A series of high-resolution crystal structures of RIG-I and RIG-I:dsRNA cocrystals has recently been reported. Comparison of these structures provides considerable insight into how this innate immune pattern recognition receptor is activated upon detecting and binding a certain class of viral RNAs.
Keywords: RIG-I activation; dsRNA PAMPs; crystal and cocrystal X-ray structures
Abstract
A series of high-resolution crystal structures of RIG-I and RIG-I:dsRNA cocrystals has recently been reported. Comparison of these structures provides considerable insight into how this innate immune pattern recognition receptor is activated upon detecting and binding a certain class of viral RNAs.
INTRODUCTION
The innate immune system is the first line of defense against viral infections and mounts an antiviral response within hours of their detection. It functions not necessarily to prevent the infection, but to slow it and to prime the adaptive immune system whose heavy guns (T cells and antibodies) can clear the infection. The innate immune system does not respond specifically to a particular pathogen, like the adaptive arm. Rather, it reacts “generically,” via pathogen recognition receptors (PRRs) that detect pathogen-associated molecular patterns (PAMPs) shared by most or all of a given class of infectious agents. The essential feature of PAMPs is that they can be used to distinguish between self and nonself, and thus signal the presence of the invader. One class of PRRs, Toll-like receptors (TLRs), is membrane bound and found at the cell surface and in endosomal compartments, whereas RIG-I-like receptors (RLRs) are in the cytoplasm. The RLRs—RIG-I (retinoic acid-inducible gene-I, DDX58), MDA5 (melanoma-differentiation-associated gene 5, IFIH1), and LGP2 (Laboratory of Genetics and Physiology 2, DHX58)—form a family within superfamily 2 (SF2) RNA helicases and are closely related to DEAD-box helicases (Fairman-Williams et al. 2010). The RIG-I family also includes Dicer, involved in the processing of small RNAs, as well as helicase-associated endonucleases (Hef) (Mosedale et al. 2005; Nishino et al. 2005). This review focuses on RIG-I and the new insight that a series of high-resolution crystal structures provide about how RIG-I is activated upon detecting and binding a certain class of viral RNAs (Civril et al. 2011; Jiang et al. 2011; Kowalinski et al. 2011; Luo et al. 2011, 2012).
RIG-I AND INNATE IMMUNITY
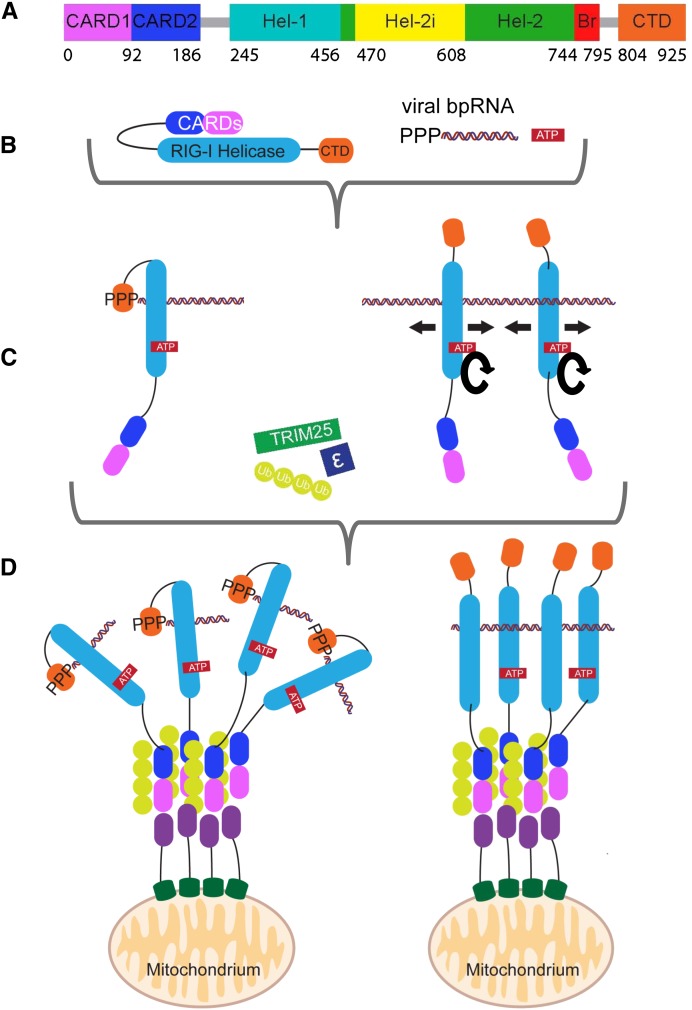
RIG-I was first reported as a retinoic acid–inducible gene in 1997 (GenBank: AF038963) and later rediscovered in the pig as a helicase (denoted RHIV-1) induced upon infection with porcine reproductive and respiratory syndrome virus (Zhang et al. 2000). Subsequently it was characterized as a key PRR that triggers type 1 interferon expression upon detection of viral RNA (Yoneyama et al. 2004). It is now known that RIG-I is activated by diverse positive- and negative-strand RNA viruses including influenza, Rift Valley fever, measles, Ebola, vesicular stomatitis (VSV), and hepatitis C viruses (Loo and Gale 2011). It is a large, multidomain protein (925 residues for human RIG-I, hRIG-I) (Fig. 1A) that is highly conserved among vertebrates. RLR-like molecules are also found in several invertebrates including nematodes, sea anemones, and sponges (Zou et al. 2009). It comprises N-terminal tandem caspase recruitment domains (CARDs), a central DECH-box type RNA helicase, composed of two Rec A domains (Hel-1 and Hel-2; the SF2 helicase core) but with a family-specific large insertion within Hel-2 (referred to as Hel-2i). Finally, there is a C-terminal regulatory domain (CTD) characteristic of the RLR family, and which contains a structural zinc ion. The CARDs, which are small helical bundles of the death-domain superfamily, are responsible for transmitting the activation signal downstream. The two helicase domains Hel-1 and Hel-2 create at their interface an active site for ATP binding and hydrolysis, as well as jointly forming an extended RNA-binding surface. The CTD binds the blunt ends of 5′OHdsRNA or 5′pppdsRNA (Lu et al. 2010; Wang et al. 2010). The CTD is connected to Hel-2 by a unique, elbow-shaped helical extension, denoted as the bridging helices (Br, Fig 1A; Kowalinski et al. 2011) or pincer domain (Luo et al. 2011). MDA-5 has a similar architecture, whereas LGP2 lacks the CARDs. RIG-I and MDA-5 complement each other by responding to distinct types of RNAs from different viruses (Yoneyama et al. 2005; Kato et al. 2006; Loo and Gale 2011). The function of LGP2 is less clearly defined, because it can act positively or negatively upon activation by different viruses (Saito et al. 2007; Satoh et al. 2010).
FIGURE 1.
Current view of the RIG-I activation pathway. (A) A color-coded schematic representation of the domain structure of RIG-I, as used throughout. Bare lines represent flexible linkers. (B) Cartoon of auto-inhibited RIG-I and its substrates, 5′ppp-bpRNA and ATP. (C) RIG-I binds via its high-affinity 5′ppp blunt-end composite binding site even on relatively short dsRNAs, or to lower-affinity sites on longer dsRNAs independent of their 5′ ends, releasing the CARDs. In the latter case, cycles of ATP hydrolysis (round arrows) can lead to translocation (straight arrows). (D) The free CARDs are covalently or noncovalently bound by K63-linked polyubiquitin chains generated by E3-ligase TRIM25. This promotes tetramerization of RIG-I via its ubiquitin-bound CARDs. Tetramers are shown either with RIG-Is bound to different 5′ppp-bpRNAs (left) or the same long dsRNA (right). The mitochondrial membrane chaperone 14-3-3ɛ directs the RNA-bound multimers to mitochondrial membranes, enabling MAVS and RIG-I CARDs to interact. This promotes MAVS aggregation and propagation of the activation signal downstream.
The current understanding of the mechanism of action of RIG-I is depicted in Figure 1. The CARDs of ligand-free RIG-I, which is monomeric, are unavailable for signal transduction due to their interaction with other domains (Fig. 1B). Upon binding viral RNA and ATP, conformational changes in RIG-I occur that expose the CARDs, allowing E3-ligase tripartite motif-containing 25 (TRIM25)–dependent K63 polyubiquitination of Lys172 (Gack et al. 2007) or unanchored K63-linked polyubiquitin chain binding (Fig. 1C,D; Zeng et al. 2010). This, in turn, induces RIG-I multimerization (Jiang et al. 2012), which promotes RIG-I association via CARD–CARD interactions with the membrane-bound mitochondrial antiviral signaling protein (MAVS, also known as IPS-1, CARDIF, or VISA), the central adaptor of the IFNβ activation pathway (Fig. 1D; Kawai et al. 2005; Meylan et al. 2005; Seth et al. 2005; Xu et al. 2005). Recently the protein 14-3-3ɛ has also been implicated in MAVS targeting by activated RIG-I (Liu et al. 2012). RNA- and polyubiquitin-bound RIG-I-induced aggregation of MAVS on the mitochondrial membrane (Hou et al. 2011; Jiang et al. 2012), aided by the regulator of mitochondrial fusion, MFN1 (Onoguchi et al. 2010), leads to activation of kinases IKK and TBK1, which, in turn, mobilize the transcription factors NF-κB and IRF3/7. Selective proteasomal degradation of the larger isoform of MAVS following TRIM25-induced ubiquitination has recently been shown to favor phosphorylation of IRF3 (Castanier et al. 2012). Signaling finally results in the expression of pro-inflammatory cytokines and type I interferons and the establishment of an intracellular antiviral state.
dsRNA AND ITS 5′ppp-ENDS AS VIRAL PAMPS
It took several years for the RIG-I-activating RNA ligands to be defined satisfactorily in vitro, using assays in which RNAs are transfected into cells and IFN expression is determined. Even so, there is still considerable controversy as to how such RNAs become available to trigger RIG-I in virally infected cells. The consensus now is that the minimal requirement for RIG-I activation is a blunt-ended base-paired RNA 10–20 bp long with a 5′ triphosphate and free of mismatches near the blunt end (denoted 5′pppbpRNA, since it could arise from ssRNA with complementary ends or dsRNA) (Schlee et al. 2009; Schmidt et al. 2009; Marq et al. 2010). The new RIG-I RNA cocrystal structures are consistent with this (see below). However, in some studies (Lu et al. 2010) but not others (Marq et al. 2010; Binder et al. 2011), it was reported that short, blunt-ended 5′OHbpRNAs are also active; this discrepancy may partly be explained by the stability of various transfected 5′OHbpRNAs within different cells. On the other hand, some bunyavirus genomes appear to escape RIG-I detection because they contain blunt-ended dsRNA panhandles that contain only 5′ monophosphates (Habjan et al. 2008). This suggests that the higher affinity and significantly longer off-rate of 5′pppbpRNA to the CTD, as compared with 5′OHbpRNA or 5′pbpRNA (Lu et al. 2010; Wang et al. 2010), is sufficient to discriminate active and inactive ligands. It has also been reported that much longer dsRNAs (>200 bp), including poly(I:C), not necessarily bearing a 5′ppp-end or even being blunt-ended, can also induce IFN via RIG-I (Strahle et al. 2007; Kato et al. 2008; Binder et al. 2011). This is more difficult to explain from a structural and biochemical point of view because of the much lower intrinsic affinity of RIG-I for such RNAs. However, it could be related to the ability of RIG-I to translocate along dsRNA (Myong et al. 2009) or the need for RNA-mediated RIG-I multimerization to promote interaction with MAVS (see below). It was also reported that RIG-I could be activated by 5′pppssRNA (Hornung et al. 2006; Kato et al. 2006; Pichlmair et al. 2006). Although in the light of the new crystal structures this now seems highly unlikely, it took a while for the explanation of these observations to be found—namely, the fact that the bacteriophage T7 RNA polymerase used to synthesize the 5′pppssRNA has a tendency to also coproduce dsRNA (Cazenave and Uhlenbeck 1994). Indeed, when 5′pppssRNAs were synthesized chemically or the T7 products were purified to remove all traces of dsRNA, these 5′pppssRNAs were unable to activate RIG-I until they were annealed with a complementary ssRNA (Schlee et al. 2009; Marq et al. 2010). Uncontaminated ssRNA is also unable to stimulate the RIG-I or MDA5 ATPases (Peisley et al. 2011).
Because intracellular RNAs must be deproteinized before they can be assayed for PAMP activity by transfection, the nature and origin of the RNA species relevant for RIG-I activation in infected cells have been difficult to pin down unambiguously. Nevertheless, panhandle structures corresponding to the blunt-ended 5′pppbpRNAs that act as RIG-I PAMPs in vitro occur in many ssRNA virus systems and have been implicated (Strahle et al. 2007; Schlee et al. 2009; Baum et al. 2010; Rehwinkel et al. 2010). These panhandles are also a special feature of copy-back defective-interfering (DI) RNA genomes. Notably, the ability of the commonly used Sendai virus (Cantell strain) to induce IFN depends on the presence of these DI genomes in the virus stock. Their elimination by plaque purification eliminates IFN induction by the Sendai virus infection because this virus, like others, expresses IFN antagonists that counteract the innate immune response (Strahle et al. 2007). For review of viral countermeasures against the RIG-I and other innate immune pathways, see Bowie and Unterholzner (2008).
RIG-I STRUCTURES WITH BOUND dsRNA
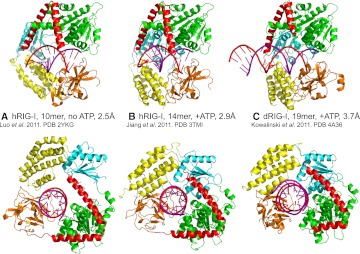
Despite the obvious need for structural information on RIG-I to give insight into its mechanism of action, for a long time the protein remained intractable to crystallization. Finally, in a short space of time in 2011, four groups independently reported high-resolution structures of RIG-I and RIG-I RNA complexes that extend previous structural studies that revealed how the CTD preferentially bound 5′pppbpRNA (Lu et al. 2010; Wang et al. 2010). These new results considerably refine our understanding of how RNA-induced activation works. Two studies used human RIG-I (hRIG-I) (Jiang et al. 2011; Luo et al. 2011) and one mouse RIG-I (mRIG-I) (Civril et al. 2011), but the most comprehensive results were obtained with duck RIG-I (dRIG-I) (Kowalinski et al. 2011). Three structures of CARD-deleted RIG-I complexed with blunt-ended 5′OHbpRNAs were determined: (1) the helicase + CTD domains of hRIG-I (hRIG-IΔCARDs, residues 230–925) with a 10-mer dsRNA but no nucleotide at 2.5 Å resolution (Fig. 2A; Luo et al. 2011); (2) hRIG-IΔCARDs (residues 232–925) complexed with the transition-state analog ADP:BeF3 and a 14-mer bpRNA at 2.9 Å resolution (Fig. 2B; Jiang et al. 2011); and (3) the helicase domain of dRIG-I without the CTD (dRIG-IΔCARDsΔCTD, 242–794) in complex with the transition-state analog ADP:AlF3 and a 19-mer bpRNA at 3.7 Å resolution (Fig. 2C; Kowalinski et al. 2011). Interestingly, the resolution is inversely correlated with the length of the dsRNA used. Very recently, CARD-deleted hRIG-I has been crystallized with a 5′-triphosphorylated stem–loop RNA (8 bp with a UUCG tetra-loop) and ADP, giving a structure at 2.8 Å resolution (Luo et al. 2012).
FIGURE 2.
The three different structures of RIG-I bound to dsRNA. (A) Human RIG-IΔCARDs with a 10-mer dsRNA but no nucleotide at 2.5 Å resolution (PDB entry 2YKG) (Luo et al. 2011). (B) Human RIG-IΔCARDs complexed with the transition-state analog ADP:BeF3 and a 14-mer dsRNA at 2.9 Å resolution (PDB entry 3TMI) (Jiang et al. 2011). (C) Duck RIG-IΔCARDsΔCTD in complex with the transition-state analog ADP:AlF3 and a 19-mer dsRNA at 3.7 Å resolution (PDB entry 4A36) (Kowalinski et al. 2011).
Despite differences due to species, presence or absence of nucleotide, and RNA length and sequence, all structures give a consistent picture of how RIG-I binds to bpRNA. RIG-I helicase + CTD when bound to dsRNA is disk shaped (approximate diameter 75 Å, length 45 Å) with a central, negatively charged cavity (approximate diameter 25 Å, length 30 Å) that accommodates ∼9 bp of bpRNA. The protein thus completely surrounds the dsRNA with the CTD capping one blunt end, the other end of the dsRNA being free to extend (Figs. 3A, 4D). Interestingly, in the dRIG-I helicase–bpRNA structure, the register of the blunt end with respect to the helicase is maintained even in the absence of the CTD. Indeed, an essentially correct model of dRIG-IΔCARDs binding to bpRNA could be constructed by simply superposing separate helicase–bpRNA and the CTD–bpRNA structures (cf. Fig. 2B and 2C). In the ATP-analog-bound forms of the complex, all four RIG-I domains (Hel1, Hel2, Hel2i, CTD) make complementary contributions to the elaborate network of mainly polar interactions made with both strands of the dsRNA (Fig. 3C). In the nucleotide-free and ADP complexes (Luo et al. 2011, 2012), the helicase is an open state (see below) with the disordered Hel-2 not contributing to RNA binding and Hel2i making different contacts due to its slightly rotated position (Fig. 2A). In all cases, the bpRNAs maintain the canonical A-form conformation, and there is no evidence of distortions or partial unwinding as seen with nucleic acid binding to functional helicases. Thus, RIG-I appears to be a helicase in name only, although ATP-dependent translocation along bpRNA has been described (Myong et al. 2009).
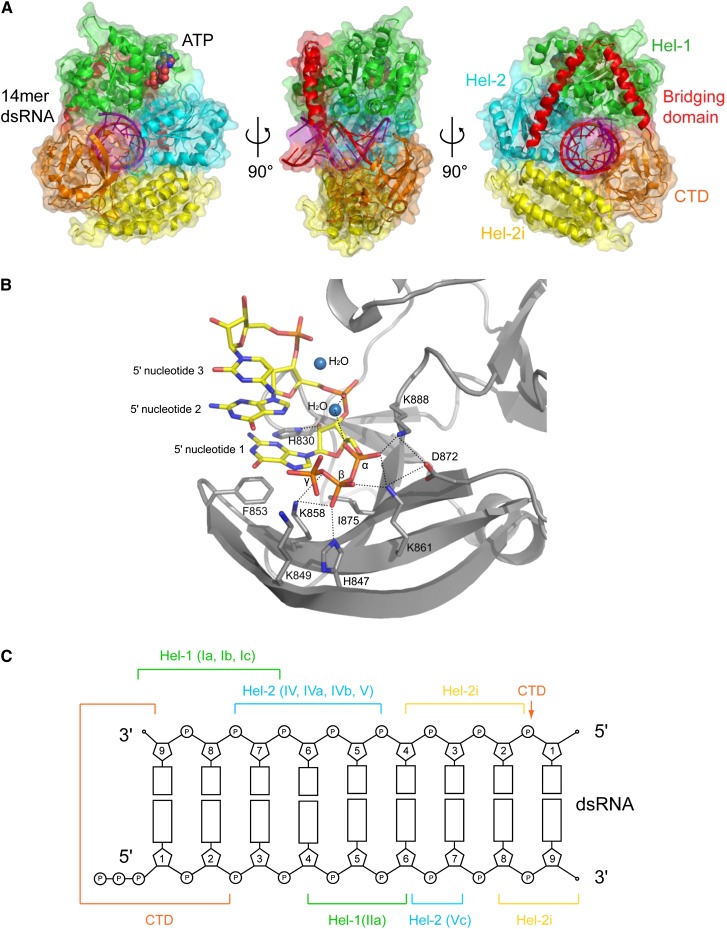
FIGURE 3.
5′ppp-dsRNA binding by RIG-I. (A) Ribbon and surface representation of human RIG-I bound to 14-mer dsRNA and transition-state analog ADP:BeF3 (indicated as ATP) in three orientations (PDB entry 3TMI) (Jiang et al. 2011) color coded as in Figure 1A. (B) Details of the binding of 5′ppp-dsRNA to the human RIG-I CTD (PDB entry 3LRN, chain A) (Lu et al. 2010). (C) Schematic diagram of contacts of various domains and motifs of human RIG-I to dsRNA based on Jiang et al. (2011) and Kowalinski et al. (2011).
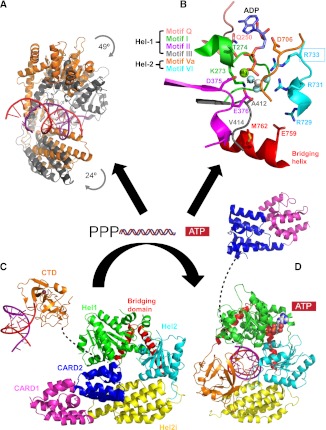
FIGURE 4.
Conformational changes associated with activation of RIG-I. (A) Binding of dsRNA and ATP coverts RIG-I helicase from a flexible open form (orange) to an ordered closed form (gray) and involves significant relative rotation of Hel-1, Hel-2, and Hel-2i; based on duck RIG-I PDB entries 4A2P and 4A36, respectively. (B) The ATP-binding site is formed at the interface of Hel-1 and Hel-2 by conserved helicase motifs; based on duck RIG-I PDB entry 4A36 with ATP analog ADP:AlF3. (C) Model of the auto-repressed state of duck RIG-I (PDB entry 4A2W). The flexibly linked CTD (invisible in the crystal structure) has been included to illustrate that it is free to detect and bind 5′ppp-bpRNA as the first step in activation. The CARDs, bound via CARD2 to Hel-2i, are unavailable for signaling. (D) Model of the activated state of human RIG-I (PDB entry 3TMI) with added CARDs on a 55-residue long flexible linker. In C and D, the Hel-2i domains (yellow) are superposed to show that CARD2 binding to Hel-2i is incompatible with the bound position of the dsRNA and CTD.
Key features of the protein–RNA contacts are as follows (Fig. 3B,C): (1) Phe853 of the CTD stacks over the bases forming the blunt end of the bpRNA. Other CTD contacts are made to the backbone of the first two 5′ nucleotides as well as via Ser906 to the backbone of base 7 of the 3′ strand. Some contacts previously described in isolated CTD–dsRNA structures are no longer observed due to a slight rotation of the bpRNA relative to the CTD. The very recent cocrystal structure of CARD-deleted hRIG-I with a 5′ppp stem–loop RNA (Luo et al. 2012) shows that the CTD contacts the α- and β-phosphates via His847, K858, K861, and K888 as previously described for the isolated CTD domain complex (Fig. 3B; Lu et al. 2010; Wang et al. 2010). Interestingly, no direct contacts are observed to the γ-phosphate itself although a favorable electrostatic interaction with nearby K849 and K851 is likely. (2) The backbone of the first five 3′-strand nucleotides binds across the two helicase subdomains making contacts, most often via phosphates to main-chain amides, with canonical helicase Motifs Ia, Ib, and Ic (Hel-1) and Motifs V and VI (Hel-2) as previously observed in several DEAD-box RNA helicase–ssRNA cocrystal structures, e.g., Vasa (Sengoku et al. 2006), Dpb5 (Montpetit et al. 2011). (3) Contacts to the 5′ strand occur between nucleotides 4 and 7. Most interestingly, novel contacts are made by Motif IIa of Hel-1 (379-KQHPY, just downstream from the critical ATP-interacting Motif II, 372-DECH), to 5′-strand nucleotides 4–6, as previously observed in SWI2/SNF2 ATPases bound to dsDNA (Du¨rr et al. 2005). (4) The helical insertion domain Hel-2i makes additional contacts to the 3′-strand backbone at nucleotides 6–8 and the 5′-strand backbone at nucleotides 8–9. The pattern of protein–RNA contacts observed thus explains why RIG-I specifically binds to duplex RNA (Fig. 3C). It is likely that several features, including the roles of Motif IIa and the helicase insertion domain in specifying the substrate as bpRNA, will be conserved in other RLH subgroup members Dicer and Hef. Indeed, a recent electron microscopy study shows that the helicase moiety of human Dicer is well modeled by RIG-I and likely undergoes the same conformational changes upon dsRNA binding (Lau et al. 2012).
LIGAND-FREE RIG-I STRUCTURES
Structures of the helicase domain in the absence of RNA are available for mRIG-I (with bound AMPPNP) (Civril et al. 2011) and dRIG-I (Kowalinski et al. 2011). So far the only crystal structures that include the CARDs are ligand-free dRIG-I constructs (Kowalinski et al. 2011). All these structures reveal that the RNA-free helicase is in an open, flexible conformation with many of the connecting loops of Hel2, in particular, being disordered. Whereas Hel2 and Hel2i form a rigid unit, relative to this, the position of Hel1 in the various structures has a large angular distribution due to the flexibility of the two interdomain connections (Kowalinski et al. 2011). The difficulty in crystallizing full-length, unliganded RIG-I is likely due to the intrinsic inter- and intradomain flexibility of the molecule in the absence of RNA. Success with dRIG-I rather than hRIG-I may be due to the slightly enhanced thermal stability of avian RIG-I (E Kowalinski and S Cusack, unpubl.), a property that correlates with crystallizability (Dupeux et al. 2011). The two CARD-containing dRIG-I structures, full-length and ΔCTD, are actually very similar even though derived from crystals of different space-groups. This is because the CTD is not visible in the electron density of the full-length crystals, although gel analysis of dissolved crystals shows the RIG-I to be intact. This suggests that the CTD is flexibly linked to the end of the bridging helices, consistent with the extended chain linker visible in the RNA-bound structures (Fig. 4C) and the absence of significant interaction between the isolated CTD and the complementary part of RIG-I in vitro (Kowalinski et al. 2011). Thus the hitherto common designation of the CTD as a “repressor” domain (RD) is a misnomer, based on early work in which the RD not only included the CTD but also the bridging helices and part of Hel2 (Saito et al. 2007). Recent work confirms that deletion of only the CTD does not activate RIG-I and has refined the notion of the RD to implicate only the bridging helices, whose integrity is required to maintain the auto-repressed state (Kageyama et al. 2011).
The ligand-free dRIG-I structure reveals that the tandem CARD domains form a rigid, head-to-tail unit (Fig. 4C). CARD1 is largely solvent exposed and its C terminus links directly to the extended N-terminal helix of CARD2. CARD2 is bound by its other end to the helicase insertion Hel-2i via a substantial interface notably mediated by conserved Phe540 (Kowalinski et al. 2011). The 55 amino acid linker between CARD2 and Hel-1 is disordered. The sequestering of CARD2 by its binding to Hel-2i is probably the key feature that maintains ligand-free RIG-I in an auto-repressed conformation (see below). RNA and ATP binding (Fig. 4B) induce a major conformational change (Fig. 4A) that closes the two halves of the helicase core, producing a much more ordered structure. In this conformation, simultaneous RNA and CARD2 binding to Hel-2i is impossible and the CARDs are ejected (Fig. 4D).
A STRUCTURE-BASED MODEL FOR RIG-I ACTIVATION
Comparison of the ligand-free and dsRNA/ATP-bound forms of RIG-I reveals the induced conformational rearrangements that accompany RIG-I activation, explaining its functional conversion from an auto-repressed state into a signaling competent configuration (Fig. 4). In the ligand-free state, RIG-I helicase is in an open and flexible conformation with low affinity for ATP and bpRNA (Fig. 5A). In this state, binding of CARD2 to Hel-2i, and possibly the presence of the long CARD2–helicase linker, sterically hinders dsRNA binding to the helicase. This also hinders the access of ubiquitination enzymes and polyubiquitin binding to the CARDs, thus inhibiting downstream signaling via MAVS. The substantial CARD2–Hel2i interface is mediated by salt bridges and hydrophobic interactions and is therefore energetically stable. This interface has significant overlap with the dsRNA–Hel2i interaction interface, suggesting that during the RNA-triggered transition, extra energy (possibly provided by ATP binding) is required to disrupt the CARD2–Hel2i interface and enable dsRNA to enter its binding site on the helicase. This is consistent with the 24-fold weaker binding of dsRNA to full-length RIG-I than to RIG-IΔCARDs (Jiang et al. 2011). The CTD, in contrast, is flexibly linked to the helicase and thus remains available for sensing and capturing 5′pppbpRNA with high affinity (Figs. 4C, 5A). bpRNA binding to the CTD increases its local concentration in the vicinity of the helicase, favoring the cooperative binding of ATP and dsRNA and inducing the high-ligand-affinity, closed form of the helicase (Fig. 5C). An intermediate in this activation process may be an open helicase with RNA but no ATP bound (Fig. 5B; Luo et al. 2011). The associated conformational switch forces the release of the tandem CARDs, the continued binding of which is sterically incompatible with tight dsRNA binding to the helicase and the repositioned CTD (Fig. 5B,C). In support of this mechanism, a point mutation at Phe540, located at the CARD2–Hel2i interface that is predicted to weaken this interaction, renders RIG-I constitutively active (Kowalinski et al. 2011). The CTD, in contrast to the CARDs and contrary to previous suggestion (see above), does not appear to play a role in maintaining RIG-I in an auto-inhibited state. Release of the CARDs on a 55-residue-long, presumably flexible linker makes them freely accessible for K63-linked polyubiquitination or polyubiquitin binding (Figs. 1C,D, 4D).
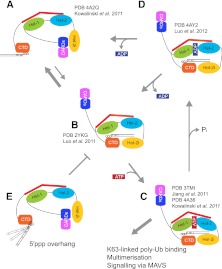
FIGURE 5.
A structure-based model of RIG-I activation. (A) In the auto-inhibited state, the CARDs are bound to Hel-2i and thus unavailable for signaling. The CTD, tethered to the red bridging helix by a flexible linker and thus available to sense RNA PAMPs, is shown bound to blunt-ended 5′ ppp-bpRNA. (B) The CTD-bound 5′ppp-bpRNA is pre-oriented to form a network of interactions with the helicase domains Hel-1 and Hel-2i, but not Hel-2, leading to displacement of the CARDs bound to Hel-2i. The CARDs now become potentially available for downstream interactions. In the absence of ATP, this state, visualized by Luo et al. (2011), could revert to the auto-inhibited state (A). (C) ATP binding to its pocket formed at the interface of Hel-1 and Hel-2 tightens and stabilizes the closed form of the structure. Jiang et al. (2011) visualized an “almost closed” version of this state with bound ADP:BeF3 in which the ATP binding motif VI is not engaged. Kowalinski et al. (2011) used the transition-state analog ADP:AlF3 and observed a fully closed form with all ATP-binding motifs configured as required for ATP hydrolysis (Fig. 4B). (D) The RNA and ADP bound state, following ATP hydrolysis and phosphate release, has very recently been visualized by Luo et al. (2012) and is in the semi-open conformation similar to the nucleotide free state (B). Upon ADP release, this state most likely reverts to B. (E) In the case in which the nucleotide bearing the 5′ppp is not base-paired (e.g., arenavirus genomes), the RNA helix is not constrained by its interaction with the CTD and is no longer pre-oriented to form a network of interactions with the helicase domains, and is thus inactive.
The state depicted in Figure 5C may not have a long lifetime because it is competent for ATP hydrolysis. However, even with bound ADP (Fig. 5D; Luo et al. 2012) or with no nucleotide (Fig. 5B; Luo et al. 2011), the crystal structures show that RNA remains bound to the CTD and helicase. Thus, ATP-consuming cycling between the states b, c, and d could create a time window for polyubiquitination and downstream signaling to occur, rather than reversion to the auto-inhibited state. As long as the CARDs stay polyubiquitinated, for steric reasons, they cannot be rebound to the helicase to reconstitute the auto-repressed state, even if RNA disassociation opens the helicase. ATP thus plays a double role in RIG-I signaling. ATP binding helps drive the conformational change that disengages the CARDs from Hel2i. This allows 5′pppbpRNA that is initially bound to the CTD to also bind the helicase. Once the RNA is bound with high affinity in its composite site, ATP hydrolysis can occur without necessarily impeding the signaling potential of the activated complex, as long as the CARDs remain released. However, helicase opening due to ATP hydrolysis and ADP release could with some probability result in disengagement of the RNA from the helicase (but not necessarily from its association with the CTD) and reversion to the auto-repressed state if the CARDs are not polyubiquitinated. In support of this view, mutants that can bind ATP but cannot hydrolyze it lead to constitutively active RIG-I and MDA5 (Bamming and Horvath 2009).
IMPORTANCE OF BLUNT-ENDED 5′ppp-BPRNA
In the above model of RIG-I activation, the CTD that is flexibly tethered to the helicase first binds the 5′ppp-end of bpRNA and then guides the helix to its final high-affinity binding site by interacting with Hel1, Hel2, and Hel2i, such that RIG-I completely surrounds the RNA and encloses it within an elaborate network of interactions (Figs. 3, 4). This model also offers a plausible explanation of why activation requires that the nucleotide bearing the 5′ppp be base-paired (Schlee et al. 2009; Marq et al. 2010). Cocrystal structures of the CTD bound to blunt-end 5′pppbpRNA show that the CTD tightly binds the 5′ppp, stacks on the first base pair, and interacts as well with the adjoining phosphodiester backbone of the 5′ strand (Fig. 3B,C), such that the orientation of the bpRNA helix has only very limited flexibility relative to the CTD. This preformed configuration of CTD with bound 5′pppbpRNA is directly compatible, subject to minor adjustments (Luo et al. 2011; Jiang and Chen 2012), with the final high-affinity RIG-I RNA complex. However, if the nucleotide bearing the 5′ppp is not base-paired, the binding to the CTD is not only weaker due to the altered interaction with the first base pair, but also the orientation of the bpRNA helix is no longer constrained because there is free rotation on both sides of the terminal phosphodiester linkage (Fig. 5E). Thus optimal interaction of the bpRNA with the helicase is severely impaired. In support of this argument, dsRNAs with an unpaired 5′ppp-nucleotide very poorly stimulate the RIG-I ATPase activity relative to blunt-end 5′pppbpRNA (Marq et al. 2010).
The strict requirement for blunt-end 5′pppbpRNA in RIG-I activation may in part explain why arenaviruses such as Tacaribe virus initiate genome RNA synthesis by an unusual mechanism (dubbed “prime and realign”), which results in an unpaired (overhanging) 5′pppG when the complementary genome ends anneal to form dsRNA panhandles (Garcin and Kolakofsky 1990, 1992). This panhandle does not induce IFN upon transfection and, moreover, appears to act as a RIG-I decoy, presumably by binding but not activating RIG-I (Marq et al. 2011). Alternative processing strategies are used by other negative-strand RNA viruses to generate genomic RNAs that fail to activate RIG-I. Notably Hantaan and Crimean-Congo hemorrhagic fever viruses (hanta- and nairo-virus genera of the Bunyaviridae, respectively) as well as Borna disease virus (Bornaviridae) process the 5′ ends of their genomes to leave only a 5′ monophosphate that does not activate RIG-I (Habjan et al. 2008; Martin et al. 2011).
RIG-I MULTIMERIZATION AND SIGNALING
Despite initial indications that dimerization or oligomerization of RIG-I would be essential for its activation (Saito et al. 2007) and that unlikely cartoons for the RIG-I dimer have been proposed (Cui et al. 2008; Hou et al. 2011), there is no indication from structural and biophysical studies that RIG-I forms higher-order structures based on protein–protein interactions, nor is dimerization required for RNA binding. Indeed, RNase footprinting of RIG-I/dsRNA complexes showed that the minimum length of RNA protected by RIG-I is 10–15 bp, consistent with both the cocrystal structures and the functional titration and microscopy experiments (Binder et al. 2011). Multiple copies of RIG-I can thus bind to dsRNA depending on its length. Both RIG-I and MDA5 bind cooperatively to dsRNA in the absence of ATP, and visualization of these filaments by atomic force microscopy (AFM) and negative stain electron microscopy, respectively, indicate that these helicases can bind in a tightly packed manner over long stretches of dsRNAs (Binder et al. 2011; Peisley et al. 2011). Oligomerization of RIG-I was critically dependent on the presence of 5′ppp blunt ends for short (40 bp) dsRNAs. For dsRNAs longer than 200 bp (leaving aside the origin of such RNAs in infected cells), however, the multiple possibilities for internal initiation of binding could apparently compensate effectively for the absence of 5′-terminal triphosphates. However, it should be stressed that internal binding of RIG-I to dsRNA almost certainly occurs with much lower affinity than blunt-end binding. This is because, for internal binding, only the helicase can be involved; the CTD, for steric reasons, cannot bind. Evidence for this is the significantly lower affinity of the helicase domain alone for dsRNA compared to full-length RIG-I (Jiang et al. 2011; Vela et al. 2012), and that 2:1 complexes of RIG-I on 60-mer dsRNA have been detected, most likely with one RIG-I capping each end (Kowalinski et al. 2011). Nevertheless, internal binding may be sufficient to displace the CARDS and also allow translocation of RIG-I along dsRNA.
Although there is now clear evidence that RIG-I itself does not oligomerize, it can appear to do so when several RIG-I molecules are bound to the same RNA (see previous paragraph and Fig. 1C,D). Perhaps more interesting from a functional point of view is that isolated RIG-I and MDA5 CARD domains form heterotetramers upon binding of unanchored K63-polyubiquitin chains, with a stoichiometry of 4:4 (Jiang et al. 2012). Furthermore, this ubiquitin-chain-mediated multimerization of RIG-I appears to be of functional importance. Mutation of conserved residues of RIG-I CARDs that disrupt their ubiquitin binding also abrogated their ability to activate IRF3 in an in vitro system (Jiang et al. 2012). Full-length RIG-I was also shown to form tetrameric complexes upon binding K63-polyubiquitin, but only after having bound dsRNA (Jiang et al. 2012). It is not clear whether the RIG-I molecules forming these complexes would be bound to the same RNA or different RNAs (Fig. 1D). In either case, polyubiquitin-mediated multimerization of RNA-bound RIG-I and MDA5 has now emerged as a key requirement for functional interaction with MAVS. Indeed, as for other receptors in the immune system that cluster for effective signaling, clustering of activated RIG-I and MDA5 may aid the aggregation of MAVS, which was recently shown to form prion-like fibrillar structures during antiviral signaling (Hou et al. 2011).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.035949.112.
REFERENCES
- Bamming D, Horvath CM 2009. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J Biol Chem 284: 9700–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, Garcia-Sastre A 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci 107: 16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder M, Eberle F, Seitz S, Mucke N, Huber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R 2011. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid inducible gene-I (RIG-I). J Biol Chem 286: 27278–27287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie AG, Unterholzner L 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 8: 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanier C, Zemirli N, Portier A, Garcin D, Bidere N, Vazquez A, Arnoult D 2012. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol 10: 44 doi: 10.1186/1741-7007-10-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazenave C, Uhlenbeck OC 1994. RNA template-directed RNA synthesis by T7 RNA polymerase. Proc Natl Acad Sci 91: 6972–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Bennett M, Moldt M, Deimling T, Witte G, Schiesser S, Carell T, Hopfner KP 2011. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep 12: 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell 29: 169–179 [DOI] [PubMed] [Google Scholar]
- Dupeux F, Rower M, Seroul G, Blot D, Marquez JA 2011. A thermal stability assay can help to estimate the crystallization likelihood of biological samples. Acta Crystallogr D Biol Crystallogr 67: 915–919 [DOI] [PubMed] [Google Scholar]
- Du¨rr H, Ko¨rner C, Mu¨ller M, Hickmann V, Hopfner KP 2005. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121: 363–373 [DOI] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E 2010. SF1 and SF2 helicases: Family matters. Curr Opin Struct Biol 20: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446: 916–920 [DOI] [PubMed] [Google Scholar]
- Garcin D, Kolakofsky D 1990. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J Virol 64: 6196–6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D, Kolakofsky D 1992. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J Virol 66: 1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Muhlberger E, et al. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE 3: e2032 doi: 10.1371/journal.pone.0002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997 [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146: 448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang QX, Chen ZJ 2012. Structural insights into the activation of RIG-I, a nanosensor for viral RNAs. EMBO Rep 13: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M Jr, Patel SS, Marcotrigiano J 2011. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479: 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ 2012. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36: 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama M, Takahasi K, Narita R, Hirai R, Yoneyama M, Kato H, Fujita T 2011. 55 amino acid linker between helicase and carboxyl terminal domains of RIG-I functions as a critical repression domain and determines inter-domain conformation. Biochem Biophys Res Commun 415: 75–81 [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105 [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205: 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6: 981–988 [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S 2011. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147: 423–435 [DOI] [PubMed] [Google Scholar]
- Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ 2012. The molecular architecture of human Dicer. Nat Struct Mol Biol 19: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HM, Loo YM, Horner SM, Zornetzer GA, Katze MG, Gale M Jr 2012. The mitochondrial targeting chaperone 14-3-3ɛ regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe 11: 528–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M Jr 2011. Immune signaling by RIG-I-like receptors. Immunity 34: 680–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P 2010. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure 18: 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM 2011. Structural insights into RNA recognition by RIG-I. Cell 147: 409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Kohlway A, Vela A, Pyle AM 2012. Visualizing the determinants of viral RNA recognition by innate immune sensor RIG-I. Structure 20: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marq JB, Kolakofsky D, Garcin D 2010. Unpaired 5′ ppp-nucleotides, as found in arenavirus double-stranded RNA panhandles, are not recognized by RIG-I. J Biol Chem 285: 18208–18216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marq JB, Hausmann S, Veillard N, Kolakofsky D, Garcin D 2011. Short double-stranded RNAs with an overhanging 5′ ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. J Biol Chem 286: 6108–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Hoefs N, Tadewaldt J, Staeheli P, Schneider U 2011. Genomic RNAs of Borna disease virus are elongated on internal template motifs after realignment of the 3′ termini. Proc Natl Acad Sci 108: 7206–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K 2011. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472: 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, Johnson M, Langevin F, Pace P, Patel KJ 2005. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol 12: 763–771 [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T 2009. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323: 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K 2005. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure 13: 143–153 [DOI] [PubMed] [Google Scholar]
- Onoguchi K, Onomoto K, Takamatsu S, Jogi M, Takemura A, Morimoto S, Julkunen I, Namiki H, Yoneyama M, Fujita T 2010. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog 6: e1001012 doi: 10.1371/journal.ppat.1001012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S 2011. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci 108: 21010–21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314: 997–1001 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, et al. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140: 397–408 [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M Jr 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci 104: 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci 107: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 2009. 5′-Triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci 106: 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S 2006. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125: 287–300 [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Strahle L, Marq JB, Brini A, Hausmann S, Kolakofsky D, Garcin D 2007. Activation of the β interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J Virol 81: 12227–12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela A, Fedorova O, Ding SC, Pyle AM 2012. The thermodynamic basis for viral RNA detection by the RIG-I innate immune sensor. J Biol Chem doi: 10.1074/jbc.M112.385146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. 2010. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol 17: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB 2005. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M Jr, Akira S, et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175: 2851–2858 [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141: 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang C, Schook LB, Hawken RJ, Rutherford MS 2000. An RNA helicase, RHIV -1, induced by porcine reproductive and respiratory syndrome virus (PRRSV) is mapped on porcine chromosome 10q13. Microb Pathog 28: 267–278 [DOI] [PubMed] [Google Scholar]
- Zou J, Chang M, Nie P, Secombes CJ 2009. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol 9: 85 doi: 10.1186/1471-2148-9-85 [DOI] [PMC free article] [PubMed] [Google Scholar]