Abstract
The differentiation of several T and B cell effector programs in the immune system is directed by signature transcription factors that induce rapid epigenetic remodeling. We report that PLZF, the BTB-ZF transcription factor directing the innate-like effector program of NKT thymocytes 1,2 was prominently associated with cullin 3 (Cul3), an E3 ubiquitin ligase previously shown to use BTB domain-containing proteins as adaptors for substrate binding 3–7. PLZF transported Cul3 to the nucleus where the two proteins were associated within a chromatin modifying complex. Furthermore, PLZF expression resulted in selective changes of ubiquitination of multiple components of this complex. Cul3 was also found associated with another BTB-ZF transcription factor, Bcl6, which directs the B cell germinal center and the T follicular helper programs. Conditional deletion in mice demonstrated an essential role of Cul3 for the development of PLZF- and Bcl6-dependent lineages. We conclude that distinct lineage-specific BTB-ZF transcription factors recruit Cul3 to alter the ubiquitination pattern of their associated chromatin modifying complex. We propose that this novel function is essential to direct the differentiation of several T and B lymphocyte effector programs, and may also be involved in the oncogenic role of PLZF and Bcl6 in leukemias and lymphomas 8,9.
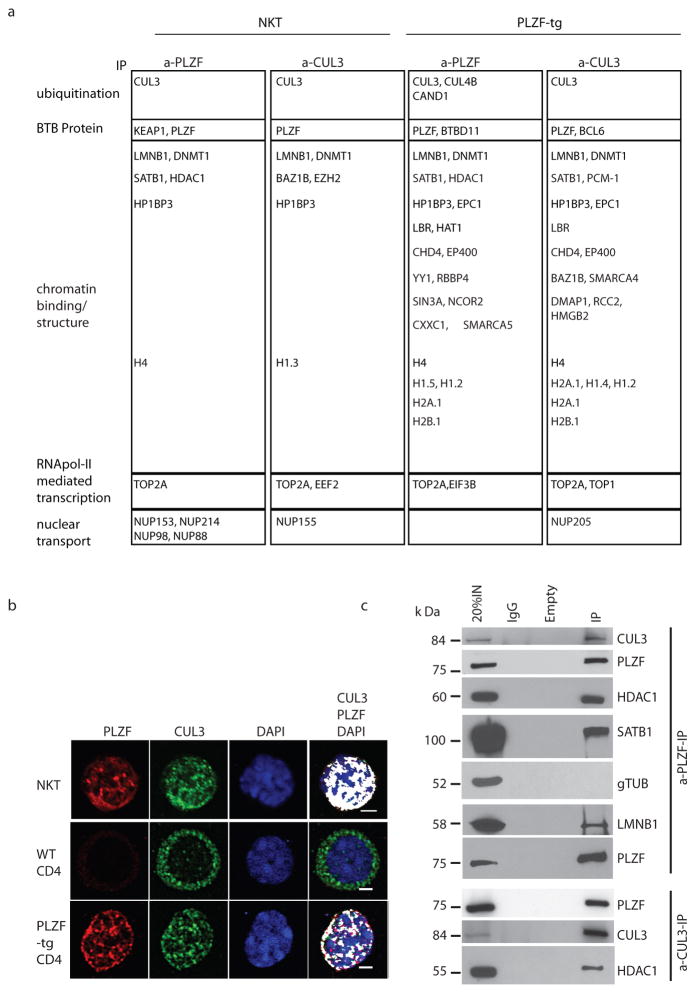
To investigate the molecular mechanisms that PLZF employs to regulate the innate-like NKT cell differentiation program during development, we examined its protein interaction partners. NKT thymocytes were purified from Vα14-Jα18 transgenic mice and, after immunoprecipitation with anti-PLZF antibody, associated proteins were submitted to mass spectrometry analysis (Figure 1a, column 1; Fig. S1). A major group was composed of nuclear proteins involved in binding and modifying chromatin, including HDAC1 and DNMT1, which were previously reported to interact with PLZF in myeloid cells 9,10, as well as special AT-rich binding protein 1 (SATB1) and lamin B1, which anchor specific DNA sequences to nuclear compartments associated with gene activation and repression, respectively 11–14. We focused on the E3 ubiquitin ligase Cul3 because previous reports had established that the BTB domain of several proteins, including the BTB-ZF protein BAZF, could serve as ‘adaptors’ for Cul3-mediated ubiquitination by binding both Cul3 and its substrates 3–7,15. Reciprocal immunoprecipitation of Cul3-associated proteins brought down PLZF as major protein along with an overlapping set of proteins (Fig. 1a, column 2; Fig. S1). Furthermore, confocal microscopic analysis of NKT thymocytes demonstrated colocalization of the two proteins in a speckled nuclear pattern (Fig. 1b, top row).
Figure 1. PLZF-Cul3 interactions.
a, Mass spectrometric analysis of proteins immunoprecipitated by anti-PLZF and anti-Cul3 from indicated thymocyte populations (data from two to three independent experiments). Additional proteins that did not belong to the indicated categories are shown with the complete datasets in Fig. S1–S2. Analysis of gene ontogeny term enrichment demonstrates p values ranging from 10−5 to 10−8 for nuclear transcriptional and chromatin organization proteins. b, Confocal microscopic analysis of fresh NKT thymocytes and splenic CD4 cells from WT and PLZF-Tg mice, as indicated. White color indicates colocalization (bar, 2 μm). c, Western blot analysis of anti-Cul3 and anti-PLZF immunoprecipitates from PLZF-tg thymocytes. Data are representative of at least three independent experiments.
In contrast, in the major lineage of CD4 T lymphocytes, Cul3 was mainly found in the cytosol with only a faint presence in nuclear speckles (Fig. 1b, middle row). However, upon expression of a CD4-promoter driven PLZF transgene, which induces developmental acquisition of the NKT lineage effector program 1,16, Cul3 was mostly in the nucleus, colocalizing with PLZF in nuclear speckles (Fig. 1b, bottom row). A similar binding and transport of Cul3 from the cytoplasm to the nucleus was previously demonstrated upon cotransfection with the nuclear BTB protein SPOP in HeLa cells 17. Mass spectrometric analysis of anti-PLZF and anti-Cul3 immunoprecipitates from PLZF-transgenic thymocytes identified a similar set of proteins as in NKT thymocytes (Fig. 1a, columns 3 and 4, Fig. S2) including additional known partners of PLZF such as Ncor and Sin3a 9. Western blot analyses confirmed that a fraction of PLZF co-precipitated with Cul3 and that chromatin binding and modifying proteins such as HDAC1, SATB1 and Lamin B1 were associated with the PLZF-Cul3 complex (Fig. 1c). The specificity of the interaction between PLZF and Cul3 was further tested using in vitro translated proteins, and shown to depend on Cul3 residues L52 and E55 (Fig. S3), as reported for other BTB proteins 3,18, athough direct binding remains to be formally established 19.
Of note, the BTB-ZF transcription factor Bcl6, which characterizes the germinal center B cell 8 and the follicular helper T cell responses 20 but is also transiently expressed by cortical thymocytes 21, was immunoprecipitated by anti-Cul3 in thymocytes (Fig. 1a, column 4). Analysis by western blot in transfected Hela cells confirmed this association (Fig. S4).
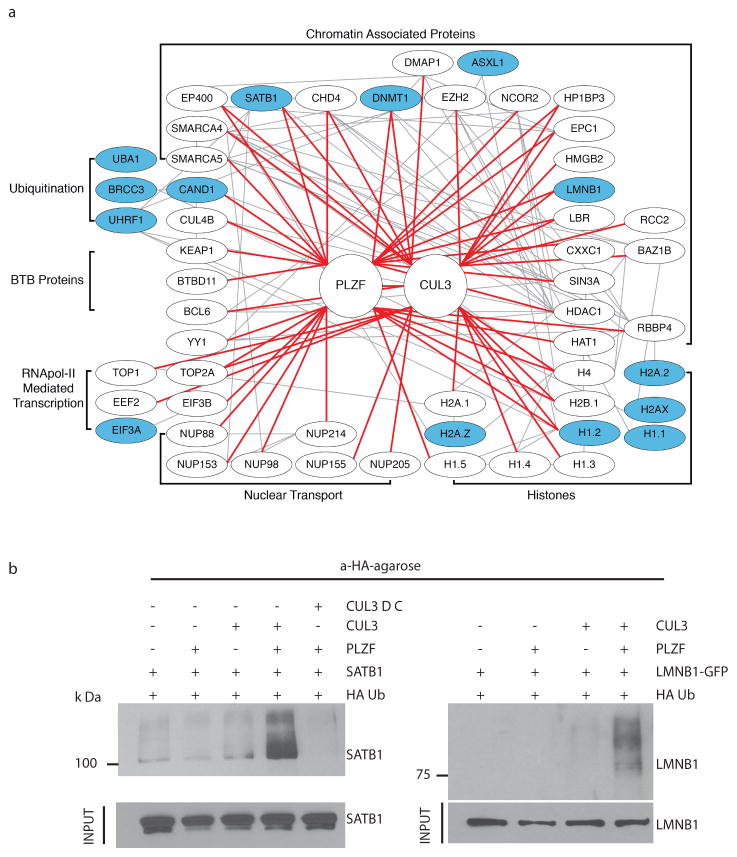
Rapid changes in ubiquitination pattern have recently been reported in chromatin remodeling situations and are thought to regulate gene expression 22–24. By bringing Cul3 from the cytosol to chromatin modifying complexes in the nucleus, PLZF might be expected to induce changes in ubiquitination. This was tested using an unbiased ubiquitination proteomics method (UbiscanR) comparing whole cell lysates of thymocytes from PLZF-transgenic and wild type littermates. Independent experiments with different batches of mice identified 48 proteins showing concordant changes, most of which consisted of increased ubiquitination in PLZF-tg cells (Fig. 2a and Fig. S5). Strikingly, 14 of these 48 proteins were either components of the Cul3-PLZF complex identified in our prior immunoprecipitation experiments, or were well-known interaction partners of one or several of the proteins identified in the complex. The former included CAND1, LMNB1, DNMT1, SATB1, H1.2. The latter included, for example, UHRF1, which regulates DNMT1 through ubiquitylation 25; H2A.1 which is regulated by Cul3 in SPOP-Cul3 complexes 26; H2A.Z, which is loaded by EP400 onto chromatin 27; the deubiquitinase BRCC3 interacting with H2A, H2B, H2A.X and CAND1 28; the repressor ASXL1 interacting with CBX5 and EZH2 27,29. These results are summarized in a diagram of protein interactions in Fig. 2a.
Figure 2. Ubiquitination changes upon PLZF expression.
a, Mass spectrometric analysis of proteins immunoprecipitated by anti-ubiquitin branch K-GG antibodies (UbiscanR) from WT and PLZF-tg thymocytes revealed differential ubiquitination of the proteins depicted in blue-filled ovals in a global diagram of PLZF and Cul3 interactions partners, with enrichment for proteins involved in chromatin organization (p<10−6). Peptides from two independent experiments are listed in Fig. S5. Red lines link proteins identified by coprecipitation in this study; grey lines represent previously established protein interactions (Ingenuity PathwayR). b, Western blot analysis of anti-HA-Ub immunoprecipitates from lysates of 293T cells transfected with indicated plasmids and immunoblotted with anti-SATB1 or anti-lamin B1. Cul3 ΔC is a mutant Cul3 lacking the E2 binding domain. Molecular weight of lamin B1-GFP fusion protein is 75kDa. Data are representative of at least three experiments..
Increased ubiquitination of the key nuclear proteins SATB1 and lamin B1 was directly confirmed by immunoprecipitation and western blot analyses in transfected 293T cells. In these experiments, SATB1 and lamin B1 were overexpressed because technical limitations made it impossible to determine the ubiquitination pattern of endogenous proteins (Fig. 2b). Furthermore, ubiquitination was shown to require the E2-binding domain of Cul3, supporting the conclusion that Cul3 can directly ubiquitinate these PLZF-associated proteins in vivo. In confocal microscopy experiments, PLZF and Cul3 were colocalized at lamin B1 positive sites in the nuclear lamina and at SATB1 sites of the nuclear matrix in a typical cage-like pattern (Fig. S6). Furthermore, Cul3 was present at PLZF-bound promoters as shown by ChIP-qPCR (Fig. S7), suggesting that PLZF-Cul3 complexes associate at PLZF-binding promoters across the genome and in nuclear subcompartments involved in gene expression or repression.
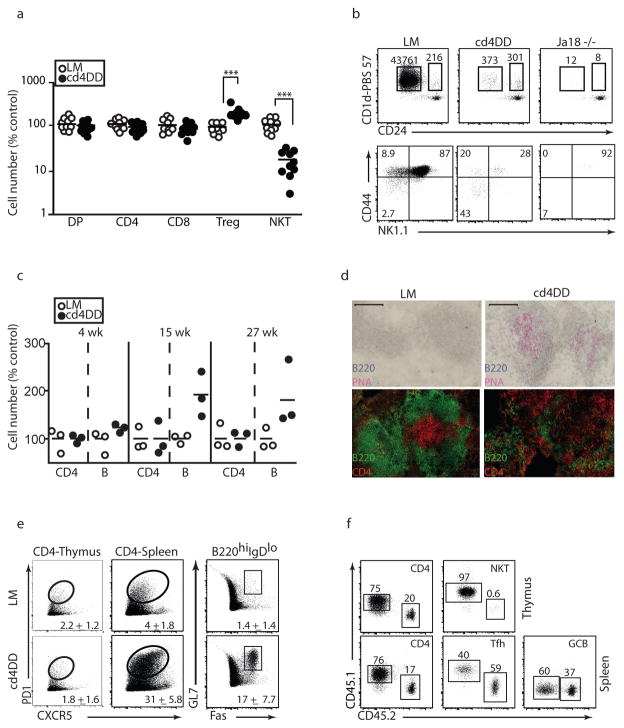
To further explore the functional role of Cul3 in lymphocyte development and function, where BTB-ZF transcription factors such as PLZF and Bcl-6 are of major importance, we bred Cul3fl/fl mice 30 to CD4-Cre and CD19-Cre deleter strains. Mice lacking Cul3 in T cells exhibited a thymus with normal cellularity and representation of CD4 and CD8 lineages (Fig. 3a). Regulatory T cells showed a modest but significant 2-fold increase (Fig. 3a). In contrast, the number of NKT cells was massively decreased, with a sharp developmental block occurring at the CD24loCD44loNK1.1− stage 1, similar to the block described in mice lacking PLZF 1,2 (Fig. 3b, Fig. S8). These results support the importance of the PLZF-Cul3 interaction in NKT cell development.
Figure 3. Lymphocyte development and function in Cul3cd4Δ/Δ mice.
a, Thymic subsets in Cul3cd4Δ/Δ and littermate controls. b, CD1d-PBS57 tetramer MACS-enriched NKT thymocytes in Cul3cd4Δ/Δ, WT littermate and in Jα18−/ − lacking NKT precursors. Top, absolute numbers of early CD24hi stage 0 precursors and maturing CD24lo cells recovered per thymus after MACS. Bottom, percentages of CD24lo stages 1, 2 and 3 based on sequential acquisition of CD44 and NK1.1. c, Splenic CD4 and B cell counts at different ages. d, Immunohistochemical analysis of spleen from 8 wk-old mice stained as indicated (bar, 100 μm) e, FACS analysis of splenic subsets, as indicated, in 8 wk-old mice. Numbers indicate mean percentage±SEM. f, Lethally irradiated recipients of a 3:1 mixture of WT:Cul3cd4Δ/Δ analysed as indicated. Tfh, CD4+PD1+CXCR5+; GCB, B220hiIgDloGL7+Fas+. Data representative of six independent experiments (e-f, n=18–20).
Intriguingly, in older mice lacking Cul3 in T cells, the spleen and lymph nodes became enlarged, the result of a net increase in B cell numbers with spontaneous formation of germinal centers made of PNA+ B cells (Fig. 3c–d). Whereas thymic CD4+8− T cells exhibited a normal phenotype, a population of splenic CD4 T cells expressing a PD1+CXCR5+ follicular helper phenotype progressively accumulated in aging mice, concomitantly with GL7+Fas+ germinal center B cells (Fig. 3e). Consistent with these findings, immunohistological analysis demonstrated large germinal centers with penetration of the B cell follicles by CD4 T cells (Fig. 3d). In radiation chimeras reconstituted with a 3:1 mixture of WT (CD45.1) and Cul3cd4Δ/Δ (CD45.2) T cells, the Cul3-deficient compartment showed absence of NKT cells and increased follicular helper CD4 T cells, demonstrating the cell-intrinsic nature of these defects (Fig. 3f). As expected, germinal center B cells of both compartments were indiscriminately expanded. Other effector programs available to CD4 T cells, however, appeared unperturbed as Cul3-deficient CD4 cells normally expanded and differentiated towards Th1, Th2 or Th17 effector cells in vitro (Fig. S8).
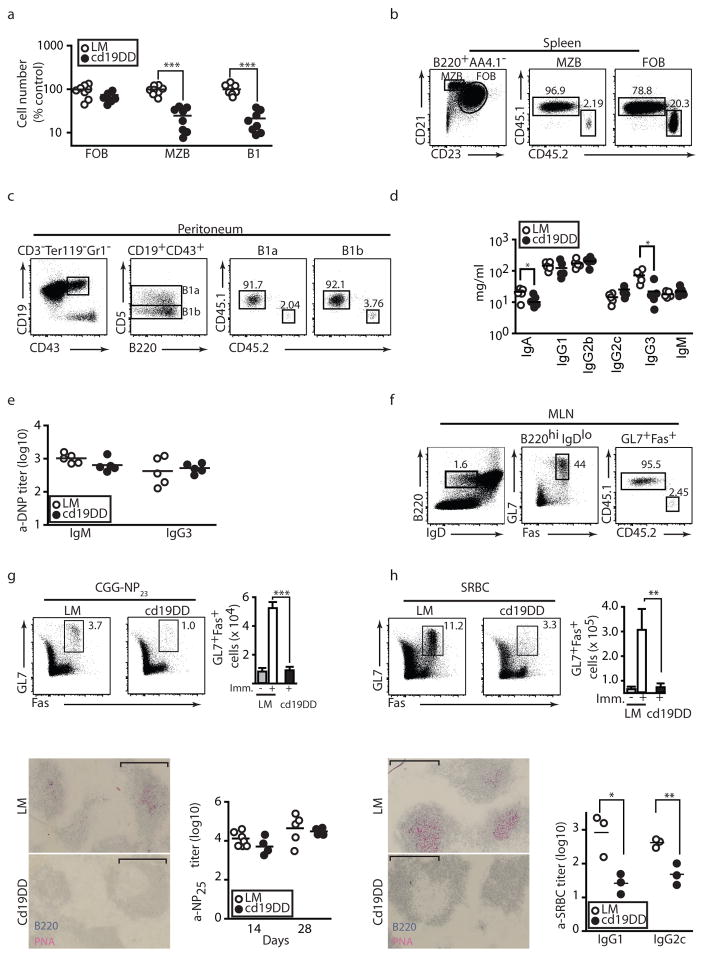
Mice lacking Cul3 in B cells showed normal development of follicular B cells but exhibited a selective 4- to 5-fold reduction of marginal zone B cells in the spleen and of B1 B cells in the peritoneum (Fig. 4a). These cell-intrinsic defects were considerably amplified in the competitive environment of mixed bone marrow chimeras (Fig. 4b–c). While circulating levels of immunoglobulins were normal or modestly decreased (Fig. 4d) and the antibody response to the T-independent antigen DNP-Ficoll appeared conserved (Fig. 4e), T-dependent B cell responses exhibited various defects. We noted that the germinal center B cells that spontaneously develop in the mesenteric lymph nodes of unimmunized mice were reduced, particularly in competitive bone marrow chimeras (Fig. 4f).. The germinal center responses observed after immunization against the T-dependent antigens CGG-NP23 and SRBC were drastically impaired, as assessed by immunohistochemical staining of PNA+ B cells and FACS staining of GL7+Fas+ B cells (Fig. 4g–h). The antibody response to SRBC was depressed, whereas the serum antibody response to highly polyvalent NP appeared conserved (Fig. 4g–h). These defects are similar to those reported in Bcl6-deficient mice 8.
Figure 4. Lymphocyte development and function in Cul3cd19Δ/Δ mice.
a, Splenic follicular B cells (FOB) and marginal zone B cells (MZB) and peritoneal B1 B cells in 8 wk-old Cul3cd19Δ/Δ and littermate controls. b and c, FACS analysis of splenic FOB and MZB (b) and peritoneal B1a/B1b subsets (c), gated as indicated, in the WT (CD45.1) and cd19Δ/Δ (CD45.2) compartments of 1:1 mixed bone marrow chimeras (data representative of n=9). d, Serum immunoglobulin isotypes in 10–11 wk old mice (n=5). e, Serum antibody response to DNP-Ficoll at day 14. f, FACS analysis of spontaneous GCB cells in mesenteric lymph nodes of 1:1 mixed bone marrow chimeras (representative of n=9). g, Splenic GCB cells (day 28) after immunization at days 0 and 21 with CGG-NP23. Upper panels show FACS staining and summary bar graph (mean & SEM, n=8). Lower panels, immunohistochemical staining of spleen at day 28 (bar, 100 μM) and serum anti-NP25 titers at days 14 and 28. h, Similar analysis 7 days after immunization with SRBC. Summary bar graph (mean and SEM, n=15).
Our study suggests that Cul3 is an essential partner of key BTB-ZF transcription factors in the lymphoid lineage. The different impact of Cul3 on the follicular helper T cell and the germinal center responses suggests that Cul3 regulates distinct components of these two Bcl6-driven programs 20. In addition, the defect in MZB and B1 cells may hint at the existence of yet unidentified BTB-ZF factors controlling these enigmatic populations.
Although proteomic analysis of ubiquitination was performed at the whole cell level, a significant proportion of changes induced by PLZF expression were concentrated on the PLZF-Cul3 associated complex itself, including key nuclear matrix proteins such as SATB1 and Lamin B1 which target specific DNA sequences for chromatin remodeling and gene regulation 11–14. The precise nature and role of these changes remain to be elucidated, but the emerging evidence of the importance of ubiquitination in chromatin regulation 22–24 suggests that they specify key aspects of the transcriptional programs directed by these transcription factors. A similar function of Cul3 may regulate the oncogenic properties of PLZF and Bcl6 in leukemias and lymphomas 8,9.
ADDITIONAL METHODS
Mice
C57BL/6 and B6.SJL-Ptprca Pep3b/BoyJ (CD45.1), Cd19-Cre (B6.129P2(C)-Cd19tm1(cre)Cgn), Cd4-Cre (B6 Tg(cd4-cre)1Cwi) were obtained from Jackson laboratories. B6.Cul3fl/fl mice 1 and B6.Jα18−/− mice were bred in our colony. Among the littermates of Cul3cd4Δ/Δ and Cul3cd19Δ/Δ mice, we used both Cre+Cul3wt/wt and Cre−Cul3fl/fl mice as controls. All mice were raised in a specific pathogen-free environment at the university of Chicago and experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee.
Cell Culture
HeLa cells, 293T cells were maintained in DMEM (GIBCO) supplemented with 10% FBS and 1% Penicillin/Streptomycin. KG1a cells were maintained in IMDM media (GIBCO) supplemented with 10% FBS, 1% Penicillin/Streptomycin. All cell lines were purchased from American Type Culture Collection (ATCC).
Plasmids and antibodies
Plasmids used in this study are kind gifts from investigators or were generated in the laboratory: human pcDNA3-myc-Cul3 (Dr. Yue Xiong, University of North Carolina at Chapel Hill); pcDNA3-DN-hCul3Flag2 (Addgene: plasmid 15820) lacking sequences 418–760 at the carboxy-terminus and serving as a catalytically inactive mutant (Cul3ΔC); pcDNA3.1-SATB1myc-his (Dr. Rudolf Grosschedl, Max Planck Institute of Immunobiology and Epigenetics); pEGFP-C3-LMNB1 (Dr. Ying-Hui Fu, UCSF); pFlag-CMV2-PLZF (where PLZF was PCR amplified from C57BL/6 thymic NKT cDNA and cloned into a Sal1 site of pFlag-CMV2).
Anti-Cul3 Ab was C-0871 from Sigma or A301-109A from Bethyl Laboratories, anti-PLZF (monoclonal 2A9 from Calbiochem or polyclonal AF2944 from R&D systems), anti-Bcl6 (polyclonal ab19011 from Abcam, C-19 and N-3 from Santa Cruz Biotechnologies, monoclonal G1191E from eBioscience), antiSATB1 (L745 from Cell Signaling Technology or monoclonal 14 from BD Bioscience), anti-lamin B1 (monoclonal 4E4 from Sigma, M-20 and S-20 from Santa Cruz), anti-HA (TA-150034 from Origene), anti-HDAC1 (ab-7028 from abcam). Mouse, goat or rabbit IgG (Abcam: mIgG2a-ab18413, gIgG-ab37373, rIgG-ab37415-5). Secondary antibodies were anti-rabbit IgG-HRP (GE healthcare or eBioscience), donkey anti-goat IgG-HRP (Santa Cruz Biotechnologies).
Immunoprecipitation of PLZF- or Cul3-associated proteins
15×106 NKT cells or 50–75×106 thymocytes were lysed on ice for 30 min in 0.5–1 ml lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA pH 8.0: 0.5% (vol/vol) Nonidet P-40, 1 mM dithiothreitol, protease inhibitor mix (Roche). Lysates were cleared by centrifugation, pre-cleared with Protein A/G sepharose beads (Invitrogen) at 4 °C for 1 h, and incubated at 4 °C for 2 h with anti-PLZF or anti-Cul3 bound Protein A/G Sepharose (Invitrogen) beads. Beads were washed 3 times with lysis buffer followed by 3 washes with PBS + 0.05% Triton. Bound proteins were eluted by boiling for 5 min and resolved on 10% SDS-PAGE (BioRad). For immunoblot, the gel was transferred to nitrocellulose membrane (Transblot transfer medium, 0.45mm, Biorad) and blotted using specific antibodies.
Mass spectrometry
For the identification of coimmunoprecipitated proteins, slices of SDS-polyacrylamide gels stained with Colloidal Blue (NuPAGE, Invitrogen) were destained using 100 mM ammonium bicarbonate pH 7.5 in 50% acetonitrile. A reduction step was performed by addition of 100 μl 50 mM ammonium bicarbonate pH 7.5 and 10 μl of 10 mM TCEP (Tris(2-carboxyethyl)phosphine HCl) at 37 oC for 30 min. The proteins were alkylated by adding 100 μl 50 mM iodoacetamide and allowed to react in the dark at 20 °C for 30 min. Gel slices were washed in water, then in acetonitrile and dried by SpeedVac for 30 min. Trypsin digestion was carried out overnight at 37 oC using sequencing grade modified trypsin (Promega) at 1:50 enzyme to protein ratio in 50 mM ammonium bicarbonate pH 7.5 and 20 mM CaCl2. Peptides were extracted from the gel pieces with 5% formic acid and dried by SpeedVac. The peptide samples were analyzed by a liquid chromatography-electrospray tandem mass spectrometry (LC-MS/MS) on a Thermo LTQ Orbitrap Hybrid FT Mass Spectrometer. Spectra were then analyzed with Mascot (Matrix Science, London, UK; version3, Mascot) and Sequest (ThermoFinnigan, San Jose, CA; version v.27, rev. 11) set up to search the mus_musculus database. Peptide identifications were accepted if they could be established at greater than 95.0% probability. Protein identifications were accepted if they could be established at greater than 90.0% probability and contained at least two unique identified peptides.
UbiScan® analysis
3×108 thymocytes were submitted to Cell Signaling Technology (CST) for UbiScan® analysis using the Ubiquitin Branch Antibody (CST cat # 1990) following a method modified from 3. Lysates were sonicated, cleared by centrifugation, reduced and carboxamidomethylated. Total protein for each lysate was normalized prior to digestion. Lysates were digested with trypsin. Peptides were separated from non-peptide material by solid-phase extraction with Sep-Pak C18 classic cartridges (Waters cat #WAT051910). Lyophilized peptides were re-dissolved, and ubiquitinated peptides were isolated using slurries of the Ubiquitin Branch Antibody. Peptides were eluted from antibody-resin into a total volume of 100 μL in 0.15% trifluoracetic acid. Eluted peptides were concentrated with C18 spin tips immediately prior to LC-MS analysis. The samples were run in duplicate to generate analytical replicates and increase the number of MS/MS identifications from each sample. Peptides were loaded directly onto a 10 cm × 75 μm PicoFrit capillary column packed with Magic C18 AQ reversed-phase resin. The column was developed with a 72-min linear gradient of acetonitrile in 0.125% formic acid delivered at 280 nl/min. Tandem mass spectra were collected with an LTQ-Orbitrap Velos hybrid mass spectrometer (Thermo), a top 20 method, a dynamic exclusion repeat count of 1 and a repeat duration of 30 s. MS spectra were collected in the Orbitrap component of the mass spectrometer, and MS/MS spectra were collected in the LTQ. MS/MS spectra were evaluated using SEQUEST 3G and the SORCERER 2 platform from Sage-N Research (v4.0, Milpitas CA). Peptide assignments were obtained using a 5% false positive discovery rate. Searches were performed against the mouse NCBI database updated on 9/6/10. Cysteine carboxamidomethylation was specified as a static modification, oxidation of methionine residues was allowed, and ubiquitination was allowed on lysine residues. Each MS/MS spectrum arises from a parent ion observed during a survey MS scan and can be linked to the intensity of that parent ion at its chromatographic apex, essentially measuring the abundance of the peptide in the sample. Parent ion intensities were extracted from each sample’s ion chromatogram file using proprietary software and are reported in the quantification tables. Changes in ubiquitinated peptide levels were measured by taking the ratio of raw intensities. Raw intensity values were used to calculate average values and raw ratios between samples. The raw ratios were normalized based on the median ratio found, and normalized ratios and fold-changes are reported.
Flow cytometry
CD1d-PBS57 tetramers were obtained from the NIH tetramer facility. Fluorochrome labeled monoclonal antibodies (clones indicated in bracket) against CD4 (GK1.5), CD8a (53-6.7), TCRβ (H57-597), CD24 (M1/69), CD25 (PC61), Foxp3 (FJK-16), CD44 (IM7), NK1.1 (PK136), B220 (RA3-6B2), CxCR5, PD1 (29F.1A12), ICOS (C398.4A), IgD (11.26c.2a), Fas (JO2), GL7, CD3e, CD1d (1B1), CD21/35 (7G6), CD23(B3B4), CD45.1 (A20), CD45.2 (104), CD5 (53-7.3), CD43 (S7), CD93 (AA4.1), CD19 (ID3), IgM (11/41), CD69 (H1.2F3), γδTCR (GL3), were purchased from e-Bioscience, BD Biosciences or Biolegend. For Foxp3 intracellular flow cytometry, cells were fixed using the permeabilization and fixation buffer “Foxp3 Staining Buffer Set” from eBioscience. Samples were analysed on an LSRII (Becton Dickinson), or sorted on a FACS Aria (Becton Dickinson) or MoFlo (Dako Cytomation). Data was analyzed using FlowJo (Tree Star).
Confocal Microscopy
Purified NKT thymocytes or CD4 splenocytes were attached to slides (Superfrost plus microscope slides, Fisherbrand) by cytospin and fixed for 15 min with 4% Paraformaldehyde in PBS followed by three washes in PBS. Cells were permeabilized with 0.5% Triton in PBS for 10 min, washed and blocked with 10% donkey serum and 1% BSA for an hour at room temperature before staining with anti-PLZF or anti-Cul3 for 2 h at room temperature in a humidifying chamber. After washes with PBS, cells were stained with donkey anti-rabbit Alexa 488 (invitrogen), donkey anti-goat Alexa 555 antibodies (Invitrogen) or donkey anti-mouse 647 (Invitrogen) for 30 min at room temperature. Cells were washed with PBS 0.005% triton, then PBS, and mounted with prolong gold mounting solution (Invitrogen). Control staining included rabbit IgG and goat IgG followed by corresponding secondary antibody or secondary antibody alone. Images were captured on Leica SPII-STED-CW super resolution laser scanning confocal (100x/1.4 oil) and Olympus 1X81 laser scanning microscope and were analyzed with Image J software.
IP/Western blot detection of ubiquitinated proteins
293T cells grown in 6-well plate dishes were lipofectamine-transfected with plasmids pHA-Ub (2 μg), pSATB1-myc-his (2 μg), pLMNB1-GFP (2 μg), pPLZF-Flag (0.5 μg), pCul3-myc (2 μg) or pCul3ΔC (2 μg) as indicated. Plasmid concentration was kept constant by adding pmaxGFP (Amaxa). 24h later, 20μM MG132 was added and cells were incubated for another 4 – 6 h. Cells were harvested with gentle scraping and resuspended in 300 μl RIPA buffer (25 mM Tris-Cl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS & 1 × protease inhibitor). Cells were lysed by sonication (Bioruptor: Diagenode) for 12m in, with 30 s on and 30 s off. Lysates were centrifuged at 12,000 rpm at 4°C for 10 min to remove cell debris. 100 μl of the lysate was diluted 1:4 with RIPA buffer supplemented with 1 × protease inhibitors, and incubated with 25 μl anti-HA agarose (Sigma: HA-7) for 4h. The beads were washed 4x with RIPA buffer and boiled for 5 min in 50 μl SDS gel-loading buffer containing 50mM β-mercaptoethanol at 95°C. Samples were separated by SDS-PAGE and immunoblotted with antibodies against SATB1 or lamin B1.
Immunohistochemistry
For immunofluorescence studies, frozen OCT (Tissue-Tek, CA)-embedded 5-μm sections of spleens were dried overnight and stained with biotinylated rat anti-B220 (RA3-6B2) (BD Biosciences, NJ) and Alexa-Fluor 488-conjugated rat anti-CD4 (RM4–5) (Invitrogen, CA) antibodies, followed by Cy3-streptavidin (Invitrogen, CA) and visualized using a SP5 II microscope (Leica, Germany). Data were analyzed using ImageJ (Bitplane, MN) software.
For immunohistochemical studies, frozen OCT-embedded sections were dried overnight, fixed with ice-cold acetone and incubated with methanol and 0.3% hydrogen peroxide to neutralize endogenous peroxidase activity and blocked with 5% rat serum, then streptavidin-biotin blocking (vector Laboratories). Sections were stained with biotinylated peanut agglutin (Vector Laboratories, CA), followed by Vectastain ABC-alkaline phosphatase kit (Vector Laboratories, CA) and the Vector Red Alkaline Phosphatase Substrate Kit (Vector Laboratories, CA). After treatment with serum and streptavidin-biotin block, sections were then stained with biotinylated rat anti-B220 (RA3-6B2) antibody (BD Biosciences, NJ), followed by reaction with the Vectastain ABC kit (Vector Laboratories, CA) and ImmPACT SG peroxidase substrate kit (Vector Laboratories, CA) according to manufacturer’s instructions. Sections were dehydrated, cleared with xylenes and mounted using Permount solution (Fischer Scientific, NJ). Micrographs were taken with the FSX-100 microscope camera system (Olympus, PA) and data were analyzed using ImageJ (Bitplane, MN) software.
Generation of Bone Marrow Chimeras
6–8 week-old B6 (CD45.1) mice were subjected to irradiation with 1000 Rads using a gamma cell 40 irradiator with a cesium source and were iv injected 3–6 h later with 2–5×106 bone marrow cells obtained from the femurs of donor mice. Bone marrow reconstituted mice were analyzed 8–10 weeks after irradiation
NKT cell enrichment
NKT cells were labeled with APC conjugated CD1d-PBS57 tetramers, bound to anti-APC magnetic beads, and enriched on an MACS cell separator (Miltenyi Biotech) as described 4.
Mouse immunizations
6–8 week-old mice were immunized intraperitoneally with 100 μg of DNP-FICOLL (Biosearch Technologies) in PBS, 50μg CGG-NP23 (Biosearch Technologies) mixed 1:1 with alum, or 2×108 SRBC (cat#7249007, Lampire Biological Laboratories). Mice injected with CGG-NP23 were boosted on day 21 with the same inoculum and killed on day 28. Levels of anti-DNP and anti-NP antibodies were determined by ELISA against BSA-DNP7 and BSA-NP25, respectively (Biosearch technologies) and anti-SRBC antibodies were measured by FACS using an indirect isotype-specific immunofluorescence assay (eBiosciences, Southern Biotech).
Statistical analysis
Unpaired student’s t test was performed with Prism (Graph Pad Software). *p<0.05, **p<0.001, ***p<0.0001.
Supplementary Material
Acknowledgments
We thank Aaron Dinner, Jonathan Licht, Gilbert Prive, Alexander Ruthenburg, Roger Sciammas, Harinder Singh and Patrick Wilson for discussions, Jeffrey C. Silva and Matt Stokes for Ubiscan analysis, Catherine Labno and Vytas Bindokas for help with confocal microscopy, and Katie Block, Lauren Roach, David Zabner, Fanyong Meng and Li Bai for help with experiments. This work was supported by NIH grants 5RO1GM082940 (JDS), RO1AI038339 (AB), Irvington Institute postdoctoral fellowship from the Cancer Research Institute (RM). AB is a Howard Hughes Medical Institute Investigator
Footnotes
Author Contributions: R.M. designed research, performed experiments and analyzed data. M.P.S., S.T.S., A.M., M.G.C., C.B.-V. performed experiments and analyzed data. J.D.S. helped design experiments and provided the Cul3fl/fl mice and Cul3 constructs. R.M. and A.B. co-wrote the paper. A.B. supervised the research.
References
- 1.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu L, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 4.Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 5.Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- 6.Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 9.McConnell MJ, Licht JD. The PLZF gene of t (11;17)-associated APL. Curr Top Microbiol Immunol. 2007;313:31–48. doi: 10.1007/978-3-540-34594-7_3. [DOI] [PubMed] [Google Scholar]
- 10.Guidez F, et al. RARalpha-PLZF overcomes PLZF-mediated repression of CRABPI, contributing to retinoid resistance in t(11;17) acute promyelocytic leukemia. Proc Natl Acad Sci U S A. 2007;104:18694–18699. doi: 10.1073/pnas.0704433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 12.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nature genetics. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 13.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 14.Zullo JM, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 15.Ohnuki H, et al. BAZF, a novel component of cullin3-based E3 ligase complex, mediates VEGFR and Notch cross-signaling in angiogenesis. Blood. 2012;119:2688–2698. doi: 10.1182/blood-2011-03-345306. [DOI] [PubMed] [Google Scholar]
- 16.Savage AK, Constantinides MG, Bendelac A. Promyelocytic leukemia zinc finger turns on the effector T cell program without requirement for agonist TCR signaling. J Immunol. 2011;186:5801–5806. doi: 10.4049/jimmunol.1100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon JE, et al. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J Biol Chem. 2006;281:12664–12672. doi: 10.1074/jbc.M600204200. [DOI] [PubMed] [Google Scholar]
- 18.Wimuttisuk W, Singer JD. The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Molecular biology of the cell. 2007;18:899–909. doi: 10.1091/mbc.E06-06-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Errington WJ, et al. Adaptor Protein Self-Assembly Drives the Control of a Cullin-RING Ubiquitin Ligase. Structure. 2012;20:1141–1153. doi: 10.1016/j.str.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 21.Hyjek E, Chadburn A, Liu YF, Cesarman E, Knowles DM. BCL-6 protein is expressed in precursor T-cell lymphoblastic lymphoma and in prenatal and postnatal thymus. Blood. 2001;97:270–276. doi: 10.1182/blood.v97.1.270. [DOI] [PubMed] [Google Scholar]
- 22.Braun S, et al. The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell. 2011;144:41–54. doi: 10.1016/j.cell.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 24.Bosch-Presegue L, et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell. 2011;42:210–223. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Du Z, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez-Munoz I, et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci U S A. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 30.McEvoy JD, Kossatz U, Malek N, Singer JD. Constitutive turnover of cyclin E by Cul3 maintains quiescence. Mol Cell Biol. 2007;27:3651–3666. doi: 10.1128/MCB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.McEvoy JD, Kossatz U, Malek N, Singer JD. Constitutive turnover of cyclin E by Cul3 maintains quiescence. Mol Cell Biol. 2007;27:3651–3666. doi: 10.1128/MCB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin J, Ang XL, Shirogane T, Wade Harper J. Identification of substrates for F-box proteins. Methods Enzymol. 2005;399:287–309. doi: 10.1016/S0076-6879(05)99020-4. [DOI] [PubMed] [Google Scholar]
- 3.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 4.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages in thymic NKT cell development. Journal of Experimental Medicine. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.