner H, Sefc KM, Reisch BI. 2000. A gene controlling sex in grapevines placed on a molecular marker-based genetic map Genome 43 333–340 [PubMed] [Google Scholar]
- D’Amato F. 1997. Role of somatic mutations in the evolution of higher plants Caryologia 50 1–15 [Google Scholar]
- Dermen H. 1960. Nature of plant sports American Horticultural Magazine July 123–173 [Google Scholar]
- Einset J, Pratt C. 1954. ‘Giant’ sports of grapes Proceedings of the American Society for Horticultural Science 63 251–256 [Google Scholar]
- Fernandez L, Doligez A, Lopez G, Thomas MR, Bouquet A, Torregrosa L. 2006. Somatic chimerism, genetic inheritance, and mapping of the fleshless berry (flb) mutation in grapevine (Vitis vinifera L.) Genome 49 721–728 [DOI] [PubMed] [Google Scholar]
- Fernandez L, Torregrosa L, Segura V, Bouquet A, Martinez-Zapater JM. 2010. Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine The Plant Journal 61 545–557 [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Lacombe T, Le Cunff L, Boursiquot JM, This P. 2010. Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.) Heredity 104 351–362 [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Le Cunff L, Gomez C, Doligez A, Ageorges A, Roux C, Bertrand Y, Souquet J-M, Cheynier V, This P. 2009. Quantitative genetic bases of anthocyanin variation in grape (Vitis vinifera L. ssp. sativa) berry: a quantitative trait locus to quantitative trait nucleotide integrated study Genetics 183 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T, Botta R, Thomas MR. 2002. Chimerism in grapevines: implications for cultivar identity, ancestry and genetic improvement Theoretical and Applied Genetics 104 192–199 [DOI] [PubMed] [Google Scholar]
- Furiya T, Suzuki S, Sueta T, Takayanagi T. 2008. Molecular characterization of bud sport of Pinot Gris bearing white berries American Journal of Enology and Viticulture 59 66–73 [Google Scholar]
- Galet P. Dictionnaire encyclopédique des cépages. Paris: Hachette; 2000. [Google Scholar]
- Garfinkel DJ. 2005. Genome evolution mediated by Ty elements in Saccharomyces Cytogenetic and Genome Research 110 63–69 [DOI] [PubMed] [Google Scholar]
- Giannetto S, Velasco R, Troggio M, Malacarne G, Storchi P, Cancellier S, De Nardi B, Crespan M. 2008. A PCR-based diagnostic tool for distinguishing grape skin color mutants Plant Science 175 402–409 [Google Scholar]
- Hartmann HT, Kester DE. 1975. Plant propagation: principles and practices 3rdedn. Prentice-Hall International; [Google Scholar]
- Hocquigny S, Pelsy F, Dumas V, Kindt S, Heloir MC, Merdinoglu D. 2004. Diversification within grapevine cultivars goes through chimeric states Genome 47 579–589 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury J-M, Noel B, et al. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla Nature 449 463–465 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H. 2004. Retrotransposon-induced mutations in grape skin color Science 304 982–982 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Yamamoto NG, Hirochika H. 2005. Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin—color mutants Journal of the Japanese Society for Horticultural Science 74 196–203 [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways Trends in Plant Science 10 236–242 [DOI] [PubMed] [Google Scholar]
- Lacombe T, Audeguin L, Boselli M, et al. 2011. Grapevine European Catalogue: towards a comprehensive list Vitis 50 65–68 [Google Scholar]
- Levadoux L. 1956. Les populations sauvages et cultivées de Vitis vinifera L Annales de l’Amélioration des Plantes 6 59–118 [Google Scholar]
- Lodhi MA, Ye GN, Weeden NF, Reisch BI. 1994. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species Plant Molecular Biology Reports 12 6–13 [Google Scholar]
- Marcotrigiano M, Bernatzky R. 1995. Arrangement of cell-layers in the shoot apical meristems of periclinal chimeras influences cell fate The Plant Journal 7 193–202 [Google Scholar]
- Marguerit E, Boury C, Manicki A, Donnart M, Butterlin G, Nemorin A, Wiedemann-Merdinoglu S, Merdinoglu D, Ollat N, Decroocq S. 2009. Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine Theoretical and Applied Genetics 118 1261–1278 [DOI] [PubMed] [Google Scholar]
- Mattivi F, Guzzon R, Vrhovsek U, Stefanini M, Velasco R. 2006. Metabolite profiling of grape: flavonols and anthocyanins Journal of Agricultural and Food Chemistry 54 7692–7702 [DOI] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology. 2008;8:83. doi: 10.1186/1471-2229-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1984. The significance of responses of the genome to challenge Science 226 792–801 [DOI] [PubMed] [Google Scholar]
- Neilson-Jones W. Plant chimeras. London: Methuen and Co. Ltd; 1969. [Google Scholar]
- Pelsy F. 2010. Molecular and cellular mechanisms of diversity within grapevine varieties Heredity 104 331–340 [DOI] [PubMed] [Google Scholar]
- Pindo M, Vezzulli S, Coppola G, Cartwright DA, Zharkikh A, Velasco R, Troggio M. SNP high-throughput screening in grapevine using the SNPlex™ genotyping system. BMC Plant Biology. 2008;8:12. doi: 10.1186/1471-2229-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regner F, Stadlbauer A, Eisenheld C, Kaserer H. 2000. Genetic relationships among Pinots and related cultivars American Journal of Enology and Viticulture 51 7–14 [Google Scholar]
- Roeder GS, Fink GR. 1980. DNA rearrangements associated with a transposable element in yeast Cell 21 239–249 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers Methods in Molecular Biology 132 365–386 [DOI] [PubMed] [Google Scholar]
- Shamel AD, Pomeroy CS. 1936. Bud mutations in horticultural crops Journal of Heredity 27 486–494 [Google Scholar]
- Shimazaki M, Fujita K, Kobayashi H, Suzuki S. Pink-colored grape berry is the result of short insertion in intron of color regulatory gene. PLoS One. 2011;6:e21308. doi: 10.1371/journal.pone.0021308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp SHG, Becker MS, Hill BHE, Blaich R, Forneck A. 2009. Clonal variation and stability assay of chimeric Pinot Meunier (Vitis vinifera L.) and descending sports Euphytica 165 197–209 [Google Scholar]
- This P, Lacombe T, Cadle-Davidson M, Owens CL. 2007. Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1 Theoretical and Applied Genetics 114 723–730 [DOI] [PubMed] [Google Scholar]
- Thomas MR, Scott NS. 1993. Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as Sequence-Tagged Sites (STSS) Theoretical and Applied Genetics 86 985–990 [DOI] [PubMed] [Google Scholar]
- Thompson MM, Olmo HP. 1963. Cytohistological studies of cytochimeric and tetraploid grapes American Journal of Botany 50 901–906 [Google Scholar]
- Tobler AR, Short S, Andersen MR, et al. 2005. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping Journal of Biomolecular Techniques 16 398–406 [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One. 2007;2:e3216. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli S, Micheletti D, Riaz S, Pindo M, Viola R, This P, Walker MA, Troggio M, Velasco R. A SNP transferability survey within the genus Vitis . BMC Plant Biology. 2008;8:128. doi: 10.1186/1471-2229-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala P, Vermorel V. Ampélographie Tome 1. Paris: Masson et Cie; 1909. [Google Scholar]
- Vitte C, Bennetzen JL. 2006. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution Proceedings of the National Academy of Sciences, USA 103 17638–17643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Lee E, Robinson SP. 2006. Two new grape cultivars, bud sports of Cabernet Sauvignon bearing pale-coloured berries, are the result of deletion of two regulatory genes of the berry colour locus Plant Molecular Biology 62 623–635 [DOI] [PubMed] [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, Robinson SP. 2007. White grapes arose through the mutation of two similar and adjacent regulatory genes The Plant Journal 49 772–785 [DOI] [PubMed] [Google Scholar]
- Whitham TG, Slobodchikoff CN. 1981. Evolution by individuals, plant–herbivore interactions, and mosaics of genetic variability. The adaptive significance of somatic mutations in plants Oecologia 49 287–292 [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, et al. 2007. A unified classification system for eukaryotic transposable elements Nature Reviews Genetics 8 973–982 [DOI] [PubMed] [Google Scholar]
- Yakushiji H, Kobayashi S, Goto-Yamamoto N, Jeong ST, Sueta T, Mitani N, Azuma A. 2006. A skin color mutation of grapevine, from black-skinned Pinot Noir to white-skinned Pinot Blanc, is caused by deletion of the functional VvmybA1 allele Bioscience, Biotechnology, Biochemistry 70 1506–1508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.