Abstract
The green alga Chlamydomonas reinhardtii mutant 76–5EN lacks photosynthesis because of a nuclear-gene mutation that specifically inhibits expression of the chloroplast gene encoding the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco; EC 4.1.1.39). Photosynthesis-competent revertants were selected from mutant 76–5EN to explore the possibility of increasing Rubisco expression. Genetic analysis of 10 revertants revealed that most arose from suppressor mutations in nuclear genes distinct from the original 76–5EN mutant gene. The revertant strains have regained various levels of Rubisco holoenzyme, but none of the suppressor mutations increased Rubisco expression above the wild-type level in either the presence or absence of the 76–5EN mutation. One suppressor mutation, S107–4B, caused a temperature-conditional, photosynthesis-deficient phenotype in the absence of the original 76–5EN mutation. The S107–4B strain was unable to grow photosynthetically at 35°C, but it expressed a substantial level of Rubisco holoenzyme. Whereas the 76–5EN gene encodes a nuclear factor that appears to be required for the transcription of the Rubisco large-subunit gene, the S107–4B nuclear gene may be required for the expression of other chloroplast genes.
Rubisco (EC 4.1.1.39) catalyzes both the carboxylation and oxygenation of ribulose-1,5-bisphosphate. Carboxylation initiates photosynthetic carbon assimilation, but oxygenation initiates photorespiration, a nonessential process that leads to the loss of CO2. Because an increase in carboxylation or a decrease in oxygenation would increase net photosynthetic CO2 fixation, there has been significant interest in engineering an improved Rubisco (for reviews, see Spreitzer, 1993; Hartman and Harpel, 1994). Eukaryotic Rubisco is a chloroplast-localized protein assembled from eight copies each of the large and small subunits. The 55-kD large subunits are coded by the chloroplast rbcL gene, whereas a family of nuclear RbcS genes encodes the 15-kD small subunits (for review, see Spreitzer, 1993). The enzyme active sites are formed at the interfaces between the large subunits (for review, see Schneider et al., 1992).
Analysis of chloroplast rbcL mutations in the green alga Chlamydomonas reinhardtii has identified a number of large-subunit structural interactions that affect the stability and catalysis of the Rubisco holoenzyme (Chen et al., 1991; Thow et al., 1994; Spreitzer et al., 1995; Zhu and Spreitzer, 1996; Hong and Spreitzer, 1997). Nuclear mutations that affect the stability, catalysis, and expression of Rubisco have also been recovered in C. reinhardtii (Spreitzer et al., 1992), but the molecular basis for these effects remains unknown. The 68–11AR mutation decreases the thermal stability and carboxylase/oxygenase ratio of Rubisco (Spreitzer et al., 1988; Gotor et al., 1994). However, the S52–2B suppressor mutation restores the thermal stability and reduced carboxylase/oxygenase ratio of Rubisco caused by a chloroplast rbcL mutation (Chen et al., 1988, 1990, 1993). These two nuclear mutations do not reside within the RbcS genes and are not allelic to each other. Instead, the 68–11AR and S52–2B mutations affect Rubisco at posttranslational steps (Chen et al., 1990, 1993; Gotor et al., 1994). The genes in which these mutations arose may prove useful as targets for exploring the production of a better Rubisco.
Another C. reinhardtii nuclear mutant, named 76–5EN, differs from the other Rubisco mutants in that it fails to accumulate significant levels of rbcL mRNA (Hong and Spreitzer, 1994). Nuclear mutations have previously been recovered that inhibit the expression of chloroplast genes at posttranscriptional steps (for review, see Mayfield et al., 1995). However, the 76–5EN mutation is unique. It appears to block large-subunit expression at the level of rbcL transcription (Hong and Spreitzer, 1994).
Genetic selection for photosynthesis-competent revertants of C. reinhardtii rbcL mutants has proven fruitful for understanding the structure/function relationships of Rubisco (Chen and Spreitzer, 1989; Chen et al., 1990, 1991; Thow et al., 1994; Spreitzer et al., 1995; Hong and Spreitzer, 1997). Because mutant 76–5EN has a photosynthesis-deficient phenotype (Hong and Spreitzer, 1994), it is also possible to select photosynthesis-competent revertants from this mutant. In this study 10 photosynthesis-competent suppressor mutations have been analyzed to better understand the nature of the 76–5EN mutant gene. Suppressors of 76–5EN were tested to see whether any might increase the expression of wild-type Rubisco, considering that an increase in the amount of Rubisco would also be beneficial for increasing net CO2 fixation.
MATERIALS AND METHODS
Strains and Culture Conditions
Chlamydomonas reinhardtii wild-type 2137 mt+ (Spreitzer and Mets, 1981), mutant 76–5EN (Hong and Spreitzer, 1994), and revertant and suppressor strains were maintained on solid medium containing 10 mm sodium acetate and 1.5% Bacto agar (Difco, Detroit, MI) at 25°C in darkness (Spreitzer and Mets, 1981). The light-sensitive, acetate-requiring mutant 76–5EN was recovered from mutagenized 2137 mt+ cells in a previous study (Hong and Spreitzer, 1994). Mutant 76–5EN lacks Rubisco holoenzyme due to a nuclear gene mutation that inhibits rbcL mRNA accumulation (Hong and Spreitzer, 1994). For biochemical analysis, cells were grown with liquid acetate medium on a rotary shaker at 220 rpm in darkness until they reached a density of 5 × 106 cells mL−1.
Genetic Selection and Analysis
Photosynthesis-competent revertants were selected from independent clones of mutant 76–5EN by plating 2 × 106 cells per 100-mm Petri dish of minimal medium (without acetate) at a light intensity of 45 to 80 μmol photons m−2 s−1. Growth phenotypes were assessed by performing spot tests on solid media at 25 and 35°C (Spreitzer and Mets, 1981; Spreitzer et al., 1988). Genetic crosses were performed as described previously (Spreitzer and Mets, 1981; Spreitzer et al., 1988). Phenotypes of dark-grown progeny were scored by replica plating tetrads to minimal medium in the light and acetate medium in the dark at 25 and 35°C.
SDS-PAGE and Western Analysis
Cells from 500-mL cultures were harvested by centrifugation and sonicated in 1 mm DTT, 10 mm MgCl2, 10 mm NaHCO3, and 50 mm Bicine, pH 8.0, at 0°C for 3 min. Cell extract was centrifuged at 30,000g for 15 min, and the supernatant of soluble proteins was prepared for SDS-PAGE (Thow et al., 1994). Protein was quantified by the method of Bradford (1976). Samples were fractionated by SDS-PAGE on 7.5 to 15% polyacrylamide gradient gels (Laemmli, 1970). After electrophoresis, proteins were either stained with Coomassie blue or transferred to nitrocellulose for western analysis (Chen et al., 1993). Rubisco subunits were detected (Gotor et al., 1994) with anti-tobacco Rubisco IgG and visualized via chemiluminescence (Amersham).
RNA Hybridization
Total RNA was isolated from dark-grown cells and subjected to northern analysis as described previously (Hong and Spreitzer, 1994). For dot hybridizations, 2 μg of total RNA was immobilized on nylon membranes according to the procedure described by Sambrook et al. (1989), using a Hybri-Dot manifold (BRL). The following DNA subfragments of chloroplast genes were nick translated with [32P]dCTP and used as probes in northern or dot hybridization experiments: 0.9-kb EcoRI-PstI fragment of the atpA gene that encodes the α-subunit of ATP synthase (Leu et al., 1992), 1.5-kb HindIII-PstI fragment of the atpB gene that encodes the β-subunit of ATP synthase (Woessner et al., 1986), 0.4-kb HindIII-PstI fragment of the petD gene that encodes subunit IV of the Cyt b6/f complex (Yu and Spreitzer, 1991), 0.9-kb BamHI-DraI fragment of the psaB gene that encodes the B protein of PSI (Kück et al., 1987), 1.5-kb HincII fragment of the psbD gene that encodes the D2 polypeptide of PSII (Rochaix et al., 1984), 0.8-kb HindIII fragment of the rbcL gene (Dron et al., 1982), 0.3-kb HindIII-HincII fragment of the rpl20 gene that encodes the L20 protein of the ribosomal large subunit (Yu et al., 1992), and 1.3-kb HindIII-EcoRI fragment of the tufA gene that encodes the Tu elongation factor (Baldauf and Palmer, 1990).
RESULTS
Recovery and Genetic Analysis of Revertants
Photosynthesis-competent revertants were recovered from mutant 76–5EN at a frequency of 2.7 × 10−7 cells. Ten genetically independent revertants were chosen for further analysis. Whereas revertants R107–4A, R118–7A, and R118–25G have essentially wild-type phenotypes, the revertants R107–4D, R118–1E, R118–6G, R118–8A, R118–26G, and R107–4B were found to have temperature-conditional, acetate-requiring phenotypes. These latter revertants died on minimal medium at 35°C but were indistinguishable from the wild type on minimal medium in the light at 25°C or on acetate medium in the dark at 25 and 35°C. Revertant R118–5E also died on minimal medium at 35°C. However, this strain also grew poorly in comparison with the wild type on minimal medium at 25°C and on acetate medium in the dark at 35°C, indicating that the R118–5E reversion event may exert pleiotropic effects on cell functions. The observed diversity of revertant phenotypes indicates that there is more than one way to genetically complement the original 76–5EN mutation.
Each revertant was crossed with a wild-type mt− tester strain to determine the genetic basis of reversion. One revertant, R107–4A, produced only wild-type progeny in 19 analyzed tetrads. Thus, R107–4A arises from true reversion, intragenic suppression, or intergenic suppression via a closely linked gene. Although zygotes were produced in crosses with revertant R118–5E, these failed to germinate. The remaining eight revertants produced parental-ditype (all wild-type progeny), nonparental-ditype (two wild-type and two acetate-requiring progeny), and tetratype (three wild-type and one acetate-requiring progeny) tetrads when 7 to 35 tetrads per strain were analyzed at 25°C. In other words, each of these eight revertants results from an intergenic suppressor mutation that segregates from and is unlinked to the original 76–5EN mutation. Strains containing only the suppressor mutation were recovered from nonparental-ditype tetrads and saved for further study. These suppressor mutations and strains were named S118–7A, S118–25G, S107–4D, S118–1E, S118–6G, S118–8A, S118–26G, and S107–4B. Except for S107–4B, all of these strains have wild-type phenotypes. Suppressor S107–4B has a temperature-conditional, acetate-requiring phenotype. It died on minimal medium at 35°C but was indistinguishable from the wild type on minimal medium at 25°C or on acetate medium in the dark at 25 and 35°C. Analysis of 20 complete tetrads with respect to the suppressor and 76–5EN mutant phenotypes (7 parental ditype, 1 nonparental ditype, and 12 tetratype tetrads) allowed the unambiguous recovery of an S107–4B strain and confirmed that the suppressor and temperature-conditional phenotypes are genetically linked.
Rubisco Levels in Revertants and Suppressors
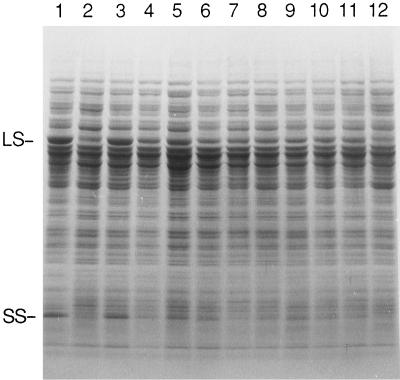
When dark-grown cells were analyzed by SDS-PAGE, mutant 76–5EN was found to lack Rubisco subunits (Fig. 1, lane 2), as observed previously (Hong and Spreitzer, 1994). The revertants were found to express various amounts of Rubisco, but none had a higher level of Rubisco subunits than the wild type (Fig. 1, lanes 3–12). Revertant R107–4A has a wild-type level of Rubisco (Fig. 1, lane 3), which would favor the idea that it results from true reversion. The temperature-conditional revertants R107–4D, R118–1E, R118–6G, R118–8A, R118–26G, R107–4B, and R118–5E (Fig. 1, lanes 6–12) were found to have less Rubisco, in general, than those revertants that have wild-type phenotypes (R107–4A, R118–7A, and R118–25G; Fig. 1, lanes 3–5). Perhaps this accounts for their temperature sensitivity (Chen and Spreitzer, 1992).
Figure 1.
SDS-PAGE of total soluble cell proteins from the wild type (lane 1), mutant 76–5EN (lane 2), and revertants of 76–5EN (lanes 3–12). Cells were grown in the dark at 25°C prior to extraction. Each lane received 60 μg of soluble protein, and the gel was stained with Coomassie blue after electrophoresis. Extracts of revertants R107–4A, R118–7A, R118–25G, R107–4D, R118–1E, R118–6G, R118–8A, R118–26G, R107–4B, and R118–5E were fractionated in lanes 3 through 12, respectively. LS, Large subunit; SS, small subunit.
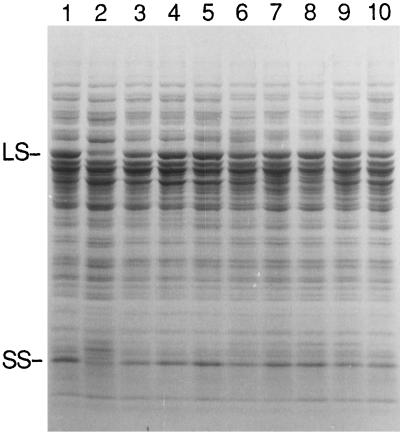
Eight of the revertants had been found to arise from intergenic suppressor mutations that could be separated from the original 76–5EN mutation. We were curious to see whether these suppressors in otherwise wild-type cells might increase the expression of wild-type Rubisco. However, when cell extracts of the suppressor strains (grown at 25°C in darkness) were fractionated by SDS-PAGE, none appeared to have more Rubisco than the wild type (Fig. 2).
Figure 2.
SDS-PAGE of total soluble cell proteins from the wild type (lane 1), mutant 76–5EN (lane 2), and the 76–5EN suppressors (lanes 3–10). The suppressor mutations were separated from the original 76–5EN mutation by performing genetic crosses. Cells were grown in the dark at 25°C prior to extraction. Each lane received 60 μg of soluble protein, and the gel was stained with Coomassie blue after electrophoresis. Extracts of suppressor strains S118–7A, S118–25G, S107–4D, S118–1E, S118–6G, S118–8A, S118–26G, and S107–4B were fractionated in lanes 3 through 10, respectively. LS, Large subunit; SS, small subunit.
Suppressor S107–4B
Suppressor mutation S107–4B is the only suppressor that causes a temperature-conditional, acetate-requiring phenotype in the absence of the original 76–5EN mutation. We reasoned that this phenotype may have been the result of either a specific defect in rbcL expression or alterations in the expression of other chloroplast genes. To discriminate between these possibilities, revertant R107–4B (76–5EN/S107–4B) and suppressor S107–4B were analyzed further.
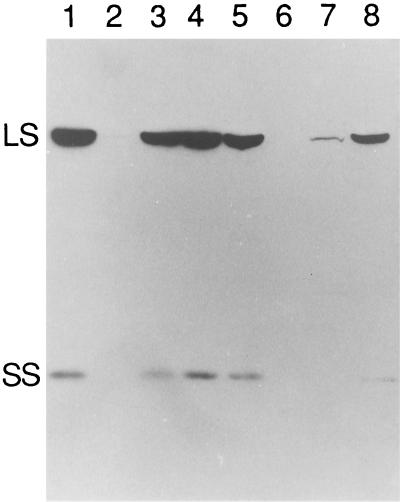
Western analysis of total soluble proteins from dark-grown cells showed that both the revertant and suppressor strains had substantial amounts of Rubisco subunits when grown at 25°C in the dark (Fig. 3, lanes 3 and 4). When the revertant R107–4B (76–5EN/S107–4B) was grown at 35°C, significant decreases in the large and small subunits were observed (Fig. 3, lane 7). Thus, revertant strain R107–4B appeared to lack photosynthetic ability at 35°C because the S107–4B suppressor failed to efficiently restore Rubisco expression at this restrictive temperature. However, suppressor strain S107–4B, which does not contain the 76–5EN mutation, also lacked photosynthetic ability at 35°C. Western analysis revealed the presence of substantial amounts of Rubisco subunits when S107–4B was grown at this restrictive temperature (Fig. 3, lane 8). Thus, the temperature-conditional photosynthesis deficiency of S107–4B does not appear to result from a lack of Rubisco.
Figure 3.
Western analysis of Rubisco subunits in cell extracts of the wild type (lanes 1 and 5), mutant 76–5EN (lanes 2 and 6), revertant R107–4B (lanes 3 and 7), and suppressor S107–4B (lanes 4 and 8). Cells were grown in darkness at 25°C (lanes 1–4) or 35°C (lanes 5–8) prior to extraction. Each lane received 60 μg of soluble protein. The gel was blotted after electrophoresis, and the filter was probed with rabbit anti-tobacco Rubisco-holoenzyme IgG. LS, Large subunit; SS, small subunit.
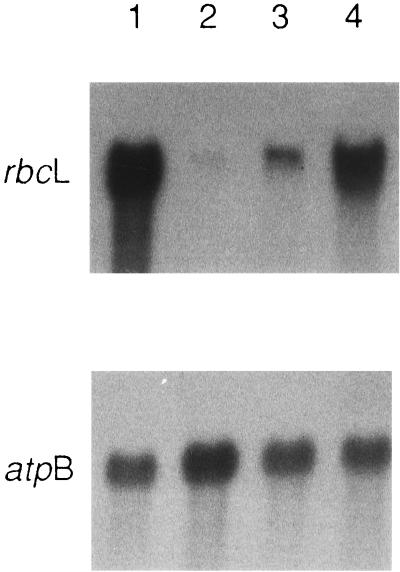
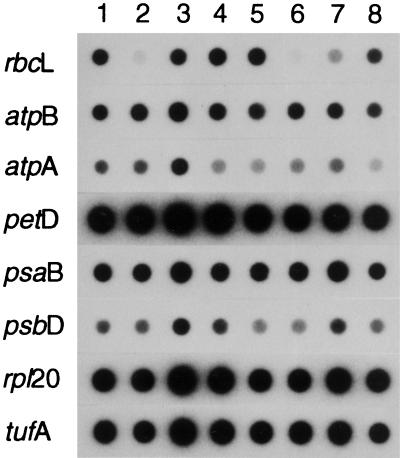
Northern analysis of 35°C dark-grown cells was performed to investigate whether the levels of Rubisco subunits observed at 35°C (Fig. 3) correspond to those of rbcL mRNA. As shown in Figure 4, the S107–4B mutation increased the level of rbcL mRNA when present with the original 76–5EN mutation (Fig. 4, compare lanes 2 and 3) but slightly decreased the level of rbcL mRNA when present alone (Fig. 4, compare lanes 1 and 4). There is a direct correlation between the levels of Rubisco subunits and rbcL mRNA in 35°C-grown cells (compare Fig. 3, lanes 5–8, with Fig. 4, lanes 1–4). Because the S107–4B suppressor strain maintains substantial levels of Rubisco subunits and rbcL mRNA at 35°C (Figs. 3 and 4), its photosynthesis-deficient phenotype must result from a deficiency in some other photosynthetic component. However, with respect to chloroplast gene expression, dot hybridization failed to detect any substantial difference in the amount of atpA, atpB, petD, psaB, psbD, rpl20, or tufA mRNA when 25°C- and 35°C-grown wild-type, mutant 76–5EN, revertant R107–4B, and suppressor S107–4B strains were compared (Fig. 5).
Figure 4.
Northern analysis of rbcL and atpB mRNA in the wild type (lane 1), mutant 76–5EN (lane 2), revertant R107–4B (lane 3), and suppressor S107–4B (lane 4) grown at 35°C in darkness. Top, Total RNA (10 μg) was separated by electrophoresis on a formaldehyde/agarose gel, blotted to nylon membrane, and hybridized with an rbcL gene probe. The blot was exposed to x-ray film with an intensifying screen for 24 h at −70°C. Bottom, After an autoradiogram was obtained, the same filter was treated with 0.1× Denhardt's reagent (Denhardt, 1966), 1 mm EDTA, and 1 mm Tris, pH 8.0, at 75°C to partially remove the rbcL DNA probe. The filter was then hybridized with an atpB gene probe. Autoradiography was performed with an intensifying screen for 24 h at −70°C.
Figure 5.
Dot-hybridization analysis of chloroplast mRNAs in the wild type (columns 1 and 5), mutant 76–5EN (columns 2 and 6), revertant R107–4B (columns 3 and 7), and suppressor S107–4B (columns 4 and 8). Cells were grown in darkness at 25°C (columns 1–4) or 35°C (columns 5–8) prior to extraction. Total RNA (2 μg) was denatured with 50% formamide and 7% formaldehyde and immobilized at each spot on nylon membranes. Individual membranes were then hybridized with each of the designated chloroplast gene probes. Samples were exposed to x-ray film with an intensifying screen at −70°C for either 24 h (rbcL, atpB, atpA, and petD) or 72 h (psaB, psbD, rpl20, and tufA).
DISCUSSION
The 76–5EN nuclear mutant was characterized previously (Hong and Spreitzer, 1994). It lacks Rubisco holoenzyme (Fig. 3) because it fails to accumulate a significant amount of rbcL mRNA (Figs. 4 and 5). The mutant strain synthesizes only a trace of rbcL mRNA during a 10-min pulse labeling with 32Pi, but this trace of rbcL mRNA is stable during a 1-h chase (Hong and Spreitzer, 1994). Thus, the 76–5EN mutation appears to block large-subunit expression at the level of transcription. It does not destabilize the mRNA at a posttranscriptional step. As shown previously (Hong and Spreitzer, 1994) and in the present study (Figs. 4 and 5), the 76–5EN mutation does not affect the accumulation of other chloroplast-encoded mRNAs, and it does not block chlorophyll production or the function of photosynthetic electron transport (Hong and Spreitzer, 1994). Based on these observations, the 76–5EN mutation appears to be quite specific in its inhibition of rbcL expression. Other nuclear mutations that block chloroplast gene expression act posttranscriptionally and sometimes affect the expression of a number of genes (for review, see Mayfield et al., 1995).
Because the 76–5EN mutation is specific for large-subunit expression, we reasoned that the 76–5EN gene might serve as a target for increasing the amount of Rubisco without disrupting the expression of other chloroplast genes (Hong and Spreitzer, 1994). To this end, photosynthesis-competent revertants of 76–5EN were selected. However, none of these revertant strains (Fig. 1) and none of the extragenic suppressor strains obtained from genetic crosses (Fig. 2) was found to have levels of Rubisco above the wild-type amount. Considering that a recent study with C. reinhardtii has indicated that the amount of small subunits controls the amount of large subunits at the level of translation (Khrebtukova and Spreitzer, 1996), we believe that it may have been unreasonable to attempt to increase the amount of Rubisco by increasing expression of the large subunits. However, the nuclear suppressors may still prove valuable if small subunit expression can be increased.
The photosynthesis-competent revertants of 76–5EN were recovered at a relatively high frequency. One of them, R107–4A, likely results from true reversion or intragenic suppression, indicating that the original 76–5EN mutation is not a deletion or gene rearrangement. However, most of the revertants arose from mutations in a number of other genes (extragenic suppressors), and these revertants have various levels of Rubisco (Fig. 1). Furthermore, more than one-half of the revertants have temperature-conditional, photosynthesis-deficient phenotypes. It thus appears that there are a number of biochemical mechanisms by which the original 76–5EN mutation can be suppressed. These suppressors may improve the expression or function of the 76–5EN mutant gene product directly, or they may complement for a lack of function in a variety of ways. Because all but one of the extragenic suppressor strains (without 76–5EN present) have no visible or biochemical phenotype (Fig. 2), they may be difficult to study further.
One of the extragenic suppressor strains, S107–4B, has a temperature-conditional, acetate-requiring phenotype. It was surprising to find that this strain accumulates considerable amounts of Rubisco subunits (Fig. 3) and rbcL mRNA (Figs. 4 and 5) at the 35°C restrictive temperature. Thus, it seems likely that the wild-type allele of the S107–4B mutant gene does not normally play a role in the expression of rbcL. One must assume that the S107–4B mutation disrupts some other cellular component at 35°C and that this component may be involved in the expression of chloroplast genes. Because the suppressor strain can survive in the dark at the restrictive temperature, it is unlikely that the suppressor mutation exerts its lethal effect on cellular metabolism in general. Perhaps the wild-type allele of the suppressor gene encodes a transcription factor required for the expression of one or more chloroplast genes (Tiller et al., 1991; Troxler et al., 1994; Kim and Mullet, 1995). The S107–4B mutation may alter the specificity of this factor so that it can now, in addition, recognize the rbcL promoter, thereby suppressing the effects of the original 76–5EN mutation. At 35°C, the mutant protein may be thermally unstable. Some of this mutant protein may still be present to partially suppress the 76–5EN mutation (Figs. 3, lane 7, and 4, lane 3) but not enough to serve its usual role for the expression of one or more other photosynthetic genes.
Because of the similarities between the sequences and regulation of the C. reinhardtii rbcL and atpB promoters (Klein et al., 1992, 1994), we suspected that the S107–4B suppressor mutation might also affect the expression of the atpB gene. However, the amount of atpB mRNA was found to be the same in the wild type, mutant 76–5EN, revertant R107–4B, and suppressor S107–4B (Fig. 4). In fact, analysis of four additional photosynthetic genes (atpA, petD, psaB, and psbD) and two protein-synthesis genes (rpl20 and tufA) failed to detect a block in mRNA accumulation at the 35°C restrictive temperature (Fig. 5, lanes 7 and 8).
Although there is evidence for the existence of transcription factors and RNA polymerases that may favor the expression of sets of chloroplast genes (Tiller et al., 1991; Iratni et al., 1994; Kim and Mullet, 1995), the S107–4B mutation does not affect the expression of a diverse sample of chloroplast genes (Fig. 5). Perhaps S107–4B affects the expression of only one or a few genes, and further screening by northern hybridization would ultimately identify the lesion. Alternatively, S107–4B may affect one of many posttranscriptional steps in protein maturation, and a number of other models could be proposed for complementing a defective transcription factor but disrupting the function of other chloroplast proteins. Nonetheless, because the 76–5EN and S107–4B strains have acetate-requiring phenotypes, it should be possible to isolate the wild-type alleles of the mutant genes via genomic complementation (Funke et al., 1997). The nature of these genes may clarify the action of the mutations, and the mutant strains may be valuable for further understanding the mechanisms of transcription in the chloroplast.
ACKNOWLEDGMENTS
We gratefully acknowledge receiving plasmid clones of atpB from N.W. Gillham, psbD from E.H. Harris, and atpA, psaB, and tufA from D.P. Weeks. We thank Carolyn M. O'Brien for assisting with the preparation of the figures.
Abbreviation:
- mt
mating-type locus
Footnotes
This work was supported by a U.S. Department of Agriculture/National Research Initiative Competitive Grant (no. 94-37306-0349) and published as paper no. 12,011 of the Nebraska Agricultural Research Division journal series.
LITERATURE CITED
- Baldauf SL, Palmer JD. Evolutionary transfer of the chloroplast tufA gene to the nucleus. Nature. 1990;344:262–265. doi: 10.1038/344262a0. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen Z, Chastain CJ, Al-Abed SR, Chollet R, Spreitzer RJ. Reduced CO2/O2 specificity of ribulose-1,5-bisphosphate carboxylase/oxygenase in a temperature-sensitive chloroplast mutant of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1988;85:4696–4699. doi: 10.1073/pnas.85.13.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Green D, Westhoff C, Spreitzer RJ. Nuclear mutation restores the reduced CO2/O2 specificity of ribulosebisphosphate carboxylase/oxygenase in a temperature-conditional chloroplast mutant of Chlamydomonas reinhardtii. Arch Biochem Biophys. 1990;283:60–67. doi: 10.1016/0003-9861(90)90612-3. [DOI] [PubMed] [Google Scholar]
- Chen Z, Hong S, Spreitzer RJ. Thermal instability of ribulose-1,5-bisphosphate carboxylase/oxygenase from a temperature-conditional chloroplast mutant of Chlamydomonas reinhardtii. Plant Physiol. 1993;101:1189–1194. doi: 10.1104/pp.101.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Spreitzer RJ. Chloroplast intragenic suppression enhances the low CO2/O2 specificity of mutant ribulose-bisphosphate carboxylase/oxygenase. J Biol Chem. 1989;264:3051–3053. [PubMed] [Google Scholar]
- Chen Z, Spreitzer RJ. How various factors influence the CO2/O2 specificity of ribulose-1,5-bisphosphate carboxylase/oxygenase. Photosynth Res. 1992;31:157–164. doi: 10.1007/BF00028792. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yu W, Lee JH, Diao R, Spreitzer RJ. Complementing amino acid substitutions within loop 6 of the α/β-barrel active site influence the CO2/O2 specificity of chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase. Biochemistry. 1991;30:8846–8850. doi: 10.1021/bi00100a017. [DOI] [PubMed] [Google Scholar]
- Denhardt DT. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966;23:641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dron M, Rahire M, Rochaix JD. Sequence of the chloroplast DNA region of Chlamydomonas reinhardtii containing the gene of the large subunit of ribulose bisphosphate carboxylase and parts of its flanking genes. J Mol Biol. 1982;162:775–793. doi: 10.1016/0022-2836(82)90547-2. [DOI] [PubMed] [Google Scholar]
- Funke RP, Kovar JL, Weeks DP. Intracellular carbonic anhydrase is essential to photosynthesis in Chlamydomonas reinhardtii at atmospheric levels of CO2. Demonstration via genomic complementation of the high-CO2-requiring mutant ca-1. Plant Physiol. 1997;114:237–244. doi: 10.1104/pp.114.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotor C, Hong S, Spreitzer RJ. Temperature-conditional nuclear mutation of Chlamydomonas reinhardtii decreases the CO2/O2 specificity of chloroplast ribulosebisphosphate carboxylase/oxygenase. Planta. 1994;193:313–319. [Google Scholar]
- Hartman FC, Harpel MR. Structure, function, regulation, and assembly of d-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu Rev Biochem. 1994;63:197–234. doi: 10.1146/annurev.bi.63.070194.001213. [DOI] [PubMed] [Google Scholar]
- Hong S, Spreitzer RJ. Nuclear mutation inhibits expression of the chloroplast gene that encodes the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Physiol. 1994;106:673–678. doi: 10.1104/pp.106.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Spreitzer RJ. Complementing substitutions at the bottom of the barrel influence catalysis and stability of ribulose-bisphosphate carboxylase/oxygenase. J Biol Chem. 1997;272:11114–11117. doi: 10.1074/jbc.272.17.11114. [DOI] [PubMed] [Google Scholar]
- Iratni R, Baeza L, Andreeva A, Mache R, Lerbs-Mache S. Regulation of rDNA transcription in chloroplasts: promoter exclusion by constitutive repression. Genes Dev. 1994;8:2928–2938. doi: 10.1101/gad.8.23.2928. [DOI] [PubMed] [Google Scholar]
- Khrebtukova I, Spreitzer RJ. Elimination of the Chlamydomonas gene family that encodes the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA. 1996;93:13689–13693. doi: 10.1073/pnas.93.24.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Mullet JE. Identification of a sequence-specific DNA binding factor required for transcription of the barley chloroplast blue light-responsive psbD-psbC promoter. Plant Cell. 1995;7:1445–1457. doi: 10.1105/tpc.7.9.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, De Camp JD, Bogorad L. Two types of chloroplast gene promoters in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1992;89:3453–3457. doi: 10.1073/pnas.89.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Salvador ML, Bogorad L. Activity of the Chlamydomonas chloroplast rbcL gene promoter is enhanced by a remote sequence element. Proc Natl Acad Sci USA. 1994;91:10819–10823. doi: 10.1073/pnas.91.23.10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück U, Choquet Y, Schneider M, Dron M, Bennoun P. Structural and transcriptional analysis of two homologous genes for the P700 chlorophyll alpha-apoproteins in Chlamydomonas reinhardtii: evidence for in vivo trans-splicing. EMBO J. 1987;6:2185–2195. doi: 10.1002/j.1460-2075.1987.tb02489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leu S, Schlesinger J, Michaels A, Shavit N. Complete DNA sequence of the Chlamydomonas reinhardtii chloroplast atpA gene. Plant Mol Biol. 1992;18:613–616. doi: 10.1007/BF00040681. [DOI] [PubMed] [Google Scholar]
- Mayfield SP, Yohn CB, Cohen A, Danon A. Regulation of chloroplast gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:147–166. [Google Scholar]
- Rochaix JD, Dron M, Rahire M, Malnoe P. Sequence homology between the 32 kilodalton and the D2 chloroplast membrane polypeptides of Chlamydomonas reinhardtii. Plant Mol Biol. 1984;3:363–370. doi: 10.1007/BF00033383. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schneider G, Lindqvist Y, Brändén CI. Rubisco: structure and mechanism. Annu Rev Biophys Biomol Struct. 1992;21:119–143. doi: 10.1146/annurev.bb.21.060192.001003. [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ. Genetic dissection of Rubisco structure and function. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:411–434. [Google Scholar]
- Spreitzer RJ, Al-Abed SR, Huether MJ. Plant Physiol. 1988;86:773–777. doi: 10.1104/pp.86.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ, Mets L. Photosynthesis-deficient mutants of Chlamydomonas reinhardtii with associated light-sensitive phenotypes. Plant Physiol. 1981;67:565–569. doi: 10.1104/pp.67.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ, Thow G, Zhu G. Pseudoreversion substitution at large-subunit residue 54 influences the CO2/O2 specificity of chloroplast ribulose-bisphosphate carboxylase/oxygenase. Plant Physiol. 1995;109:681–685. doi: 10.1104/pp.109.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ, Thow G, Zhu G, Chen Z, Gotor C, Zhang D, Hong S (1992) Chloroplast and nuclear mutations that affect Rubisco structure and function in Chlamydomonas reinhardtii. In N Murata, ed, Research in Photosynthesis, Vol III. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 593–600
- Thow G, Zhu G, Spreitzer RJ. Complementing substitutions within loop regions 2 and 3 of the α/β-barrel active site influence the CO2/O2 specificity of chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase. Biochemistry. 1994;33:5109–5114. doi: 10.1021/bi00183a014. [DOI] [PubMed] [Google Scholar]
- Tiller K, Eisermann A, Link G. The chloroplast transcription apparatus from mustard (Sinapis alba L.): evidence for three different transcription factors which resemble bacterial ς factors. Eur J Biochem. 1991;198:93–99. doi: 10.1111/j.1432-1033.1991.tb15990.x. [DOI] [PubMed] [Google Scholar]
- Troxler RF, Zhang F, Hu J, Bogorad L. Evidence that ς factors are components of chloroplast RNA polymerase. Plant Physiol. 1994;104:753–759. doi: 10.1104/pp.104.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner JP, Gillham NW, Boynton JE. The sequence of the chloroplast atpB gene and its flanking regions in Chlamydomonas reinhardtii. Gene. 1986;44:17–28. doi: 10.1016/0378-1119(86)90038-7. [DOI] [PubMed] [Google Scholar]
- Yu W, Spreitzer RJ. Sequences of trnR-ACG and petD that contain a tRNA-like element within the chloroplast genome of Chlamydomonas reinhardtii. Nucleic Acids Res. 1991;19:957. doi: 10.1093/nar/19.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zhang D, Spreitzer RJ. Sequences of the Chlamydomonas reinhardtii chloroplast genes encoding tRNASer and ribosomal protein L20. Plant Physiol. 1992;100:1079–1080. doi: 10.1104/pp.100.2.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Spreitzer RJ. Directed mutagenesis of chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase: loop 6 substitutions complement for structural stability but decrease catalytic efficiency. J Biol Chem. 1996;271:18494–18498. doi: 10.1074/jbc.271.31.18494. [DOI] [PubMed] [Google Scholar]