Abstract
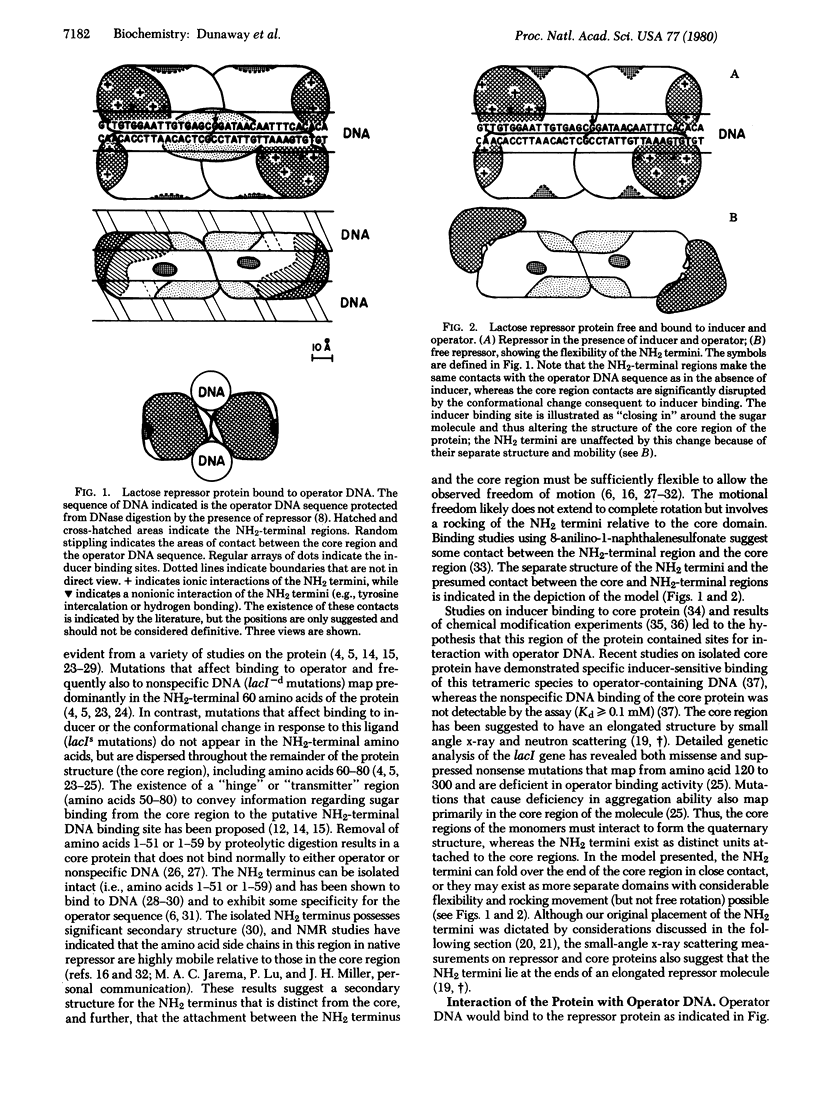
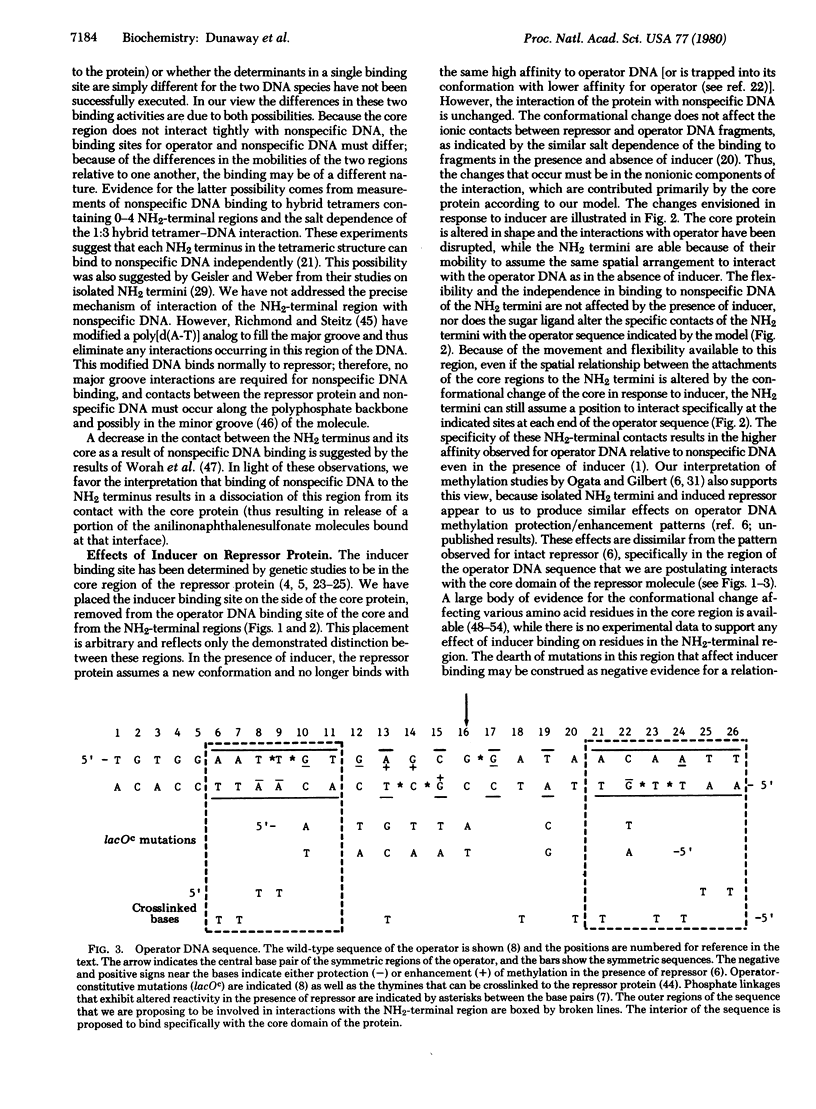
A model is presented for the structure of the lactose repressor protein and for its interaction with inducer, operator DNA, and nonspecific DNA. The proposed structure is based on experimental evidence from this laboratory and from the literature and is offered as an integration of the available data on this system. Features unique to this model include: (i) interaction of the core region of the protein with the operator, (ii) primary effects of the conformational change in response to inducer on the core-operator interaction, (iii) contacts between all four subunits of the protein and the operator DNA, and (iv) qualitative differences in operator and nonspecific DNA binding.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler K., Beyreuther K., Fanning E., Geisler N., Gronenborn B., Klemm A., Müller-Hill B., Pfahl M., Schmitz A. How lac repressor binds to DNA. Nature. 1972 Jun 9;237(5354):322–327. doi: 10.1038/237322a0. [DOI] [PubMed] [Google Scholar]
- Bahl C. P., Wu R., Stawinsky J., Narang S. A. Minimal length of the lactose operator sequence for the specific recognition by the lactose repressor. Proc Natl Acad Sci U S A. 1977 Mar;74(3):966–970. doi: 10.1073/pnas.74.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois S., Pfahl M. Repressors. Adv Protein Chem. 1976;30:1–99. doi: 10.1016/s0065-3233(08)60478-7. [DOI] [PubMed] [Google Scholar]
- Buck F., Rüterjans H., Beyreuther K. 1H NMR study of the lactose repressor from Escherichia coli. FEBS Lett. 1978 Dec 15;96(2):335–338. doi: 10.1016/0014-5793(78)80430-x. [DOI] [PubMed] [Google Scholar]
- Burgum A. A., Matthews K. S. Lactose repressor protein modified with fluorescein mercuric acetate. J Biol Chem. 1978 Jun 25;253(12):4279–4286. [PubMed] [Google Scholar]
- Charlier M., Maurizot J. C., Zaccai G. Neutron scattering studies of lac repressor. Nature. 1980 Jul 24;286(5771):423–425. doi: 10.1038/286423a0. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Dunaway M., Matthews K. S. Hybrid tetramers of native and core lactose repressor protein. Assessment of operator and nonspecific DNA binding parameters and their relationship. J Biol Chem. 1980 Nov 10;255(21):10120–10127. [PubMed] [Google Scholar]
- Dunaway M., Olson J. S., Rosenberg J. M., Kallai O. B., Dickerson R. E., Matthews K. S. Kinetic studies of inducer binding to lac repressor.operator complex. J Biol Chem. 1980 Nov 10;255(21):10115–10119. [PubMed] [Google Scholar]
- Friedman B. E., Matthews K. S. Inducer binding to lac repressor: effects of poly[d(A-T)] and trypsin digestion. Biochem Biophys Res Commun. 1978 Nov 14;85(1):497–504. doi: 10.1016/s0006-291x(78)80069-2. [DOI] [PubMed] [Google Scholar]
- Friedman B. E., Olson J. S., Matthews K. S. Kinetic studies of inducer binding to lactose repressor protein. J Biol Chem. 1976 Feb 25;251(4):1171–1174. [PubMed] [Google Scholar]
- Geisler N., Weber K. Escherichia coli lactose repressor: isolation of two different homogeneous headpieces and the existence of a hinge region between residues 50 and 60 in the repressor molecule. FEBS Lett. 1978 Mar 15;87(2):215–218. doi: 10.1016/0014-5793(78)80335-4. [DOI] [PubMed] [Google Scholar]
- Geisler N., Weber K. Isolation of amino-terminal fragment of lactose repressor necessary for DNA binding. Biochemistry. 1977 Mar 8;16(5):938–943. doi: 10.1021/bi00624a020. [DOI] [PubMed] [Google Scholar]
- Goeddel D. V., Yansura D. G., Caruthers M. H. Binding of synthetic lactose operator DNAs to lactose represessors. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3292–3296. doi: 10.1073/pnas.74.8.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeddel D. V., Yansura D. G., Caruthers M. H. How lac repressor recognizes lac operator. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3578–3582. doi: 10.1073/pnas.75.8.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovin T. M., Geisler N., Weber K. Amino-terminal fragments of Escherichia coli lac repressor bind to DNA. Nature. 1977 Oct 20;269(5630):668–672. doi: 10.1038/269668a0. [DOI] [PubMed] [Google Scholar]
- Kallai O. B., Rosenberg J. M., Kopka M. L., Takano T., Dickerson R. E., Kan J., Riggs A. D. Large-scale purification of two forms of active lac operator from plasmids. Biochim Biophys Acta. 1980;606(1):113–124. doi: 10.1016/0005-2787(80)90103-3. [DOI] [PubMed] [Google Scholar]
- Kania J., Brown D. T. The functional repressor parts of a tetrameric lac repressor-beta-galactosidase chimaera are organized as dimers. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3529–3533. doi: 10.1073/pnas.73.10.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania J., Müller-Hill B. Construction, isolation and implications of repressor-galactosidase - beta-galactosidase hybrid molecules. Eur J Biochem. 1977 Oct 3;79(2):381–386. doi: 10.1111/j.1432-1033.1977.tb11819.x. [DOI] [PubMed] [Google Scholar]
- Kolchinsky A. M., Mirzabekov A. D., Gilbert W., Li L. Preferential protection of the minor groove of non-operator DNA by lac repressor against methylation by dimethyl sulphate. Nucleic Acids Res. 1976 Jan;3(1):11–18. doi: 10.1093/nar/3.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiken S. L., Gross C. A., Von Hippel P. H. Equilibrium and kinetic studies of Escherichia coli lac repressor-inducer interactions. J Mol Biol. 1972 Apr 28;66(1):143–155. doi: 10.1016/s0022-2836(72)80012-3. [DOI] [PubMed] [Google Scholar]
- Lin S., Riggs A. D. A comparison of lac repressor binding to operator and to nonoperator DNA. Biochem Biophys Res Commun. 1975 Feb 3;62(3):704–710. doi: 10.1016/0006-291x(75)90456-8. [DOI] [PubMed] [Google Scholar]
- Lin S., Riggs A. D. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975 Feb;4(2):107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- Manly S. P., Matthews K. S. Activity changes in lac repressor with cysteine oxidation. J Biol Chem. 1979 May 10;254(9):3341–3347. [PubMed] [Google Scholar]
- Matthews K. S. Tryptic core protein of lactose repressor binds operator DNA. J Biol Chem. 1979 May 10;254(9):3348–3353. [PubMed] [Google Scholar]
- Matthews K. S. Ultraviolet difference spectra of the lactose repressor protein. II. Trypsin core protein. Biochim Biophys Acta. 1974 Aug 8;359(2):334–340. doi: 10.1016/0005-2795(74)90232-3. [DOI] [PubMed] [Google Scholar]
- Miller J. H. Genetic studies of the lac repressor. XI. On aspects of lac repressor structure suggested by genetic experiments. J Mol Biol. 1979 Jun 25;131(2):249–258. doi: 10.1016/0022-2836(79)90075-5. [DOI] [PubMed] [Google Scholar]
- O'Gorman R. B., Dunaway M., Matthews K. S. DNA binding characteristics of lactose repressor and the trypsin-resistant core repressor. J Biol Chem. 1980 Nov 10;255(21):10100–10106. [PubMed] [Google Scholar]
- O'Gorman R. B., Matthews K. S. Fluorescence and ultraviolet spectral studies of Lac repressor modified with N-bromosuccinimide. J Biol Chem. 1977 Jun 10;252(11):3572–3577. [PubMed] [Google Scholar]
- O'Gorman R. B., Matthews K. S. N-Bromosuccinimide modification of Lac repressor protein. J Biol Chem. 1977 Jun 10;252(11):3565–3571. [PubMed] [Google Scholar]
- O'Gorman R. B., Rosenberg J. M., Kallai O. B., Dickerson R. E., Itakura K., Riggs A. D., Matthews K. S. Equilibrium binding of inducer to lac repressor.operator DNA complex. J Biol Chem. 1980 Nov 10;255(21):10107–10114. [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. DNA-binding site of lac repressor probed by dimethylsulfate methylation of lac operator. J Mol Biol. 1979 Aug 25;132(4):709–728. doi: 10.1016/0022-2836(79)90384-x. [DOI] [PubMed] [Google Scholar]
- Ogata R., Gilbert W. Contacts between the lac repressor and the thymines in the lac operator. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4973–4976. doi: 10.1073/pnas.74.11.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y., Matsuura M., Horiuchi T. Conformational change of the lac repressor induced with the inducer. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1444–1450. doi: 10.1016/0006-291x(72)90234-3. [DOI] [PubMed] [Google Scholar]
- Pfahl M., Stockter C., Gronenborn B. Genetic analysis of the active sites of lac repressor. Genetics. 1974 Apr;76(4):669–679. doi: 10.1093/genetics/76.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T., Files J. G., Weber K. Lac repressor. Specific proteolytic destruction of the NH 2 -terminal region and loss of the deoxyribonucleic acid-binding activity. J Biol Chem. 1973 Jan 10;248(1):110–121. [PubMed] [Google Scholar]
- Richmond T. J., Steitz T. A. Protein-DNA interaction investigated by binding Escherichia coli lac repressor protein to poly(d(A-U-HgX)). J Mol Biol. 1976 May 5;103(1):25–38. doi: 10.1016/0022-2836(76)90050-4. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Newby R. F., Bourgeois S. lac repressor--operator interaction. II. Effect of galactosides and other ligands. J Mol Biol. 1970 Jul 28;51(2):303–314. doi: 10.1016/0022-2836(70)90144-0. [DOI] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. Sequence-specific interactions of the tight-binding I12-X86 lac repressor with non-operator DNA. Nucleic Acids Res. 1980 Feb 11;8(3):487–506. doi: 10.1093/nar/8.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Richmond T. J., Wise D., Engelman D. The lac repressor protein: molecular shape, subunit structure, and proposed model for operator interaction based on structural studies of microcrystals. Proc Natl Acad Sci U S A. 1974 Mar;71(3):593–597. doi: 10.1073/pnas.71.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade-Jardetzky N., Bray R. P., Conover W. W., Jardetzky O., Geisler N., Weber K. Differential mobility of the N-terminal headpiece in the lac-repressor protein. J Mol Biol. 1979 Feb 25;128(2):259–264. doi: 10.1016/0022-2836(79)90129-3. [DOI] [PubMed] [Google Scholar]
- Worah D. M., Gibboney K. M., Yang L. M., York S. S. Association of Escherichia coli lac repressor with poly[d(A-T)] monitored with 8-anilino-1-napthalenesulfonate. Biochemistry. 1978 Oct 17;17(21):4487–4492. doi: 10.1021/bi00614a020. [DOI] [PubMed] [Google Scholar]
- Yang D. S., Burgum A. A., Matthews K. S. Modification of the cysteine residues of the lactose repressor protein using chromophoric probes. Biochim Biophys Acta. 1977 Jul 22;493(1):24–36. doi: 10.1016/0005-2795(77)90257-4. [DOI] [PubMed] [Google Scholar]
- York S. S., Lawson R. C., Jr, Worah D. M. Binding of recrystallized and chromatographically purified 8-anilino-1-naphthalenesulfonate to Escherichia coli lac repressor. Biochemistry. 1978 Oct 17;17(21):4480–4486. doi: 10.1021/bi00614a019. [DOI] [PubMed] [Google Scholar]


