Abstract
PURPOSE
Protein kinases including Protein Kinase A (PKA) underlie myriad important signaling pathways. The ability to monitor kinase activity in vivo and in real-time with high spatial resolution in genetically-specified cellular populations is a yet unmet need, crucial for understanding complex biological systems as well as for preclinical development and screening of novel therapeutics.
METHODS
Using the hypothesis that the natural recognition sequences of protein kinases may be detected using chemical exchange saturation transfer (CEST) magnetic resonance imaging (MRI), we designed a genetically encoded biosensor composed of eight tandem repeats of the peptide LRRASLG, a natural target of PKA.
RESULTS
This sensor displays a measurable change in CEST signal following phosphorylation by PKA. The natural PKA substrate LRRASLG exhibits a CEST-MRI contrast at +1.8 and +3.6 ppm, with a >50% change after phosphorylation with minutes-scale temporal resolution. Expression of a synthetic gene encoding eight monomers of LRRASLG yielded two peaks at these CEST frequencies.
CONCLUSION
Taken together, these results suggest that this gene may be used to assay PKA levels in a biologically relevant system. Importantly, the design strategy used for this specific sensor may be adapted for a host of clinically interesting protein kinases.
Keywords: chemical exchange saturation transfer (CEST), biosensor, reporter gene, protein kinase A (PKA)
INTRODUCTION
Protein kinases play an instrumental role in almost every signaling process in the living cell, and yet, there is no means to visualize these enzymes in vivo with high spatial and temporal resolution. Kinases are drug development targets of great importance for a wide range of diseases including cancer, diabetes, and inflammation, as each disease is linked to the perturbation of protein kinase–mediated cell signaling pathways (1). There are more than 500 different protein kinases identifies in the human genome (2). Protein Kinase A (PKA), also known as cAMP-dependent protein kinase, was the first one discovered in this large family of proteins (3). Since then, PKA has been extensively studied in innumerable systems, mostly in tissue extracts using antibodies or pharmacological inhibitors, which are not always specific (4). Protein kinases catalyze the energetically irreversible step of adding a phosphoryl group (-PO4-2) to hydroxyl (-OH) targets. In particular, PKA phosphorylates the hydroxyl group of serines or threonines having the pattern RRX(S/T)Y with the highest specificity towards the peptide sequence LRRASLG (5). This sequence has allowed the development of a genetically encoded sensor for PKA based on fluorescence resonance energy transfer (FRET) (6), which exhibits extraordinary spatial and temporal resolution for cells in culture when using optical microscopy (7). However, due to limited visible light penetration and scattering in tissue, FRET cannot be easily applied in vivo. As an alternative, we sought to develop an MRI sensor that can report on phosphorylation in real-time.
Molecular MRI, with tools like chemical exchange saturation transfer (CEST) reporter genes (8,9), is attractive as it can track metabolite dynamics in vivo with better spatial resolution and anatomic co-registration than traditional techniques. In CEST MRI, a chemical group of interest is identified as one whose resonance frequency is separated sufficiently from water via a chemical shift (10-12). For example, the guanidyl and amide protons of arginine residues resonate at +1.8 and +3.6 ppm from water, respectively, with a separation from water that is sufficient for performing CEST MRI. During CEST MRI, these proton spins are saturated with radiofrequency pulses tuned to their resonance frequency. When these protons undergo chemical exchange with protons of the surrounding water, they transfer saturation to the water protons, thereby significantly reducing the signal generated by the water protons. This new contrast mechanism has been used in a range of applications, including measuring temperature (13), pH (14), enzymatic activity (15), metal ions (16), gene expression (8), glycogen (17), glycosaminoglycan (18), glutamate (19), drug delivery (20,21) and kidney function (22).
Since LRRASLG contains two arginine residues coupled with a peptide (amide) bond, we hypothesized that this peptide could serve as a good CEST agent. Moreover, a polypeptide containing multiple repeats of this sequence should yield amplification of the contrast with an overall detection sensitivity appropriate for in vivo use and a size that would yield ease of handling with standard biochemical techniques. Given that CEST contrast is highly dependent on the proton exchange rate, the presence of a strong negatively charged group, (i.e. PO4-2) should slow the exchange rate and consequently reduce the contrast, making this candidate sensor able to detect phosphorylation by PKA. Based on this hypothesis, we designed a genetically encoded biosensor whose proton exchange rates should change sufficiently with phosphorylation by PKA to be detected with CEST MRI. Eight tandem repeats of the base peptide LRRASLG was chosen for the design of the sensor based on the above considerations. Specifically, the gene is translated to a 6.8 kDa protein that allows both high CEST contrast as well as better protein detection and purification with standard biochemical techniques. Such an MRI biosensor may have applications in monitoring stem cell and organ transplants, and screening therapeutics that target this important signaling pathway.
MATERIALS AND METHODS
Compound preparation
Peptides were synthesized by NeoBioSci (Cambridge, MA) and provided as lyophilized powder. All compounds were dissolved in 10 mM PBS (pH=7.4) at the indicated concentration.
CEST-MRI
CEST MRI experiments were performed on a vertical 11.7T Bruker Avance system. Approximately 20 μl of each sample was loaded into a capillary tube and up to 20 samples were scanned at each session to minimize variability as described before (23). A modified RARE (TR/TE=8000/9.4 ms, RARE factor=16, 1 mm slice thickness, FOV=11×11 mm, matrix size=64×32, resolution=0.17×0.34mm, and NA=1-2) including a magnetization transfer (MT) module (B1=5.0μT/4000ms) was used to acquire CEST weighted images from -5 to +5 ppm (step=0.2 ppm) around the water resonance (0 ppm). To account for inhomogeneity of the B0 field, the absolute water resonant frequency shift was measured using a modified Water Saturation Shift Reference (WASSR) method (24), using the same parameters as in CEST imaging except TR=1.5 sec, saturation pulse=500 ms, B1=0.5 μT, and a sweep range from -2 to +2 ppm (step= 0.1 ppm). Data processing was performed using custom-written scripts in MATLAB. After B0 correction for each voxel using the WASSR data, mean CEST-spectra were calculated from an ROI for each sample. The MTR asymmetry was quantified as MTRasym=(S-Δω – S+Δω)/ S0 (S-Δω and S+Δω are the water peak differentials due to saturation at the target frequency +ω and its negative –ω, respectively. S0 is the unsaturated signal. By using the difference in signal between saturation at the target frequency and its opposite (negative) frequency cancels any effects due to a direct, non-exchange mediated saturation of water.
Phosphorylation assay
PKA and ATP were purchased from New England Biolabs (NEB, Ipswich, MA) and provided in solution buffer. The reactions were completed with a peptide concentration of 1 mM in 1x PKA buffer and 4 mM ATP. Samples were prepared on ice. Then, following the manifacturer's protocol, samples were reacted for the indicated time at 30 °C using a bench-top S1000 Thermal Cycler (BioRad, Hercules, CA) and then heat-shocked at 65 °C for 20 minutes to inactivate the enzyme. Following heat inactivation, samples were stored at 4 °C until further analysis.
Plasmid construction
Plasmids were generated by custom synthesis of a gene encoding 8 tandem repeats of the base sequence LRRASLG, with a 5-histidine C-terminal tag, whose coding sequence is optimized for expression in E. coli (Genscript, Piscataway, NJ). This gene was then subcloned into the pET expression system using TOPO cloning (Invitrogen, Carlsbad, CA).
Protein expression
E. coli: BL21(DE3) chemically competent cells (Invitrogen) were transformed with pET-LRRASLG8-His6. After induction in Magic Media™ (Invitrogen) at 30 °C for 18 hours, the total protein was extracted using Cell Lytic B reagent (Sigma, St. Louis, MO). Lysate buffer was exchanged by passage through a Zeba 7K MWCO spin column (Pierce, Rockford, IL) with PBS (pH=7.4) as the elution buffer.
RESULTS
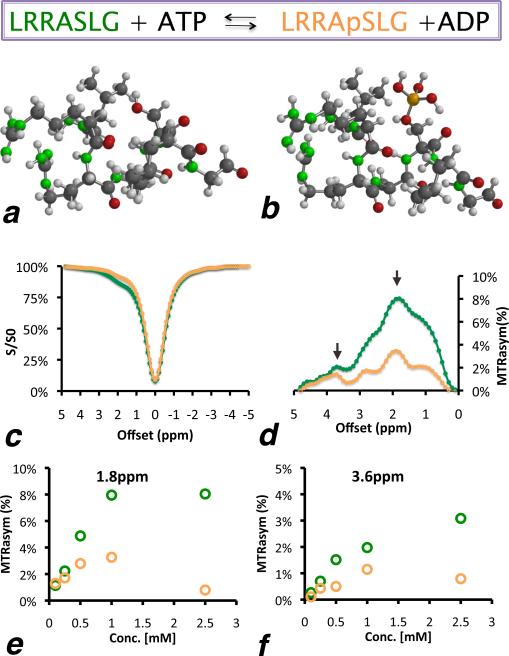
CEST contrast was measured for the PKA consensus sequences LRRASLG (Figure 1a) and its phosphorylated form, LRRApSLG, which has a phosphoryl group synthetically conjugated to the hydroxyl group of the serine, modeling the product of PKA action on the unphosphorylated substrate (Figure 1b). In Figure 1c, the CEST-spectra – i.e., the normalized signal intensity of water protons – is plotted as a function of the off-resonance saturation frequency for these two peptides at a concentration of 1 mM. For both peptides, the CEST spectra are asymmetric, indicating that these peptides generate CEST contrast, with the unphosphorylated form generating significantly higher contrast. There is a considerable difference in MTRasym between the unphosphorylated and the phosphorylated peptide, with two distinguishable peaks at 1.8 ppm and 3.6 ppm (Figure 1d) frequency offsets. This difference is directly concentration-dependent down to micromolar concentrations (Figure 1e,f).
Figure 1.
Sequence visualization of (a) unphosphorylated and (b) phosphorylated LRRASLG peptide (phosphoryl group in orange). (c) CEST-spectra and (d) and MTRasym plots of both peptides at 1 mM. MTRasym at (e) 1.8 ppm and (f) 3.6 ppm. L=leucine; R=arginine; A=alanine; S=serine and G=glycine.
To test whether this effect can resolve the temporal action of PKA, recombinant PKA was incubated with the peptide LRRASLG for varying amounts of time (Figure 2). A >50% decrease in CEST contrast was seen with kinetics similar to that expected for recombinant PKA activity, with the majority of the signal difference generated within the first 15 minutes of incubation.
Figure 2.
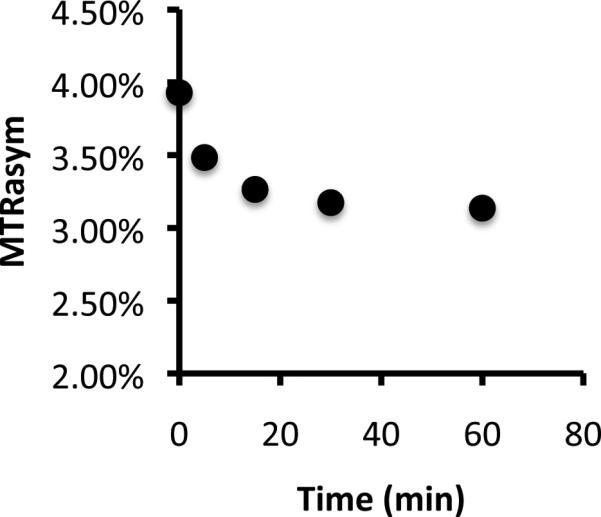
Biosensor phosphorylation by PKA. Shown is the time course of PKA phosphorylation measured with CEST MRI for 1 mM peptide.
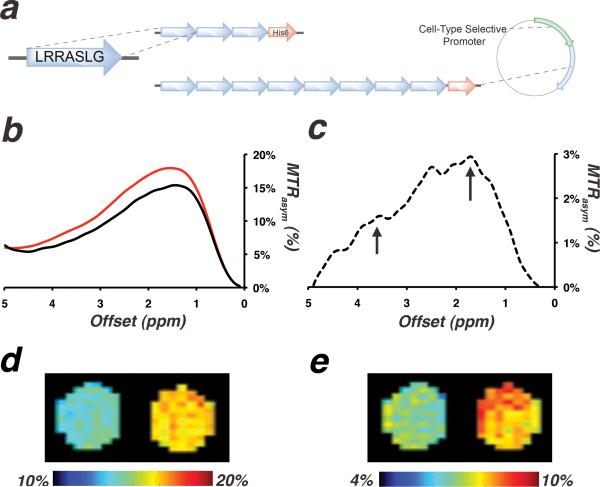
Next, an artificial gene that encodes eight tandems repeats of LRRASLG sequence was designed. The gene was cloned into a bacterial expression vector and was introduced into E. coli. Figure 3b shows that a lysate of E. coli expressing the genetically encoded PKA biosensor provides higher MTRasym compared to a lysate of E. coli expressing a control protein, cytosine deaminase, which shows no measurable CEST contrast of its own. This difference in MTRasym is more pronounced at the expected peaks of 1.8 ppm and 3.6 ppm (Figure 3c). Taken together, these results demonstrate the feasibility of detecting this genetically encoded candidate PKA biosensor in a biologically relevant environment.
Figure 3.
(a) Design of a genetically encoded biosensor for measuring PKA activity. (b) MTRasym of lysates of E. coli expressing the PKA biosensor gene (red) and a non-CEST-contrast-generating control (cytosine deaminase, black). (c) The difference between the two spectra in b (dashed-line), with arrows pointing to peaks at 1.8 and 3.6 ppm. MTRasym maps at (d) 1.8 ppm and (e) 3.6 ppm of cell extracts of E. coli overexpressing cytosine deaminase (left capillary) and extracts of E. coli overexpressing the PKA biosensor (right capillary).
DISCUSSION
Magnetic resonance has been used for detection of different kinases using genetically encoded reporters. Transgenic overexpression of creatine kinase (CK) by Koretsky et. al. (25) and of arginine kinase by Walter et. al. (26) was detected via 31P MR spectroscopy. In addition, an innovative design for an MRI sensor for PKA was described based on a transverse relaxation (T2) contrast mechanism, which requires aggregation of several proteins to generate MRI contrast (27). Here, we describe a biosensor that is encoded by a single gene that may respond directly to PKA activity. We anticipate that these reporters are uniquely suited to non-invasively monitor treatment response to small molecule and cellular therapeutics.
This sensor could potentially be further optimized using molecular tools, such as systematic mutagenesis, to improve the MRI sensitivity and the contrast between the phosphorylated and unphosphorylated state. Similar methods have been used to develop an MRI sensor for dopamine (28). This biosensor can be expressed in numerous target tissues using viral vectors for transduction (29) or in transgenic animals as was demonstrated with other genetically encoded MRI reporters (30,31).
In this study, we have capitalized on the arginine and serine content of consensus recognition sequences of proteins phosphorylated by PKA. Both of these residues are expected to provide high CEST contrast (32). In particular, peptides with basic, positively-charged lysine and arginine residues show high CEST contrast as the exchange rate of amide protons (NH) and amine/guanidyl protons (-NH3+/=NH2+) is sufficiently fast to generate contrast, without being too fast to merge their resonance peak with that of water. Hydroxyl protons (-OH) of polar serine and threonine residues can also contribute to CEST signal and perhaps enhance contrast by inducing water coordination with nearby basic residues. This suggests that phosphorylation by serine/threonine kinases could measurably alter the exchange rates of substrate protons by 1) altering the exchange of nearby guanidyl and amide protons through coordination by the negatively-charged phosphate group and 2) removing the CEST contrast generating hydroxyl proton of the phosphorylated serine or threonine. Indeed, the peptide described in our experiments exhibited a high CEST contrast in the unphosphorylated state, that is quenched by >50% in the phosphorylated state. Importantly, this effect is conserved across a range of sub-millimolar concentrations that are similar to the expected intracellular concentration of functional reporter gene expression.
As can be seen in Figures 1 and 2, the change in the CEST contrast displays both a linear and non-linear range. Thus, we anticipate that protocols for in vivo applications will have to include acquisition of baseline data prior to manipulation of PKA and are only quantitative in a linear range of concentrations. Our in vitro studies show that the sensor responds to phosphorylation on a timescale of minutes (Figure 2). This is the same timescale that is required for acquisition of the CEST data. However, in vivo, enzymes such as phosphatases can remove the phosphate group from the substrate, and may thus reduce the measured PKA activity before the end of the acquisition. In the future, faster CEST acquisition schemes on a timescale of seconds (33) should dramatically improve the temporal resolution. It also should be noted that the accuracy of in vivo quantification of CEST agents at chemical shifts close to water resonance frequency may be greatly influenced by B0 and B1 inhomogeneity. Therefore, the same WASSR protocol used here can be used to correct for B0 inhomogeneity for in vivo studies, as it is suitable for correcting inhomogeneities of up to 1000 Hz (34). The B1 inhomogeneity should also be measured separately and corrected if needed as shown previously (35).
PKA is not the only protein kinase that phosphorylates consensus sequences rich in arginine, lysine and serine residues. Among such kinases of biological importance are protein kinase C (PKC) (36) and Ca2+/cAMP-dependent protein kinase IIα (CaMKIIα) (37). Even though many kinases phosphorylate sequences with high similarity in amino acid composition, the specific ordering of these amino acids determines the kinase specificity. Thus, for instance PKC and CaMKII will not phosphorylate the substrate for PKA (6) and vice versa (36). This opens the possibility to engineer an array of CEST MRI-based sensors for detecting multiple variants of protein kinases in vivo.
ACKNOWLEDGMENTS
This study was supported by NIH grants EB008769, NS065284, EB005252, EB012590, EB006394, NS072171 and EB015032.
REFERENCES
- 1.Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303(5665):1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Walsh DA, Perkins JP, Krebs EG. An adenosine 3',5'-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243(13):3763–3765. [PubMed] [Google Scholar]
- 4.Murray AJ. Pharmacological PKA Inhibition: All May Not Be What It Seems. Sci Signal. 2008;1(22):re4. doi: 10.1126/scisignal.122re4. [DOI] [PubMed] [Google Scholar]
- 5.Kemp BE, Graves DJ, Benjamini E, Krebs EG. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. Journal of Biological Chemistry. 1977;252(14):4888–4894. [PubMed] [Google Scholar]
- 6.Zhang J, Ma Y, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci U S A. 2001;98(26):14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni Q, Ganesan A, Aye-Han NN, Gao X, Allen MD, Levchenko A, Zhang J. Signaling diversity of PKA achieved via a Ca2+-cAMP-PKA oscillatory circuit. Nat Chem Biol. 2011;7(1):34–40. doi: 10.1038/nchembio.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilad AA, McMahon MT, Walczak P, Winnard PT, Jr., Raman V, van Laarhoven HW, Skoglund CM, Bulte JW, van Zijl PC. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25(2):217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 9.Liu G, Liang Y, Bar-Shir A, Chan KW, Galpoththawela CS, Bernard SM, Tse T, Yadav NN, Walczak P, McMahon MT, Bulte JW, van Zijl PC, Gilad AA. Monitoring enzyme activity using a diamagnetic chemical exchange saturation transfer magnetic resonance imaging contrast agent. J Am Chem Soc. 2011;133(41):16326–16329. doi: 10.1021/ja204701x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J Magn Reson. 2000;143(1):79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 11.Sherry AD, Woods M. Chemical Exchange Saturation Transfer Contrast Agents for Magnetic Resonance Imaging. Annu Rev Biomed Eng. 2008;10:391–411. doi: 10.1146/annurev.bioeng.9.060906.151929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): what is in a name and what isn't? Magn Reson Med. 2011;65(4):927–948. doi: 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li AX, Wojciechowski F, Suchy M, Jones CK, Hudson RH, Menon RS, Bartha R. A sensitive PARACEST contrast agent for temperature MRI: Eu3+-DOTAM-glycine (Gly)-phenylalanine (Phe). Magn Reson Med. 2008;59(2):374–381. doi: 10.1002/mrm.21482. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, Soesbe TC, Kiefer GE, Zhao P, Sherry AD. A responsive europium(III) chelate that provides a direct readout of pH by MRI. J Am Chem Soc. 2010;132(40):14002–14003. doi: 10.1021/ja106018n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo B, Pagel MD. A PARACEST MRI contrast agent to detect enzyme activity. J Am Chem Soc. 2006;128(43):14032–14033. doi: 10.1021/ja063874f. [DOI] [PubMed] [Google Scholar]
- 16.Trokowski R, Ren J, Kalman FK, Sherry AD. Selective sensing of zinc ions with a PARACEST contrast agent. Angew Chem Int Ed Engl. 2005;44(42):6920–6923. doi: 10.1002/anie.200502173. [DOI] [PubMed] [Google Scholar]
- 17.van Zijl PC, Jones CK, Ren J, Malloy CR, Sherry AD. MRI detection of glycogen in vivo by using chemical exchange saturation transfer imaging (glycoCEST). Proc Natl Acad Sci U S A. 2007;104(11):4359–4364. doi: 10.1073/pnas.0700281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST). Proc Natl Acad Sci U S A. 2008;105(7):2266–2270. doi: 10.1073/pnas.0707666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai K, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, Detre JA, Reddy R. Magnetic resonance imaging of glutamate. Nat Med. 2012;18(2):302–306. doi: 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delli Castelli D, Dastru W, Terreno E, Cittadino E, Mainini F, Torres E, Spadaro M, Aime S. In vivo MRI multicontrast kinetic analysis of the uptake and intracellular trafficking of paramagnetically labeled liposomes. J Control Release. 2010;144(3):271–279. doi: 10.1016/j.jconrel.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Choi J, Kim K, Kim T, Liu G, Bar-Shir A, Hyeon T, McMahon MT, Bulte JW, Fisher JP, Gilad AA. Multimodal imaging of sustained drug release from 3-D poly(propylene fumarate) (PPF) scaffolds. J Control Release. 2011;156(2):239–245. doi: 10.1016/j.jconrel.2011.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinogradov E, He H, Lubag A, Balschi JA, Sherry AD, Lenkinski RE. MRI detection of paramagnetic chemical exchange effects in mice kidneys in vivo. Magn Reson Med. 2007;58(4):650–655. doi: 10.1002/mrm.21393. [DOI] [PubMed] [Google Scholar]
- 23.Liu G, Gilad AA, Bulte JW, van Zijl PC, McMahon MT. High-throughput screening of chemical exchange saturation transfer MR contrast agents. Contrast Media Mol Imaging. 2010;5(3):162–170. doi: 10.1002/cmmi.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim M, Gillen J, Landman BA, Zhou J, van Zijl PC. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med. 2009;61(6):1441–1450. doi: 10.1002/mrm.21873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koretsky AP, Brosnan MJ, Chen LH, Chen JD, Van Dyke T. NMR detection of creatine kinase expressed in liver of transgenic mice: determination of free ADP levels. Proc Natl Acad Sci U S A. 1990;87(8):3112–3116. doi: 10.1073/pnas.87.8.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walter G, Barton ER, Sweeney HL. Noninvasive measurement of gene expression in skeletal muscle. Proc Natl Acad Sci U S A. 2000;97(10):5151–5155. doi: 10.1073/pnas.97.10.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro MG, Szablowski JO, Langer R, Jasanoff A. Protein nanoparticles engineered to sense kinase activity in MRI. J Am Chem Soc. 2009;131(7):2484–2486. doi: 10.1021/ja8086938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro MG, Westmeyer GG, Romero PA, Szablowski JO, Kuster B, Shah A, Otey CR, Langer R, Arnold FH, Jasanoff A. Directed evolution of a magnetic resonance imaging contrast agent for noninvasive imaging of dopamine. Nat Biotechnol. 2010;28(3):264–270. doi: 10.1038/nbt.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iordanova B, Ahrens ET. In vivo magnetic resonance imaging of ferritin-based reporter visualizes native neuroblast migration. Neuroimage. 2012;59(2):1004–1012. doi: 10.1016/j.neuroimage.2011.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen B, Ziv K, Plaks V, Israely T, Kalchenko V, Harmelin A, Benjamin LE, Neeman M. MRI detection of transcriptional regulation of gene expression in transgenic mice. Nat Med. 2007;13(4):498–503. doi: 10.1038/nm1497. [DOI] [PubMed] [Google Scholar]
- 31.Bartelle BB, Berrios-Otero CA, Rodriguez JJ, Friedland AE, Aristizabal O, Turnbull DH. Novel genetic approach for in vivo vascular imaging in mice. Circ Res. 2012;110(7):938–947. doi: 10.1161/CIRCRESAHA.111.254375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahon MT, Gilad AA, DeLiso MA, Berman SM, Bulte JW, van Zijl PC. New “multicolor” polypeptide diamagnetic chemical exchange saturation transfer (DIACEST) contrast agents for MRI. Magn Reson Med. 2008;60(4):803–812. doi: 10.1002/mrm.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones CK, Polders D, Hua J, Zhu H, Hoogduin HJ, Zhou J, Luijten P, van Zijl PC. In vivo three-dimensional whole-brain pulsed steady-state chemical exchange saturation transfer at 7 T. Magn Reson Med. 2012;67(6):1579–1589. doi: 10.1002/mrm.23141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu G, Moake M, Har-el Y-e, Long CM, Chan KWY, Cardona A, Jamil M, Walczak P, Gilad AA, Sgouros G, van Zijl PCM, Bulte JWM, McMahon MT. In vivo multicolor molecular MR imaging using diamagnetic chemical exchange saturation transfer liposomes. Magn Reson Med. 2012;67(4):1106–1113. doi: 10.1002/mrm.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh A, Cai K, Haris M, Hariharan H, Reddy R. On B(1) inhomogeneity correction of in vivo human brain glutamate chemical exchange saturation transfer contrast at 7T. Magn Reson Med. 2012 doi: 10.1002/mrm.24290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161(5):899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondo N, Nishimura S. MALDI-TOF mass-spectrometry-based versatile method for the characterization of protein kinases. Chemistry. 2009;15(6):1413–1421. doi: 10.1002/chem.200801650. [DOI] [PubMed] [Google Scholar]