Abstract
CD4 T cells have been shown to be necessary for the prevention of encephalitis during West Nile virus infection. However, the mechanisms used by antigen-specific CD4 T cells to protect mice from West Nile virus encephalitis remain incompletely understood. Contrary to the belief that CD4 T cells are protective because they merely maintain the CD8 T cell response and improve antibody production, we here provide evidence for the direct anti-viral activity of CD4 T cells which functions to protect the host from WNV encephalitis. In adoptive transfers, naïve CD4 T cells protected a significant number of lethally infected RAG−/− mice, demonstrating the protective effect of CD4 T cells independent of B cells and CD8 T cells. To shed light on the mechanism of this protection, we defined the peptide specificities of the CD4 T cells responding to West Nile virus infection in C57BL/6 (H-2b) mice, and used these peptides to characterize the in vivo function of antiviral CD4 T cells. WNV-specific CD4 T cells produced IFN-γ and IL-2, but also showed potential for in vivo and ex vivo cytotoxicity. Furthermore, peptide vaccination using CD4 epitopes conferred protection against lethal West Nile virus infection in immunocompetent mice. These results demonstrate the role of direct effector function of antigen-specific CD4 T cell in preventing severe West Nile virus disease.
Keywords: Rodent, CD4 T cells, viral immunity
INTRODUCTION
West Nile virus (WNV) is a small, enveloped arbovirus of the Flaviviradae family that persists in an enzootic cycle between mosquitoes and birds, with humans and many other animals as incidental hosts. Since WNV appeared on the Eastern seaboard of the United States in 1999 (1, 2), it has spread through all 48 continental states, infecting more than 27,551 people and has been directly linked to the death of 1,077 people (3–6). WNV leads to systemic disease in approximately 20% infected individuals, and the most severe disease is caused by viral neuroinvasion resulting in meningitis and encephalitis (7, 8), occurring in >5% of the patients. T cells play an essential role in preventing meningitis and encephalitis upon primary infection, and limiting disease severity upon potential re-infection (9–14). It has been shown that both CD4 and CD8 T cells are required for the control and clearance of West Nile virus (11, 13–16). Still, the relative importance of each cell population at different stages of infection and the critical antiviral mechanisms employed in controlling systemic and central nervous system (CNS) infection remain to be fully elucidated.
Sitati et al. have demonstrated that CD4 T cells are required for survival following WNV infection (14), however the mechanism of protection provide by CD4 T cells during WNV infection was not explored. This group demonstrated that CD4 T cell deficient mice, generated by continued antibody depletion, exhibited high viral titers for over 50 days within the CNS, eventually leading to death (14). In these same mice, viral titers in the spleen were not altered, suggesting that CNS, but not systemic, virus control requires CD4 cells (14). Moreover, these same experiments suggested that CD4 T cells are responsible for aiding in the survival and proliferation of CD8 T cells and the priming of B cells (14) but that hypothesis was not formally tested. Prior work has identified a requirement for CD4 T cells in controlling other Flavivirus infections, including those with the Japanese Encephalitis virus (JEV) (17) and Yellow Fever virus (YFV) (18), but again a direct role for CD4 T cell effector function has not been previously investigated.
While it is well established that CD4 T cells play an accessory role providing help to both CD8 T cells and B cells there is evidence that CD4 T cells can also have a direct effector response during a viral infection (reviewed in (19)). Thus, during influenza infection CD4 T cells use perforin-mediated cytotoxicity to clear virus from the periphery, (20) whereas measles-specific CD4 T cells use IFNγ to control virus within the CNS (21, 22).
In this study we show that by themselves CD4 T cells are sufficient for the control of WNV infection in RAG-1−/− mice. WNV-specific CD4 T cells secreted cytokines and lysed infected cells following WNV infection. Since the vaccination of mice with CD4 epitopes increased protection, the direct CD4 T cell effector function may be a relevant target for future vaccine studies. We propose the potential beneficial role of CD4 T cells may be more important for the protective vaccination of the elderly, a population adversely affected by WNV infection. These results demonstrate that CD4 T cells contribute to protection during primary WNV infection via direct effector function, albeit they do not exclude other modes of CD4 T-cell action.
Materials and Methods
Mice
Adult (2–6 months old) male C57BL/6 (B6) mice were purchased from the National Cancer Institute Breeding Program (Frederick, MD). B6.Rag-1−/−, B6.Perforin−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME), and bred at the VGTI vivarium (Oregon Health & Science University); they were used at 2–4 months of age. B6.SJL-Ptprca Pepcb/BoyJ, commonly referred to as B6. Ly-5.1 congenic mice were purchased from the National Cancer Institute and were used at 2–4 months of age. B6.Fas deficient (Lpr/Lpr), B6.Fas x Perforin−/−-double-deficient and B6.CD4−/− mice were bred at the Washington University School of Medicine, St Louis, MO). All animals were housed and bred under specific pathogen-free conditions at either the Oregon Health & Science University, Portland Oregon or the Washington University School of Medicine, St Louis, MO. All WNV experiments were completed within United States Department of Agriculture (USDA, Frederick, MD)-approved Biosafety Level 3 facilities, and were approved by the Institutional Animal Care and Use Committee, and the Institutional Biosafety Committee in accordance with the applicable federal, state, and local regulations.
Virus, Peptides, and Cell Lines
West Nile virus strains 31A and 385–99 were used and both virus strains yielded similar results. West Nile virus strain 385–99 was a kind gift of Dr. Robert Tesh (University of Texas Medical Branch, Galveston, TX); strain 31A was provided by the USDA reagent program (Ames, IA). An overlapping peptide library covering the entire length of the viral polyprotein (15-mers overlapping by 10 aa) was obtained from Sigma Aldrich. Additional synthetic peptides were purchased at >95% purity from Sigma Aldrich and 21st Century Biochemicals, diluted in 10% H2O/90% DMSO, stored at −80°C and subsequently used at indicated concentrations. Virus was grown in mycoplasma-negative Vero cells, cultured under aseptic conditions as described previously (15); mycoplasma-negative IC-21 cells were used in stimulation assays. Cells were infected using variable multiplicity of infection (MOI) as indicated.
Infection and vaccination
All mice were infected subcutaneously (sc) between the shoulder blades with indicated doses (20–1200 pfu) of WNV in 100ul of 1xPBS+2%FBS. On specified days post-infection, splenocytes were isolated and subjected to FCM, ICCS or CTL assay analysis as described below.
For peptide vaccinations 20μl of 1 mg/ml peptide/PBS solution containing peptides NS31616 and NS32066were emulsified in equal volume of the adjuvant TiterMax Gold® (Sigma-Aldrich), by vortex mixing for 30 minutes. Mice were immunized twice (sc) at 21day intervals, at the base of the tail.
Adoptive transfer and virus challenge experiments
Naïve CD4+ T cells were isolated from spleens of 4-wk old B6 mice. Briefly, splenic T cells were coated with anti-CD8, B220, NK1.1 coated beads (Miltenyi Biotech), and CD4+ cells isolated to 80–95% purity; such preparations contained <0.5% of B and CD8 T-cells, and were overwhelmingly of the naïve phenotype, containing <5% CD44hi (memory) cells; most of the contaminating cells were of the macrophage/monocyte lineage. CD4 T cells were transferred i.v. (at 5–10 × 106 cells/recipient), transfers monitored and infected as described above. Virus specific CD4 T cell lines were generated by in vitro restimulation of WNV-primed spleen cells, were purified to deplete CD8+ and B220+ cells to <1%, and were injected (2–5 × 105 cells/recipient) i.v. into RAG-1−/−recipients. Engraftment success was evaluated by FCM 24 h later, at which time the animals were infected with WNV as described above. Survival was scored on a daily basis. Death occurred between days 10 and 18, and all animals surviving this period remained disease free for 60–90 days at which point the experiment was terminated.
Antibody depletion of C57BL/6 mice
C57BL/6 mice were given two injections of 0.1 mg anti-CD4 (GK1.5) antibody intraperitoneally two times, day −3 and 0. Antibody depletion was confirmed by testing peripheral blood lymphocytes for the presence of CD4 T cells (clone RM4-5) using flow cytometry as described below.
Determination of viral titer
Viral titer was determined by plaque assay where a virus sample was serially diluted onto Vero cells. After co-culture of the virus with the cells for two hours, agarose overlay was added. Two days after the initial overlay, cells were overlayed with additional agarose-containing Neutral Red (0.2%). Plaques where then counted to determine viral load.
To evaluate a potential chronic virus infection, infected mice were perfused to avoid bloodborne contamination and brains were homogenized in RPMI using a beadbeater-96 (Biospec). The cellular suspension was seeded onto a monolayer of Vero cells, in triplicate, and cultured for 7 days, while monitored for cytopathic effect (CPE). CPE was monitored visually and defined as the rounding up of cells and loss of a monolayer. At the end of the culture period, cells and supernatant were transferred to a second monolayer of Vero cells and incubated for 48 hours, after which they were trypsinized, fixed and permeabilized and stained intracellularly, for the expression of WNV envelope protein using an E16-alexa647 (23) conjugated antibody, as described below for intracellular cytokine staining.
Intracellular cytokine and surface flow cytofluorometric (FCM) staining
Cytokine-producing T cells were detected using the Cytofix-Cytoperm Kit (BD PharMingen), as described below. Single-cell splenocyte suspension were depleted of RBC using ammonium chloride, incubated with 1μM peptide or infected with WNV in the presence of 5μg/ml Brefeldin A (Sigma Aldrich) for 6 h at 37°C, except when looking at production of IL-4, where monensin (Sigma Aldrich) was substituted for Brefeldin A. After six hours the cells were washed and blocked with Fc block (anti–mouse FcγRI/III; BD PharMingen) and incubated overnight in the presence of a saturating dose of surface antibodies against CD8, CD3, CD4, CD11a, CD43 (Clone 1B11), CD44 and CD62L (BD PharMingen). After washing, the cells were fixed, permeabilized and intracellular antibodies (anti-IFN-γ, anti-TNFα, anti-IL-4 or anti- IL-2; BD PharMingen) were added for 30 minutes. For detection of Granzyme B, splenocytes were isolated and kept on ice, surface stained, fixed and permeabilized as described above without stimulation. Cells were stained with Granzyme B Alexa647 (clone: gb11, BD PharMingen). The samples were then washed and analyzed using either a FACSCalibur or LSR II cytometer (Becton Dickinson Immunocytometry Systems) instrument. FCM analysis was performed by collecting a minimum of 5 × 104 events and gates set on lymphocyte population based on forward and orthogonal light scatter, followed by marker positioning to denote fluorescence greater than that of control stained or unstained cells.
CTL assays
For ex vivo CTL assays, CD4+ T cells were isolated from spleens of B6 mice 7 days post-infection with WNV, by negative selection using anti-CD8, B220, NK1.1-coated beads (Miltenyi Biotec). CD4+ T cells were isolated at 80–95% purity with <0.5% CD8+ cells present in the isolate. Direct ex vivo CTL activity was determined using radioactively labeled peptide-coated IC21 cells as targets. 1×105 IC21 cells (1×104 cells per well) were pulsed with 51CR and peptides (1μM NS31616+2066 or 1μM OTII peptide-Ova323-339). overnight in a 96 well plate. Cells were washed 3 times with warm media (5% FBS + RPMI) and purified CD4 T cells were serially diluted, then placed into the 96 well plate at indicated effector to target ratio for six hours. After six hours, 30μl of supernatant was removed and added to a lumaplate (Packard Co., Chicago, IL). Radioactivity was measured using TopCount Packard δ/γ radioactivity reader (Packard Co). Percent specific lysis was calculated as [(E − S)/(M − S)] times 100, where E (experimental release) equals the counts per minute released from targets incubated with lymphocytes, S (spontaneous release) equals the counts per minute released from target cells incubated with no lymphocytes and M (maximal release) equals the counts per minute released from cells after lysis with 1% Nonidet P40 (USB).
In vivo CTL assays were performed as previously described (24). Briefly, splenocytes from B6. Ly-5.2 mice were isolated, then labeled with carboxymethyl-fluorescein succinimide ester (CFSE) to produce fluoresceinhi (1μM) and fluoresceinlow (10nM) labeled population using standard CFSE labeling protocol (Molecular probes). After CFSE labeling, cells with different fluorescence intensity were peptide pulsed for 1hr at 37°C with 1μM NS31616+2066 or 1μM OTII peptide (Ova323-339). Cells were counted and equal numbers of fluorescein-high or -low labeled cells, coated with OTII or NS31616, respectively, were mixed and injected intravenously (iv) into infected and naïve mice. After 12 hours mice were sacrificed and splenocytes were gated on Ly-5.2+ cells (detected using mAb clone A20) MHC class II +double positive cells. In experiments where donor mice were not on a Ly5.1 background, PKH26 (2μM) was used to label all cells using the standard labeling protocol included within the kit (Sigma aldrich), then labeled with CFSE. The percent killing was calculated as follows: (1 − (ratio immune/ratio naive)) × 100. Ratio = number of events NS31616+2066 peptide-coated target/number of events reference target (24).
Statistical analysis
All statistical significance for survival experiments was determined using Log-Rank test. All statistical significance for flow cytometry results was completed using the Mann-Whitney test. All calculations were done using the Prism (GraphPad, San Diego, CA) software.
RESULTS
Naïve CD4 T cells protect mice from WNV severe disease
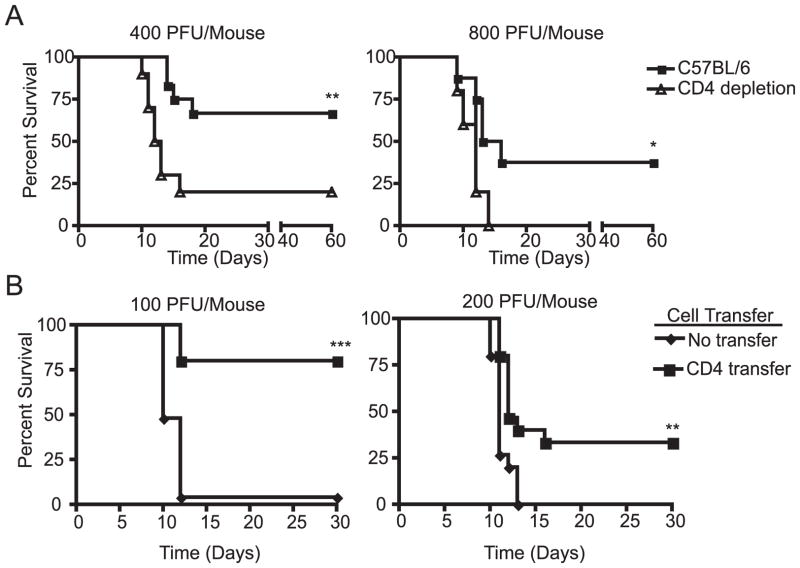
To confirm the CD4 T cell requirement for control of WNV infection, we depleted CD4 T cells in B6 mice prior to WNV infection. The CD4 T cells were depleted using the monoclonal antibody GK1.5 and depletion was confirmed by flow cytofluorometric (FCM) analysis of peripheral blood mononuclear cells, where there were fewer than 0.05% CD4 T cells remaining (data not shown). Antibody depletion of CD4 T cells in C57BL/6 mice during WNV infection with 400 plaque forming units (pfu)/mouse resulted in a significant increase in the mortality rate (p<0.005) (Figure 1A). At that dose, 66% of the C57BL/6 mice survived compared to only 20% of the CD4 T cell depleted mice. Overall mortality was higher in mice infected with 800 pfu, documenting the limits of protection against WNV. Still, no CD4 T cell-depleted mice survived when infected with 800 pfu of WNV, in contrast to 37% of the C57BL/6 control mice (p<0.04). CD4-depleted mice exhibited slightly shorter, but not statistically significant difference in the mean survival time (MST) (MST 14 days B6 vs 12 days CD4 depleted B6). Given the time of deaths, which coincided with the published action of adaptive immune system and the presence of the virus in the CNS (9–11, 13–15, 25–27), our data suggest that CD4 T cells do not alter the rate of disease, but rather may reduce the viral load below a threshold which causes high incidence of mortality(see below). From this result we concluded that within an immunocompetent animal, CD4 T cells are necessary for protection against severe WNV disease.
Figure 1. Protective effect of Naïve CD4 T cells.
A. Antibody depletion of CD4 T cells (triangles) renders C57BL/6 mice significantly (**p<0.005, *p<0.04) more susceptible to WNV induced mortality compared to controls (squares). CD4 T cells were depleted using two doses of GK1.5 antibody on day −3 and day 0, then infected with either 400pfu/mouse (left) or 800pfu/mouse (right). Results of one experiment with n=10 mice per group are shown, representative of two independent experiments. B. Adoptive transfer of naïve CD4 T cells provides protection to RAG-1−/− mice against lethal WNV infection. Splenic CD4+ T cells (5–10×106) from naive C57BL/6 mice were isolated by negative selection (80–95% purity) and transferred to C57BL/6 RAG-1−/− mice. 24 hours after transfer, mice were challenged with 100 pfu or 200 pfu WNV sc. Survival in these groups was significantly different according to the log-rank test (*** p<0.0005, left) (**p<0.002, right). Two individual experiments are shown n=25 mice per group are shown, representative of a total of 4 experiments completed.
We next sought to determine whether naïve CD4 T cells were sufficient for direct control of WNV disease in the absence of CD8 T cells or B cells. Others and we have previously shown that recombination activating gene 1−/− mice (RAG-1−/−), which contain no T and B cells, are extremely sensitive to WNV infection (9, 15). Therefore RAG-1 −/− mice can serve as an excellent host to determine the relative contribution of different lymphocyte subsets in protecting from disease upon adoptive transfer. Naïve CD4 T cells were purified by negative selection using magnetic beads. The resulting populations were 85–95% CD4+, but never contained more than 0.5% contaminating CD8+ or CD19+ cells. We transferred 5×106-1×107 CD4 T cells from naïve C57BL/6 mice into RAG-1−/− deficient mice and challenged these mice 24 h later with WNV. Adoptively transferred naïve CD4 T cells significantly protected the RAG-1−/− mice from mortality following subcutaneous WNV challenge with 100pfu (p<0.0005) or 200pfu (p<0.002) (Figure 1B). Indeed, 80% of the RAG-1−/− mice that received naïve CD4 T cells survived, as compared to only 4% of the RAG-1−/− controls when challenged with 100pfu/mse of WNV. Transferred CD4 T cells also significantly protected (p<0.0001) the RAG-1−/− mice following intra-peritoneal (ip) WNV challenge (data not shown). Moreover, 60 days post infection, we could not detect infectious WNV in the brains of RAG-1−/− mice that had received CD4 T cells by either plaque assay or co-culture (level of detection 100 and 10 pfu, respectively, in contrast to the levels of 105–7 pfu/organ detected in brains of moribund B6 or RAG-1 −/− mice).
To ascertain that the above effects can be directly ascribed to the sole activity of CD4 T cells, we had to exclude the possibility of contamination by CD8 T cells or B cells. As mentioned above, maximum potential contamination in the CD4 inoculum was <0.5%, or <2.5–5×104 of either cell subset. Given that the prevalent frequencies of antigen-specific T and B cell precursors are in the 10−5 range, it was not likely that this contamination will play a major role. To address this experimentally, we bled all mice at the peak of the immune response, 7 days post infection (8 days post transfer) and enumerated CD8 and B cell contaminants by FCM (data not shown). If a contaminating population was playing a role in protection, the cell population would have expanded by 7 days post infection. Such an expansion was detected in only 1 mouse out of 30 and that animal was eliminated from the study. Moreover, upon necropsy of a random sample of animals (4/group) we found no evidence of expansion of CD8 or B cells in the spleen, peripheral lymph node (<0.2% cells positive for CD8β, B220 or CD19), either at 24h or at 7 days post infection (not shown). Altogether, our data indicates that naïve CD4 T cells are both necessary and sufficient to protect mice from a lethal WNV challenge. Finally, we also examined surviving Rag-KO mice that received CD4 T-cell and could not find detectable infectious virus in the organs of these animals (detection limit 10 pfu; data not shown). This result suggests that CD4 T-cells are able to provide sterilizing immunity. Overall, we conclude that protection from WNV challenge in the model used above was the result of direct antiviral action of CD4 T-cells.
Ex vivo analysis of effector CD4 T cell function
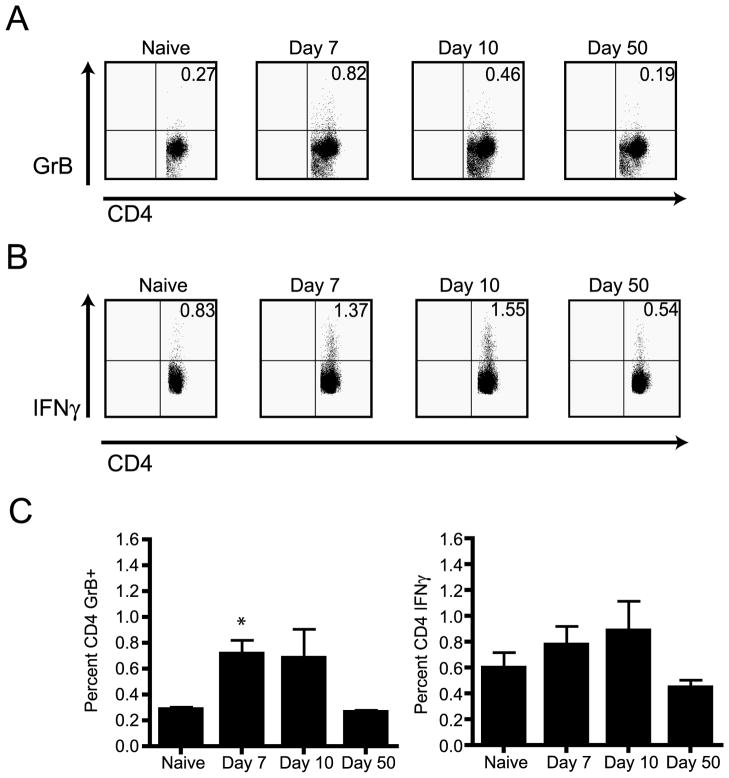
To evaluate the potential of antiviral CD4 T cells for direct anti-WNV action in the course of WNV infection of B6 mice, we chose to monitor Granzyme B (GzB) expression levels in this T-cell subset. We focused our analysis upon CD4 T-cells by selective gating; CD4 T-cells represented between 25–37% of total splenic population, and exhibited consistent response to WNV across experiments. GzB is expressed on effector T cells with lytic potential and its expression does not require in vitro stimulation, limiting any potential misinterpretation of data due to in vitro bias (28). FCM analysis revealed that a significant percentage of CD4 T cells upregulated GzB content on day 7 (p<0.02) post WNV infection compared to naïve controls (Figure 2A and 2C). By day 10 post infection we began to see a decrease in GzB content and by day 50 post infection the percentage of CD4 T cells that contained GzB returned to baseline levels (Figure 2A and 2C). The kinetics of IFNγ production by CD4 T cells was similar to what we observed for GzB during the course of WNV infection, except that the detection required brief in vitro stimulation of CD4 T cells with anti-CD3e in vitro (Figure 2B). From the results shown in Figure 2 we concluded that in response to WNV infection, CD4 T cells express molecules involved in direct effector T-cell function, including lytic granules and effector cytokine.
Figure 2. CD4 T cell response during West Nile virus infection.
A. Representative example of GzB expression by CD4 T cells after WNV infection as measured by direct ex-vivo intracellular FCM, without in-vitro stimulation. A naïve animal is shown as a control. Results are from one representative mouse of four mice from each time point. One experiment of two is shown. B. Representative example of IFNγ expression in CD4 T cells after WNV infection, measured by ICCS upon stimulation with 0.5μg/ml anti-CD3e (clone 2c11); a naïve animal is shown as a control. Results are from one mouse out of 4. Data is representative one experiment out of two. C. Left panel- Aggregate analysis of GzB expression in CD4 T cells, determined as in panel A. Percentage of CD4 GzB+ T cells for the time points given above. There is a significant induction of GzB in CD4 T cells on day 7 (p<0.02), but not on day 10 (p>0.05). Panels show average of four mice per time point (x± S.E.M.), representative of two experiments. Right panel- Aggregate analysis of IFNγ expression in CD4 T cells, determined as in panel B. Percentage of CD4 IFNγ+ T cells for the time points given above. Average of four mice per time point (x± S.E.M.), representative of two experiments.
Functional characteristics of WNV antigen-specific CD4 T cells
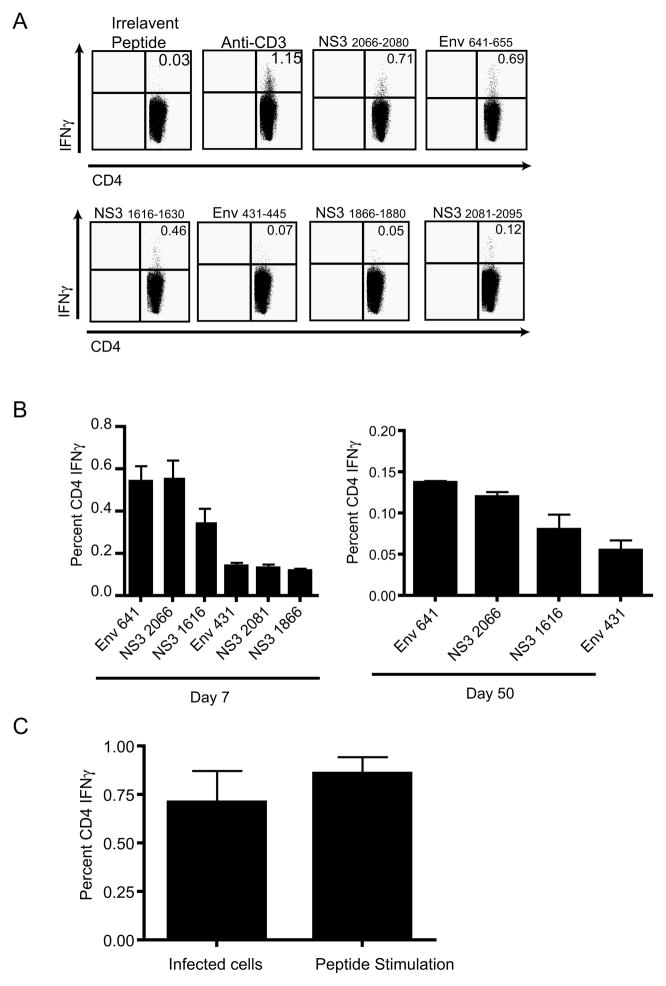
To gain a firmer understanding of the mechanism by which CD4 T cells protect WNV infected mice, we developed tools to track and study WNV-specific T cells. To that effect, we identified multiple CD4 T cell epitopes encoded by WNV using pools of overlapping peptides that cover the entire WNV polyprotein (Table I, part 1). For the strongest three epitopes, several peptide truncations were synthesized in order to determine optimal epitopes, which conformed to the expected MHC class II I-Ab binding motifs (Table I, part 2). All peptides described are cited with their inclusive amino acid numbers the first time in the text, as well as in Table I. Subsequently, abbreviated nomenclature is used based upon designation of the protein component from which the peptide is derived, e.g. NS3 (nonstructural protein 3), followed by the initial amino acid at which the peptide begins counting from the beginning of the polyprotein. Therefore, the NS3 peptide 2066–2080 is designated NS32066. Of note, none of the described peptides elicited IFNγ responses (<0.03%) by cells of any other phenotype (CD8 T-cells or DN cells- data not shown). A summary of the results for all identified CD4 T cell epitopes is listed in Table I. Through this process we identified in B6 mice three dominant epitopes, Env641-655, NS32066-2080, and NS31616-1630 (Figure 3A,B) and three sub-dominant epitopes (Figure 3A,B). Epitopes Env641 and NS32066 are each responsible for 30% of the total response, NS31616 is responsible for 20% of the total response and Env431-445, NS32081-2095, and NS31866-1880 are each responsible for less than 8% of the total response. The response to Env641, NS32066, and NS31616 peptide pool was always at least equal to, and often significantly larger than, the response to WNV-infected cells (Figure 3C), probably due to peptide competition, unequal temporal expression of all epitopes or a combination of these and other, unknown, factors. Importantly, Figure 3C demonstrates direct recognition of infected targets by CD4 T-cells, showing that CD4 T-cells react to relevant, endogenously processed viral fragments, and not just to peptides themselves. This data suggested that we have indeed identified the vast majority of WNV epitopes restricted by the I-Ab.
Table I. Identification of WNV epitopes eliciting CD4+ T-cell response in B6 mice.
Part 1- List of identified CD4 T cell 15-mer epitopes. Amino acid numbers and sequence of the CD4 T cell epitopes are listed for peptides that elicited IFNγ as measured by intracellular cytokine staining of splenocytes on day 7 post infection. Part 2. List of truncated CD4 T cell peptides. This table lists the truncated CD4 T cell peptides used to determine the optimal epitope. The optimal epitope is shown in bold. For both parts, responses were determined by ICCS for IFNγ production by splenocytes on day 7 post infection, as described in methods.
| PART 1 15-mer identification | ||||||
|---|---|---|---|---|---|---|
| Day Post Infection | Peptide identified | Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | Exp. 5 |
| # of mice | N=4 | N=3 | N=4 | N=5 | N=2 | |
| Anti-CD3 | 1.16 | 1.136 | 0.52 | ND | 0.911 | |
| E431-445 | IFVHGPTTVESHGNY | 0.105 | 0.179 | 0.03 | 0.14 | 0.152 |
| E641-655 | PVGRLVTVNPFVSVA | 0.505 | 0.326 | 0.1 | 0.41 | 0.273 |
| NS3 1616-1630 | TKPGVFKTPEGEIGA | 0.317 | 0.499 | ND | 0.30 | ND |
| NS3 1866-1880 | WFVPSVKMGNEIALC | 0.082 | 0.089 | 0.01 | 0.13 | ND |
| NS3 2066-2080 | RRWCFDGPRTNTILE | 0.527 | 0.406 | 0.487 | 0.49 | 0.176 |
| NS3 2081-2095 | DNNEVEVITKLGERK | 0.096 | 0.029 | 0.01 | 0.14 | ND |
Figure 3. Antigen-specific CD4 T cell IFNγ response to class II epitopes.
A. Representative example of CD4 T cell IFNγ ICCS response to the three immunodominant and three sub-dominant CD4 T cell epitopes as measured by 6 hour ICCS. CD4 T cells were from mice 7 days post infection and were stimulated with the peptide (10−6M) indicated above each plot. One mouse of 5 is shown for one experiment, representative of 3 experiments. B. Aggregate analysis of IFNγ expression in primary and memory responses. Left panel- Quantification of the CD4 T cell IFNγ ICCS response 7 days post infection, shown in panel A. Average of 5 mice per time point (x± S.E.M.), representative of three experiments. Right panel- Quantification of day 50 CD4 T cell IFNγ response. Average of 5 mice per time point (x± S.E.M.), representative of three experiments. C. Quantification of day 7 CD4 T cell IFNγ response (measured by ICCS) to either WNV-infected IC-21s (MOI:40) or peptide pulsed IC-21s 10−6M (Env641 and NS31616+2066) peptide. CD4 T cells were stimulated for 6 hours in the presence of BFA. Results depict average values of four mice per time point (x± S.E.M.), representative of two experiments.
Antigen-specific CD4 T cell cytokine secretion
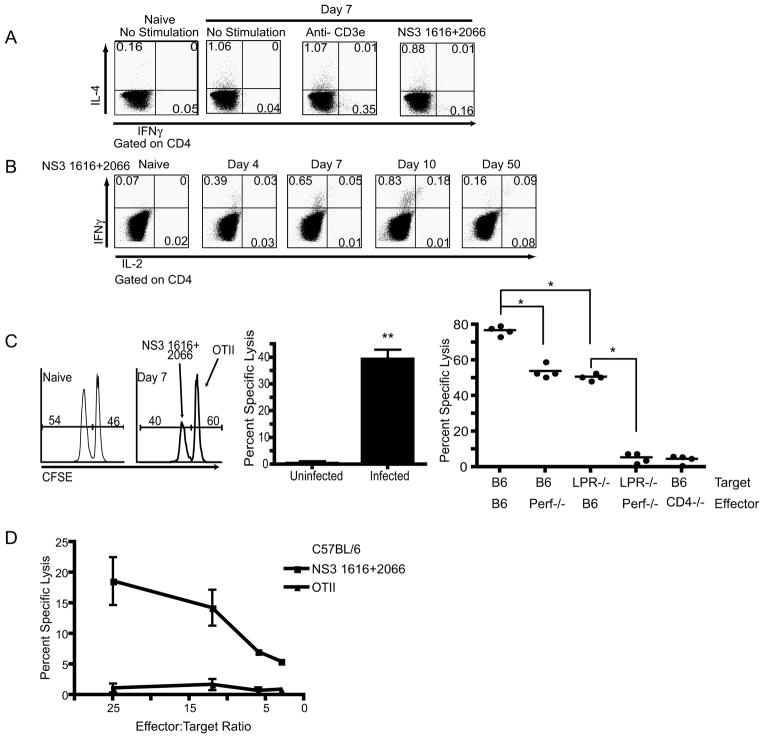
Using these newly identified epitopes we sought to determine the functional capabilities of the WNV-specific CD4 T cells. As shown in Figure 3, the peptide stimulation of splenocytes isolated from mice 7 days after infection resulted in a robust IFNγ response. We also observed spontaneous IL-4 production by CD4 T cells from day 7 infected animals (infected mice CD4 IL-4+= 0.91%, naïve mice CD4 IL-4+=0.20%, n=4 mice per group), which may be due to a bystander effect, since no increase in IL-4 production was induced following stimulation with TCR (anti-CD3ε), purified peptides (Figure 4A), or infected cell lysate (data not shown). The results of the cytokine profile produced by the WNV-specific CD4 T cells directly ex vivo suggest that the WNV CD4 T cell response shows a Th1 bias. Our data does not indicate that the IL-4 production is generated by antigen-specific CD4 T cells, however, at this point it is unclear whether this IL-4 response is targeted directly against WNV in these animals, which is possible in light of the role of CD4 T-cells as boosters of B-cell immunity and antibody production (14).
Figure 4. Functional potential of antigen specific CD4 T cells.
A. Representative example of a CD4 T cell cytokine response 7 days post infection. Following gating on CD4, IFNγ and IL-4 were measured after 6 hr stimulation in the presence of monensin. Cells were stimulated with media, 2c11 (0.5μg/ml) or NS31616+2066(10−6M), and a naïve mouse was used as a control. One mouse of four is shown, from one representative experiment of two. B. Representative example of CD4 T cell ICCS during the course of infection, as indicated above each plot. Following gating on CD4, IFNγ and IL-2 were measured after 6 hr stimulation with NS31616+2066(10−6M) in BFA. Controls and repetitions were as in A. C. In vivo CD4 T cell CFSE cytotoxicity assay. Left panel- Representative histogram of transferred (donor, Ly-5.1+) splenocytes 12 hours after adoptive transfer of target cells into naïve (left histogram) and infected (right histogram) mice. Middle panel- Aggregate quantification of in vivo CD4 T cell CFSE based cytotoxicity assay. CD4 T cells were cytolytic in vivo on day 7 post WNV infection (**p<0.008). Results represent the average of 5 mice per group, and are representative of three independent experiments. Right panel- Quantification of in vivo CD4 T cell CFSE-based cytotoxicity assay completed within WNV infected Perforin−/−, CD4−/− and B6 control mice. B6 or LPR−/− splenocytes were coated with WNV or control peptides, labeled with CFSE and used as targets with different hosts – the x-axis legend denotes which molecule(s) were missing during the interaction of CTL with their targets. B6 (targets) transferred into Perf−/− mice as well as the transfer of LPR−/− deficient targets leads to a reduction in cytotoxicity (*p<0.03). Transfer of LPR−/− targets into perf−/− mice leads to a complete ablation of cytotoxicity (p<0.03). Results represent the compilation 2 independent experiments. D. In vitro CTL activity of purified CD4 T-cells against targets coated with indicated WNV peptides or the control class II-restricted ovalbumin peptide (OT-II, Ova323-339). IC21 cells were coated with the indicated peptides and labeled with 51Cr. The cells were incubated with CD4 T-cells purified from spleens of B6 mice infected with WNV 7 days earlier, as described, with minimal purity of 89% and contaminating CD8 or B-cells at <0.5%. Chromium release assay was performed as described in Methods. Results are representative of 3 experiments.
We next used these epitopes to track the percentage of the antigen-specific CD4 T cells within the B6 spleen during the course of an infection (Figure 4B). Using ICCS, we were able to define the time course of the systemic antigen-specific CD4 T cell responses (Figure 4B). We could start to detect the antigen-specific response by day 4 and the peak of the response in the spleen occurred at day 10 (Figure 4B). As expected, upon peptide stimulation, memory CD4 T cells were capable of immediately producing both IFNγ and, to a lower extent, IL-2 (Figure 4B).
In vivo CD4 T cell cytotoxicity
Since we saw a strong induction of GzB in the CD4 T cell population during infection we next wanted to determine whether the CD4 T cells elicited in B6 mice were capable of in vivo cytotoxicity (Figure 4C). We chose to use a CFSE- based CD4 in vivo cytotoxicity assay as initially described by Jellison et al. (24). In these experiments we adoptively transferred peptide-pulsed splenocytes from B6 Ly-5.1 congenic mice (target cells), into day 6 WNV infected B6 mice, where the congenic marker was used to identify transferred cells and two concentrations of CFSE labeling were used to differentiate specific targets from the control ones. The in vivo CTL assays showed the cytotoxic capacity of the antigen-specific CD4 T cells, where 39% of the MHC class II+ targets were killed within a 12-hour time period (p<0.008) (Figure 4C middle panel).
Additional in vivo CTL assays, were used to determine the mechanism of cytolytic clearance. PKH26 was used to label transferred cells and two different concentrations CFSE was used to differentiate targets. In these experiments we showed that targets sensitized with class II-restricted peptides were not as efficiently eliminated in vivo in mice lacking perforin (p<0.03 vs B6) or targets lacking Fas (lpr/lpr) (p<0.03 vs B6) versus B6 mice. Most importantly, when Fas−/− targets were transferred into perforin deficient mice, thereby eliminating both of the main cytolytic mechanisms simultaneously, CD4 T cells were no longer able to clear peptide coated targets (p<0.03 vs LPR−/− ->B6) (Figure 4C right panel). As an additional control for specificity, when sensitized targets were transferred into CD4 deficient mice, no cytolytic activity was seen (Figure 4C right panel). We therefore conclude that both Fas/FasL and perforin mediated mechanisms are involved in CD4 cytotoxicity, and that they have a level of redundancy for CD4 T cell mediated cytolytic clearance of WNV peptide coated targets in vivo.
Furthermore, direct ex vivo CD4 51Cr release assays were also completed using peptide pulsed IC-21 cells, a macrophage cell line, where we again readily observed specific lysis of WNV-coated, but not ovalbumin-coated (OT-II) targets (Fig. 4D). This data strongly suggests that during the course of WNV infection CD4 T cells differentiate into in vivo cytotoxic effectors capable of killing infected cells.
In vivo relevance of antigen-specific CD4 T cell response
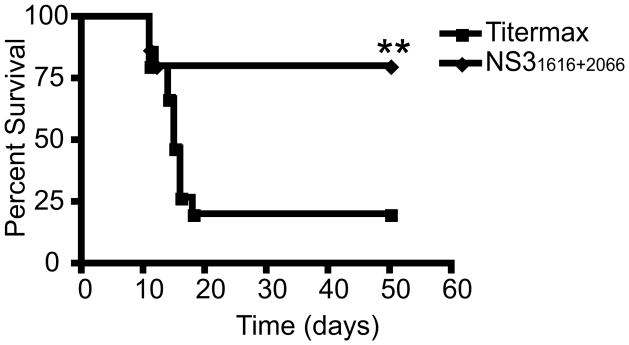
Based upon adoptive transfer of naive CD4 T cells and the direct ex vivo functions of antigen-specific T cells, we proposed that antigen-specific CD4 T cells use both their cytolytic and cytokine capacity to control WNV infection, preventing the development of encephalitis/meningitis. To look at the direct protective capacity of antigen-specific CD4 T cells we used a peptide vaccination approach. B6 mice were vaccinated two times with an emulsion of NS31616 and NS32066 peptide in adjuvant TiterMax Gold® (29). Twenty days after the last vaccination, mice were challenged with a high dose of WNV (1200pfu sc) and were observed for 60 days. The mice that received the vaccination with WNV epitopes NS31616 + NS32066 were protected significantly better then the mice vaccinated with OVA323-339 (p<0.03) (Figure 5). This data demonstrates that the dominant peptide epitopes recognized by CD4 T cells can be used as a vaccine to protect mice given a lethal dose of WNV.
Figure 5. Antigen-specific CD4 T cell responses are essential for protection against WNV.
CD4 T cell peptide vaccination leads to a significant (p<0.03) increase in protection of WNV infected mice. Mice were vaccinated using 20μg of NS31616+2066 or 20μg of OTII control peptide in Titermax Gold® emulsion. Ten mice per group were challenged with 1200 pfu of WNV. One representative experiment of three is shown.
DISCUSSION
In this study, we show that naïve CD4 T cells differentiate into primary effector cells and protect RAG-1−/− mice from lethal WNV infection. This indicates that not only do CD4 T cells have the ability to directly control WNV infection, but that they are sufficient to measurably protect RAG-1−/− mice from WNV. In this report we also defined, in the H-2b haplotype, most, if not all, CD4 T cell epitopes that develop during the course of a WNV infection. More importantly, we used these CD4 T cell epitopes as a vaccine to show that the generation of memory CD4 T cells response in intact B6 mice can be protective.
Previous reports studying anti-viral immune responses to Flaviviruses, including WNV, have indicated that T cells are required to prevent the development of encephalitis/meningitis (17, 18, 30). More recent work indicates a requirement for CD8 T cells in protection from lineage I strains of WNV (13, 15, 16, 18). While CD4 T cells are not required during primary Dengue virus infection of mice (12), there is an absolute requirement for CD4 T cells during primary WNV infection ((14) and data above). Our studies indicate that while the CD4 antigen-specific response may be important for the protection by other mechanisms, such as CD8 T-cells and antibody/B-cells, as originally described by Sitati et al. (14), they also can protect in vivo at least in part by direct effector function, and that this is sufficient to confer a significant degree of anti-WNV protection to adoptive hosts.
There has been a long-standing interest in the CD4 T cell functional response to many of the flaviviruses that infect humans. It has been shown, predominantly with T cell lines, that CD4 T cells responding to Dengue virus (DV), JEV, YF and WNV can proliferate, produce IFNγ and IL-2 and are cytotoxic in response to viral antigens (11, 30–33). It has been recently shown that during resolution of JEV infection the presence of a strong Th1 T cells response, including IFNγ production, results in reduction of neurological sequelae (34). However, the CD4 T cell response to lineage I WNV infection in real time has not been analyzed in these studies. We provide what we believe to be the first description of a CD4 T cell antigen-specific response to WNV and a list of WNV determinants recognized by CD4 T cells. Our data indicate that the antigen specific CD4 T cells respond in a Th1 fashion, including pronounced IFNγ, and low amounts of IL-2 production.
The one report that examined the direct effector role of CD4 T cells during a lineage I WNV infection did observe a protective effect upon the transfer of CD4 T cells, but did not observe a difference in the protective capacity of CD4 T cells that lacked IFNγ, perforin or Fas-FasL (14). The authors interpreted this as evidence for the lack of direct effector function. Our results suggest, that CD4 T cells can lyse peptide-coated targets in vivo using a combination of perforin and Fas-FasL mediated pathway. The use of the perforin mediated pathway would be consistent with our results on the expression of GzB. It is possible that WNV-specific CD4 effector T cells in vivo use all three effector mechanisms, IFNγ, perforin and Fas-FasL, with a level of redundancy built into the antigen-specific response to control WNV replication and spread. However, CD4 T cells deficient in one effector function may not be sufficient to reduce their antiviral activity and to observe a difference in survival in an adoptive transfer model. If so, that would explain why Sitati et al. (14) saw a similar increase in survival in mice receiving CD4 T cells, regardless of whether they were wt, IFNγ−/−, perforin−/− or FasL−/−.
Several studies have identified proper T cell trafficking as critical for protection from WNV neurological disease. CXCL10 and CD40 deficient mice have impaired trafficking of CD4 T cells into the CNS (10, 16). In B6 mice, WNV infection of the CNS begins approximately 3 to 4 days post-infection, which is the earliest time point when we could detect antigen-specific CD4 T cells that are prepared to secrete anti-viral cytokines. However, control of viral spread within the CNS requires the use of non-cytolytic mechanisms of viral clearance for host survival (35). The secretion of IFNγ by lymphocytes is required for the clearance of neurotropic viral infections, such as Sindbis virus (36) and yellow fever (18), in order to keep neurons intact. Although neurons are not known to express MHC class II in situ, they do constitutively express IFNγR (37), and upon exposure to IFNγ secreted by WNV-specific CD4 T cells can up regulate essential anti-viral molecules such as RNaseL (38). Therefore, one could expect that the secretion of IFNγ by WNV-specific CD4 T-cells would play a role in containing viral pathology/loads in the CNS. Experiments are in progress to address this issue.
In summary our experiments show that CD4 T cells are sufficient for controlling and clearing WNV from RAG-1−/− mice. Antigen-specific CD4 T cells rapidly respond to infection within the periphery by secreting a multitude of cytokines. In addition to cytokine production, our data shows that CD4 T cells have in vivo cytotoxic capabilities. These antigen-specific CD4 T cells most likely use both cytokine production and cytotoxicity as mechanisms to prevent WNV encephalitis/meningitis during viral challenge. Together with the published results on other immune mechanisms deployed against WNV (9–11, 13–15, 25–27, 32, 38), including other components of the adaptive immune system (notably CD8 T cells and B cells), the above results illustrate a very broad collaboration of diverse innate and adaptive effector mechanisms that protect against this virus.
Supplementary Material
Table II.
| PART 2 – optimal peptide identification | |||||
|---|---|---|---|---|---|
| Day Post Infection | Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | Exp. 5 |
| # of mice | N=4 | N=3 | N=4 | N=5 | N=2 |
| Anti-CD3 | 1.16 | 1.14 | 0.52 | 1.33 | 0.91 |
| E646-660 | ND | 0.32 | ND | 0.29 | 0.16 |
| E641-655 | 0.51 | 0.33 | 0.10 | 0.41 | 0.27 |
| NS3 1616-1630 | 0.32 | 0.50 | 0.15 | 0.30 | 0.14 |
| 1617-1627 | ND | 0.11 | ND | ND | 0.08 |
| 1618-1628 | ND | 0.10 | ND | 0.17 | 0.08 |
| 1619-1629 | ND | 0.15 | ND | 0.28 | 0.37 |
| 1620-1630 | ND | 0.16 | ND | ND | ND |
| NS3 2066-2080 | 0.53 | 0.41 | 0.41 | 0.49 | 0.43 |
| 2068-2078 | ND | 0.25 | ND | 0.24 | ND |
| 2070-2080 | ND | 0.20 | 0.01 | 0.34 | 0.08 |
Levels of cytokine responses (in % of total CD4 T cells) are indicated in the table as mean values from indicated numbers of mice. ND-not determined.
Acknowledgments
Supported by the USPHS awards N01 50027 (J.N-Z.), T32 AI007472 (J.B.) and RR0163 (to the ONPRC) from the National Institute of Allergy and Infectious Diseases and the National Institute for Research Resources, National Institutes of Health.
We are thankful to Dr. Elizabeth Sitati and. Dr. Vesselin Mitaksov (Washington University, St. Louis, MO) for helpful discussions and advice, and the members of the Nikolich laboratory for assistance and stimulating discussion. We appreciate that Dr. Michael Diamond (Washington University, St. Louis, MO) allowed us to complete additional experiments within his laboratory.
Abbreviations
- FCM
flow cytofluorometry
- GzB
granzyme B
- ICCS
intracellular cytokine staining
- i.v
intravenous injection
- JEV
Japanese encephalitis virus
- MOI
multiplicity of infection
- MST
mean survival time
- Perf−/−
mice carrying a genomic deletion at the perforin locus
- pfu
plaque forming units
- s.c
subcutaneous
- WNV
West Nile virus
- YFV
Yellow fever virus
References
- 1.Anderson JF, Andreadis TG, Vossbrinck CR, Tirrell S, Wakem EM, French RA, Garmendia AE, Van Kruiningen HJ. Isolation of West Nile virus from mosquitoes, crows, and a Cooper’s hawk in Connecticut. Science. 1999;286:2331–2333. doi: 10.1126/science.286.5448.2331. [DOI] [PubMed] [Google Scholar]
- 2.Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, Crise B, Volpe KE, Crabtree MB, Scherret JH, Hall RA, MacKenzie JS, Cropp CB, Panigrahy B, Ostlund E, Schmitt B, Malkinson M, Banet C, Weissman J, Komar N, Savage HM, Stone W, McNamara T, Gubler DJ. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- 3.West nile virus activity--United States, January 1-November 7, 2006. Mmwr. 2006;55:1204–1205. [PubMed] [Google Scholar]
- 4.Anonymous. West Nile Virus. Centers for Disease Control; 2003. [Google Scholar]
- 5.CDC. West Nile virus Statistics, Surveillance, and Control. 2008. [Google Scholar]
- 6.Marfin AA, Petersen LR, Eidson M, Miller J, Hadler J, Farello C, Werner B, Campbell GL, Layton M, Smith P, Bresnitz E, Cartter M, Scaletta J, Obiri G, Bunning M, Craven RC, Roehrig JT, Julian KG, Hinten SR, Gubler DJ. Widespread West Nile virus activity, eastern United States, 2000. Emerg Infect Dis. 2001;7:730–735. doi: 10.3201/eid0704.010423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers TJ, Diamond MS. Pathogenesis of flavivirus encephalitis. Adv Virus Res. 2003;60:273–342. doi: 10.1016/S0065-3527(03)60008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes C. West Nile Virus: Uganda. Ann NY Acad Sci. 2001;951:25. doi: 10.1111/j.1749-6632.2001.tb02682.x. [DOI] [PubMed] [Google Scholar]
- 9.Diamond MS, Shrestha B, Mehlhop E, Sitati E, Engle M. Innate and adaptive immune responses determine protection against disseminated infection by West Nile encephalitis virus. Viral Immunol. 2003;16:259–278. doi: 10.1089/088282403322396082. [DOI] [PubMed] [Google Scholar]
- 10.Klein RS, Lin E, Zhang B, Luster AD, Tollett J, Samuel MA, Engle M, Diamond MS. Neuronal CXCL10 directs CD8+ T-cell recruitment and control of West Nile virus encephalitis. J Virol. 2005;79:11457–11466. doi: 10.1128/JVI.79.17.11457-11466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulkarni AB, Mullbacher A, Parrish CR, Westaway EG, Coia G, Blanden RV. Analysis of murine major histocompatibility complex class II-restricted T-cell responses to the flavivirus Kunjin by using vaccinia virus expression. J Virol. 1992;66:3583–3592. doi: 10.1128/jvi.66.6.3583-3592.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shresta S, Kyle JL, Snider HM, Basavapatna M, Beatty PR, Harris E. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J Virol. 2004;78:2701–2710. doi: 10.1128/JVI.78.6.2701-2710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha B, Diamond MS. Role of CD8+ T cells in control of West Nile virus infection. J Virol. 2004;78:8312–8321. doi: 10.1128/JVI.78.15.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sitati EM, Diamond MS. CD4+ T-cell responses are required for clearance of West Nile virus from the central nervous system. J Virol. 2006;80:12060–12069. doi: 10.1128/JVI.01650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brien JD, Uhrlaub JL, Nikolich-Zugich J. Protective capacity and epitope specificity of CD8(+) T cells responding to lethal West Nile virus infection. European journal of immunology. 2007;37:1855–1863. doi: 10.1002/eji.200737196. [DOI] [PubMed] [Google Scholar]
- 16.Purtha WE, Myers N, Mitaksov V, Sitati E, Connolly J, Fremont DH, Hansen TH, Diamond MS. Antigen-specific cytotoxic T lymphocytes protect against lethal West Nile virus encephalitis. European journal of immunology. 2007;37:1845–1854. doi: 10.1002/eji.200737192. [DOI] [PubMed] [Google Scholar]
- 17.Murali-Krishna K, Ravi V, Manjunath R. Protection of adult but not newborn mice against lethal intracerebral challenge with Japanese encephalitis virus by adoptively transferred virus-specific cytotoxic T lymphocytes: requirement for L3T4+ T cells. J Gen Virol. 1996;77(Pt 4):705–714. doi: 10.1099/0022-1317-77-4-705. [DOI] [PubMed] [Google Scholar]
- 18.Liu T, Chambers TJ. Yellow fever virus encephalitis: properties of the brain-associated T-cell response during virus clearance in normal and gamma interferon-deficient mice and requirement for CD4+ lymphocytes. J Virol. 2001;75:2107–2118. doi: 10.1128/JVI.75.5.2107-2118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swain SL, Agrewala JN, Brown DM, Jelley-Gibbs DM, Golech S, Huston G, Jones SC, Kamperschroer C, Lee WH, McKinstry KK, Roman E, Strutt T, Weng NP. CD4+ T-cell memory: generation and multi-faceted roles for CD4+ T cells in protective immunity to influenza. Immunological reviews. 2006;211:8–22. doi: 10.1111/j.0105-2896.2006.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol. 2006;177:2888–2898. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 21.Tishon A, Lewicki H, Andaya A, McGavern D, Martin L, Oldstone MB. CD4 T cell control primary measles virus infection of the CNS: regulation is dependent on combined activity with either CD8 T cells or with B cells: CD4, CD8 or B cells alone are ineffective. Virology. 2006;347:234–245. doi: 10.1016/j.virol.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 22.Weidinger G, Czub S, Neumeister C, Harriott P, ter Meulen V, Niewiesk S. Role of CD4(+) and CD8(+) T cells in the prevention of measles virus-induced encephalitis in mice. J Gen Virol. 2000;81:2707–2713. doi: 10.1099/0022-1317-81-11-2707. [DOI] [PubMed] [Google Scholar]
- 23.Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jellison ER, Kim SK, Welsh RM. Cutting edge: MHC class II-restricted killing in vivo during viral infection. J Immunol. 2005;174:614–618. doi: 10.4049/jimmunol.174.2.614. [DOI] [PubMed] [Google Scholar]
- 25.Diamond MS, Shrestha B, Marri A, Mahan D, Engle M. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J Virol. 2003;77:2578–2586. doi: 10.1128/JVI.77.4.2578-2586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shrestha B, Samuel MA, Diamond MS. CD8+ T cells require perforin to clear West Nile virus from infected neurons. J Virol. 2006;80:119–129. doi: 10.1128/JVI.80.1.119-129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitati E, McCandless EE, Klein RS, Diamond MS. CD40-CD40 Ligand Interactions Promote Trafficking of CD8+ T Cells into the Brain and Protection against West Nile Virus Encephalitis. J Virol. 2007 doi: 10.1128/JVI.00941-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence CW, Ream RM, Braciale TJ. Frequency, specificity, and sites of expansion of CD8+ T cells during primary pulmonary influenza virus infection. J Immunol. 2005;174:5332–5340. doi: 10.4049/jimmunol.174.9.5332. [DOI] [PubMed] [Google Scholar]
- 29.Dyall R, Vasovic LV, Molano A, Nikolic-Zugic J. CD4-independent in vivo priming of murine CTLs by optimal MHC class I-restricted peptides derived from HIV and other pathogens. International Immunology. 1995;7:1205–1212. doi: 10.1093/intimm/7.8.1205. [DOI] [PubMed] [Google Scholar]
- 30.Mathur A, Arora KL, Chaturvedi UC. Host defence mechanisms against Japanese encephalitis virus infection in mice. J Gen Virol. 1983;64(Pt 4):805–811. doi: 10.1099/0022-1317-64-4-805. [DOI] [PubMed] [Google Scholar]
- 31.Green S, Kurane I, Edelman R, Tacket CO, Eckels KH, Vaughn DW, Hoke CH, Jr, Ennis FA. Dengue virus-specific human CD4+ T-lymphocyte responses in a recipient of an experimental live-attenuated dengue virus type 1 vaccine: bulk culture proliferation, clonal analysis, and precursor frequency determination. J Virol. 1993;67:5962–5967. doi: 10.1128/jvi.67.10.5962-5967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulkarni AB, Mullbacher A, Blanden RV. Functional analysis of macrophages, B cells and splenic dendritic cells as antigen-presenting cells in West Nile virus-specific murine T lymphocyte proliferation. Immunol Cell Biol. 1991;69(Pt 2):71–80. doi: 10.1038/icb.1991.12. [DOI] [PubMed] [Google Scholar]
- 33.Pan CH, Chen HW, Huang HW, Tao MH. Protective mechanisms induced by a Japanese encephalitis virus DNA vaccine: requirement for antibody but not CD8(+) cytotoxic T-cell responses. J Virol. 2001;75:11457–11463. doi: 10.1128/JVI.75.23.11457-11463.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar P, Sulochana P, Nirmala G, Chandrashekar R, Haridattatreya M, Satchidanandam V. Impaired T helper 1 function of nonstructural protein 3-specific T cells in Japanese patients with encephalitis with neurological sequelae. The Journal of infectious diseases. 2004;189:880–891. doi: 10.1086/381768. [DOI] [PubMed] [Google Scholar]
- 35.Rottenberg M, Kristensson K. Effects of interferon-gamma on neuronal infections. Viral Immunol. 2002;15:247–260. doi: 10.1089/08828240260066206. [DOI] [PubMed] [Google Scholar]
- 36.Burdeinick-Kerr R, Griffin DE. Gamma interferon-dependent, noncytolytic clearance of sindbis virus infection from neurons in vitro. J Virol. 2005;79:5374–5385. doi: 10.1128/JVI.79.9.5374-5385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neumann H, Schmidt H, Wilharm E, Behrens L, Wekerle H. Interferon gamma gene expression in sensory neurons: evidence for autocrine gene regulation. J Exp Med. 1997;186:2023–2031. doi: 10.1084/jem.186.12.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BR, Silverman RH, Gale M, Jr, Diamond MS. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol. 2006;80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.