This paper describes the results of a genome-wide RNAi screen designed to identify novel factors required for Drosophila snRNA 3′-end formation. The RNAi screen identified two additional core members of the fly Integrator complex as well as a previously unknown function for cyclin C/cdk8 in the processing of Drosophila small nuclear RNA.
Keywords: Integrator complex, snRNA, RNAPII, cyclin C, Cdk8, Asunder
Abstract
Formation of the 3′ end of RNA polymerase II–transcribed snRNAs requires a poorly understood group of proteins called the Integrator complex. Here we used a fluorescence-based read-through reporter that expresses GFP in response to snRNA misprocessing and performed a genome-wide RNAi screen in Drosophila S2 cells to identify novel factors required for snRNA 3′-end formation. In addition to the known Integrator complex members, we identified Asunder and CG4785 as additional Integrator subunits. Functional and biochemical experiments revealed that Asunder and CG4785 are additional core members of the Integrator complex. We also identified a conserved requirement in both fly and human snRNA 3′-end processing for cyclin C and Cdk8 that is distinct from their function in the Mediator Cdk8 module. Moreover, we observed biochemical association between Integrator proteins and cyclin C/Cdk8, and that overexpression of a kinase-dead Cdk8 causes snRNA misprocessing. These data functionally define the Drosophila Integrator complex and demonstrate an additional function for cyclin C/Cdk8 unrelated to its function in Mediator.
INTRODUCTION
The biosynthesis of snRNAs and their subsequent incorporation into functional snRNPs are critical for the expression of the majority of RNA polymerase II (RNAPII) transcripts. These snRNAs include the abundant RNAPII–transcribed snRNAs important for the function of the major and minor spliceosomes as well as the less abundant U7 snRNA involved in histone mRNA 3′-end formation (for review, see Matera et al. 2007; Marzluff et al. 2008). For most snRNAs, only a single processing step takes place in the nucleus; the 3′ end is formed by endonucleolytic cleavage of the nascent transcript from the elongating polymerase to allow release and subsequent export to the cytoplasm (for review, see Egloff et al. 2008).
Work from several laboratories established that snRNA 3′-end processing requires three features: a 3′-box element located downstream from the cleavage site, an snRNA promoter, and the C-terminal domain (CTD) of Rpb1 (de Vegvar et al. 1986; Hernandez and Weiner 1986; Ach and Weiner 1987; Medlin et al. 2003; Egloff et al. 2007, 2010). Although the 3′ box is highly tolerant of mutation (Hernandez 1985; Ach and Weiner 1987; Ezzeddine et al. 2011), it is likely to be a binding site in the RNA for the machinery that governs snRNA 3′-end formation. Studies have also shown that substituting either human or Drosophila snRNA promoters with mRNA promoters results in total loss of snRNA 3′-end formation (de Vegvar et al. 1986; Hernandez and Weiner 1986; Ezzeddine et al. 2011). In addition, the 3′-end formation of viral-encoded snRNA-like transcripts found in Herpesvirus saimiri also exhibits a dependency on an snRNA promoter (Cazalla et al. 2011). This species conservation and viral mimicry strongly suggests that the 3′-end processing machinery is loaded onto RNAPII early in the transcription cycle. Finally, the CTD of Rpb1 must be phosphorylated at serines 2 and 7, but not serine 5 of its heptad repeats to facilitate snRNA 3′-end formation (Egloff et al. 2007).
New insight to these snRNA processing events has come from the biochemical purification of the Integrator complex and establishment of its functional requirement in snRNA 3′-end formation (Baillat et al. 2005; Ezzeddine et al. 2011). Integrator subunit 9 (IntS9) and IntS11 (Albrecht and Wagner 2012) are homologous to CPSF100 and CPSF73, which are components of the cleavage factor for mRNA 3′-end formation (Mandel et al. 2006; Sullivan et al. 2009). Aside from these two subunits, the other 10 founding members of the complex display little similarity with the proteins involved in either poly(A) mRNA or histone mRNA 3′-end formation. Since the initial isolation of the core 12-member Integrator complex, subsequent high-throughput immunopurifications from mammalian cells have identified several additional factors that interact with core Integrator subunits (Malovannaya et al. 2010, 2011). This “extended Integrator complex” has been proposed to contain additional RNAPII subunits, a group of phosphatases, zinc-finger proteins, OB-fold nucleic acid binding proteins, and many others either uncharacterized and/or not previously implicated in snRNA biogenesis. Functional validation of these additional proteins has yet to be performed, making their importance to snRNA 3′-end formation unclear.
Here we describe a genome-wide RNAi screen in Drosophila S2 cells to identify genes required for snRNA 3′-end formation using our U7-GFP read-through reporter gene assay (Ezzeddine et al. 2011). In addition to confirming the functional requirement for known Integrator subunits, we identified four new factors necessary for snRNA 3′-end formation. These proteins—CG4785, Asunder (Asu), cyclin C (CycC), and Cdk8—have not previously been known to function in snRNA biogenesis. Both Asunder and CG4785 immunoprecipitates contained high amounts of endogenous Integrator subunits, suggesting that these two proteins are additional core members of the Integrator complex. In addition, we show that cyclin C/Cdk8 are specifically required for efficient snRNA 3′-end formation in both flies and humans, and also show that overexpression of a kinase-deficient Cdk8 causes snRNA misprocessing, directly implicating Cdk8 kinase activity in 3′-end processing. This function of cyclin C/Cdk8 is distinct from its function in the Mediator complex since depletion of Med12 and Med13, two members of the Mediator Cdk8 module, did not affect snRNA processing.
RESULTS AND DISCUSSION
Genome-wide RNAi screen for Drosophila snRNA 3′-end processing factors
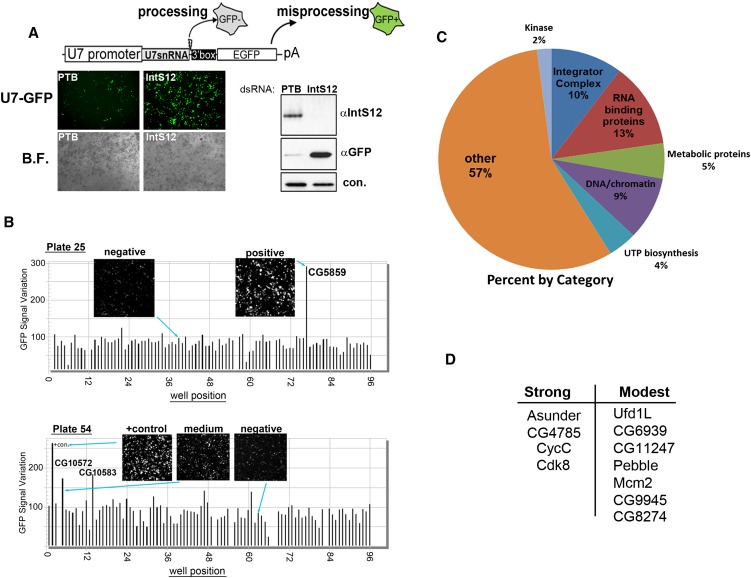
We previously developed a cell-based reporter that expresses green fluorescent protein (GFP) in response to misprocessing of U7 snRNA, allowing for non-invasive and sensitive detection of misprocessing in vivo (Fig. 1A; Ezzeddine et al. 2011). To determine whether this reporter would be suitable for use in a genome-wide RNAi screen, we treated S2 cells with dsRNA targeting either the Polypyrimidine Tract Binding Protein (PTB) (negative control) or IntS12 (positive control), and in both instances found no overt effects on growth. Three days following the dsRNA treatment, cells were transfected with the U7-GFP reporter, and fluorescence was assessed 48 h later. Robust GFP expression was clearly observed after IntS12 depletion, relative to a low level of background fluorescence in cells treated with PTB dsRNA (Fig. 1A). Western blot analysis confirmed that only after significant reduction in IntS12 expression was GFP expression observed (Fig. 1A).
FIGURE 1.
Using the U7-GFP reporter to conduct a genome-wide RNAi screen. (A) Schematic of the U7-GFP reporter and the results from transient transfection of the reporter into S2 cells treated with dsRNA targeting PTB (−control) or IntS12 (+control). (Left) Fluorescence and bright-field images; (right) Western blot analysis. (B) Bar graph representing quantification of screen results for plates 25 and 54. Insets are taken from the acquired image collection. (C) Pie graph representing results of RNAi screen categorically. (D) Trimmed-down list of non-Integrator proteins that reproducibly scored as strong or modest by secondary screening.
Using the U7-GFP reporter, we performed a genome-wide RNAi screen in Drosophila S2 cells using a library consisting of 15,881 unique dsRNAs targeting >90% of the annotated fly genome. U7-GFP reporter expression in cells grown in 96-well plates was imaged via automated high-throughput microscopy and plotted as a function of fluorescence variance. We observed that in ∼7% of the wells, the dsRNA treatment resulted in cell death, resulting in no viable cells at the time of image acquisition. The imaging of nearly all plates of the dsRNA screen revealed a relatively homogeneous level of background fluorescence in 98%–99% of the wells. Representative results of two plates are shown in Figure 1B. A small number of wells displayed strong GFP fluorescence; for example, on plate 25, cells treated with dsRNA targeting CG5859 identified the fly ortholog of IntS8 as a “strong hit.” In addition, a modest number of moderately fluorescent wells were also identified, as evidenced by the GFP signal and cell images presented for plate 54 (Fig. 1B). The levels of GFP expression after knockdown of either CG10572 or CG10583 were clearly above plate background yet were below the signal of the positive control dsRNA (targeting Drosophila IntS9).
In total, our RNAi screen identified 89 genes that, when subjected to RNAi knockdown, resulted in detectable levels of GFP expression from the U7-GFP reporter (Fig. 1C; Supplemental Fig. S1). The screen identified 10 of the 12 annotated Drosophila Integrator subunits, as well as genes involved in RNA binding, chromatin/DNA function, and UTP biosynthesis. Both IntS3 and IntS10 did not score, which is consistent with our previous observations that depletion of these two subunits did not result in appreciable GFP expression (Ezzeddine et al. 2011). To determine the reproducibility of the data, we generated the same dsRNAs used in the initial screen corresponding to each of the 89 target genes and performed sequential RNAi hits followed by transfection of the U7-GFP reporter. From this, the initial list of 89 genes was reduced down to 21 that either scored as high or modest with regard to their GFP expression (Fig. 1D). Importantly, 11 of these were genes not previously known to be involved in snRNA biogenesis. Because depletion of four of these proteins (CG4785, Asu, cyclin C, and Cdk8) produced high levels of GFP expression comparable to that observed after depletion of known Integrator subunits, we focused the remainder of our analysis on these genes.
Asunder and CG4785 are specifically required for snRNA 3′-end formation
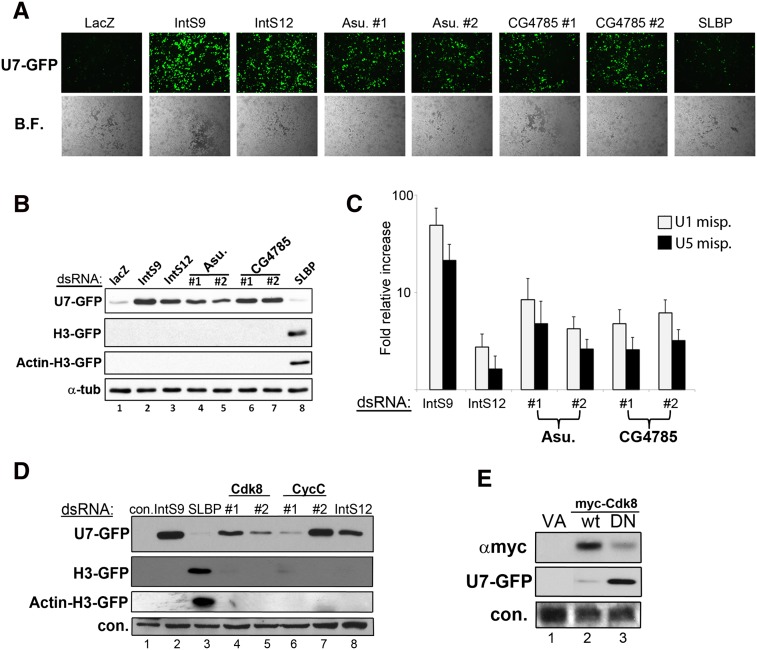
To confirm that the observed snRNA misprocessing occurs as a consequence of knockdown of CG4785 and Asu, we designed a second set of dsRNA targeting a distinct region of each open reading frame. S2 cells were then treated with dsRNA targeting LacZ or the stem–loop binding protein (SLBP) as negative controls, IntS9 and IntS12 as positive controls, and either dsRNA targeting Asu or CG4785. We observed only background levels of GFP expression in negative control dsRNA-treated cells, while both dsRNAs targeting Asu and CG4785 gave rise to robust and nearly identical levels of GFP expression comparable to those observed after depletion of IntS9 or IntS12 (Fig. 2A,B). To determine the specificity of the misprocessing phenotype, we used two related GFP reporters that we developed previously to determine defects in histone pre-mRNA processing (Wagner et al. 2007; Yang et al. 2009; Ezzeddine et al. 2011). The H3-GFP report is almost identical to the U7-GFP construct, except the histone H3 gene is used. In addition, the Act-H3-GFP reporter replaces the H3 promoter with the actin promoter, which was found to sensitize it to additional factors whose depletion normally extinguishes the histone promoter (Yang et al. 2009). We treated cells with the same series of positive control, negative control, and test dsRNA as described above followed by the transfection with either H3-GFP or Act-H3-GFP reporter. Using these other two constructs, we only observed GFP expression after depletion of SLBP, confirming that both Asu and CG4785 are specifically required for snRNA 3′-end processing.
FIGURE 2.
Validation of Asunder, CG4785, CycC, and Cdk8 as required for snRNA 3′-end formation. (A) Images from S2 cells transfected with the U7-GFP reporter after dsRNA treatment targeting Asu or CG4785. (B) Western blot analysis of cell lysates from A. (C) qRT-PCR analysis specific for misprocessed endogenous snRNA from knockdown cells. (D) Western blot analysis of S2 cells treated with dsRNAs targeting CycC or Cdk8 followed by transient transfection with reporters measuring snRNA or histone mRNA 3′-end formation. (E) Western blot analysis of cells cotransfected with U7-GFP reporter and myc-tagged Cdk8 that is wild type or catalytically inactive.
To characterize the role of Asunder and CG4785 in the processing of endogenous snRNA, we used a sensitive qRT-PCR assay designed to measure misprocessed U1 and U5 snRNAs following RNAi depletion. Total cell RNA was isolated following several days of incubation with dsRNA targeting LacZ (negative control), IntS9 or IntS12 (positive controls), or dsRNAs targeting Asu and CG4785 (two separate dsRNAs each gene). We subjected total RNA to reverse transcription followed by real-time PCR analysis using primers specific for the misprocessed U1 and U5 snRNAs (Ezzeddine et al. 2011). Relative to control treated cells, we observed a fivefold to 10-fold increase in the level of misprocessed snRNA (Fig. 2C). The levels of misprocessed snRNAs were consistent between the two individual dsRNAs for each target and comparable to that observed after depletion of IntS12.
Cyclin C and Cdk8 are required for snRNA 3′-end formation
The observation that both Cdk8 and CycC were independently isolated in our screen strongly suggests that they are involved in snRNA 3′-end formation, because they form a cognate cyclin/cdk pair (Leclerc et al. 1996). To address the possibility of any potential off-target effects and characterize their specificity toward snRNA 3′-end formation, we created a second set of dsRNAs targeting distinct regions of each ORF. Treatment of S2 cells with dsRNAs targeting Cdk8 or targeting CycC resulted in robust expression of GFP from the U7-GFP reporter (Fig. 2D). We also tested the effect of CycC or Cdk8 depletion on both the histone H3-GFP and Actin-H3-GFP reporters. Neither the H3 nor Actin H3 reporters were found to produce GFP in response to Cdk8 or CycC knockdown. In contrast, we observed robust expression of GFP from both reporters in response to treating cells with dsRNAs targeting SLBP. These data implicate that CycC and Cdk8 are specifically involved in the process of snRNA 3′-end formation.
To confirm that the kinase activity of Cdk8 is required for snRNA 3′-end formation, we cloned wild-type Drosophila Cdk8 and generated a kinase-dead (D173A) form analogous to a mutant human CDK8 shown previously to behave as a dominant negative (DN) (Akoulitchev et al. 2000). Both wild-type and DN cDNAs were expressed as N-terminal myc-tagged proteins in S2 cells cotransfected with the U7-GFP reporter. Despite the reduced accumulation of the catalytically inactive Cdk8 relative to the wild-type protein, we observed potent misprocessing of the U7-GFP reporter (Fig. 2E). These data demonstrate that the kinase activity of the CycC/Cdk8 complex is essential for correct snRNA 3′-end processing.
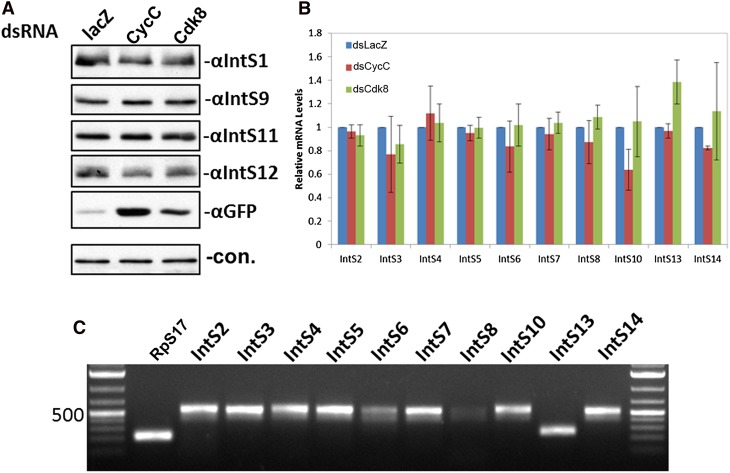
One possible explanation for the observed phenotype is that CycC/Cdk8 fulfills a general requirement in the expression of one or more Integrator subunits, and therefore CycC/Cdk8 plays an indirect role in snRNA 3′-end formation. To test this, we analyzed the expression of all 12 Integrator subunits as well as Asunder and CG4785 after depletion of CycC or Cdk8 using both Western blot analysis (Fig. 3A) and qRT-PCR (Fig. 3B,C). We found no observable differences in the levels of Integrator subunit expression following CycC or Cdk8 knockdown. Therefore, we conclude that these two proteins are likely functioning directly in the 3′-end formation of Drosophila snRNA.
FIGURE 3.
Cyclin C and Cdk8 are not involved in regulating Integrator expression. (A) Western blot analysis of protein levels of Integrator subunits after treating cells with dsRNAs targeting LacZ, CycC, or Cdk8. Cells were also transfected with the U7-GFP reporter to monitor snRNA misprocessing as a functional readout of knockdown of CycC/Cdk8. (B) Quantitative RT-PCR analysis using SYBR Green staining of PCR products. RNA was isolated from S2 cells, treated with the three dsRNAs described in A, and subjected to real-time PCR analysis using amplicons specific to Integrator subunits not tested by Western blot analysis in A. All measurements were normalized to RpS17 signals as a housekeeping internal control. (C) Ethidium bromide staining of agarose gel electrophoresis of the amplicons amplified in B.
Asunder and CG4785 associate with Integrator subunits
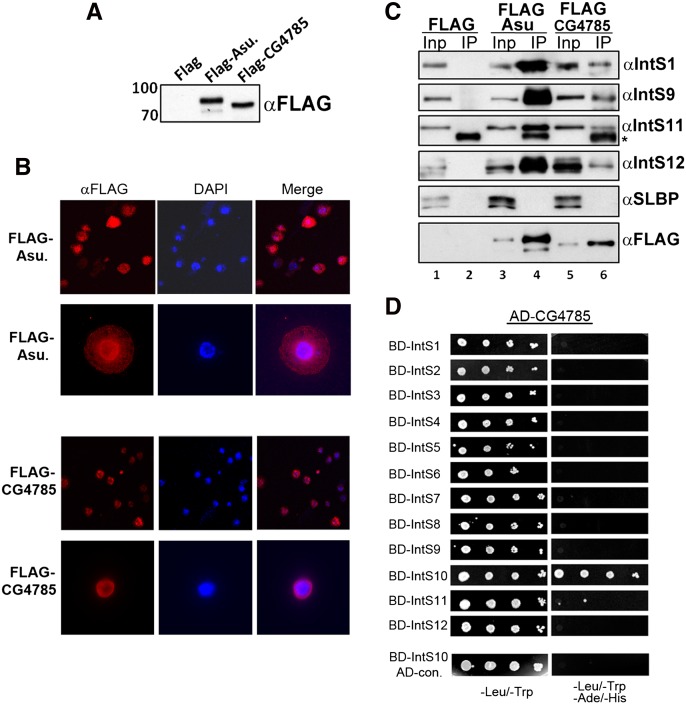
To determine if either CG4785 or Asunder biochemically associate with the other Integrator subunits, we cloned full-length cDNAs and generated stable S2 cell lines expressing either Flag-tagged Asunder or Flag-tagged CG4785. Western blot analysis of lysates from these cell lines using anti-Flag antibodies demonstrates the expression of specific bands of the predicted molecular weight for both proteins (Fig. 4A). We analyzed the subcellular localization of both of the tagged proteins and observed a nuclear localization of CG4785 and a bimodal localization of Asu in both the nucleus and the cytoplasm (Fig. 4B). Nuclear extracts were prepared from both stable cell lines from which epitope-tagged proteins were immunoprecipitated using anti-Flag agarose. A large majority of Flag-tagged protein was recovered in each precipitate from both cell lines, and we determined the levels of interacting endogenous Integrator subunits using antibodies to Drosophila IntS1, IntS9, IntS11, and IntS12 proteins (Fig. 4C). A large proportion of all four Integrator subunits were detected specifically associating with Asu and CG4785, relative to control pull-downs (Fig. 4C). We also did not detect SLBP in either immunoprecipitate, consistent with the observation that depletion of these two proteins does not affect histone mRNA 3′-end formation. To confirm potential direct interactions with Drosophila Integrator subunits, we performed binary interaction tests using pairwise, directed yeast two-hybrid analysis. This is an ideal assay to perform this analysis given the lack of endogenous yeast Integrator subunits. Under low and high stringency conditions, we did not observe any interaction between Asu and the known Integrator subunits (data not shown). In contrast, we did observe a robust interaction between CG4785 and IntS10 by Y2H (Fig. 4D). Collectively, these data demonstrate that, in addition to their functional requirement in snRNA 3′-end formation, both Asu and CG4785 biochemically associate with the previously described members of the Integrator complex (Baillat et al. 2005). While the interaction of CG4785 is most likely mediated through IntS10, the binding partner(s) of Asunder remain to be identified.
FIGURE 4.
Asunder and CG4785 biochemically associate with the fly Integrator complex. (A) Western blot analysis of Flag-tagged Asunder or CG4785 expression in stable S2 cell lines. (B) IF analysis of stable S2 cell lines expressing Flag-tagged Asunder or CG4785. (C) Western blot analysis of Flag immunoprecipitates from cell lines described in A. Input lanes represent 5% of input, and IP represents 50% of the immunoprecipitate. (D) Directed yeast two-hybrid using fly Integrator subunits and CG4785 demonstrating an interaction with IntS10.
Asu was initially given the name Mat89b, due to its maternal deposition into the Drosophila embryo (Stebbings et al. 1998). Subsequently, it was found to be a PanGu kinase substrate, which, when its expression is reduced in the spermatocyte, causes aberrant centrosome and spindle localization (Anderson et al. 2009). Asu has a predicted molecular weight of 76 kDa with no readily identifiable domains. The CG4785 gene encodes a 65-kDa protein containing a weakly defined N-terminal von Willebrand Associated (VWA)–like domain, similar to that observed in IntS6. Large-scale mass spectrometry of the human Integrator complex found evidence for association of the human orthologs of Asu and CG4785 in that complex, suggesting that these two proteins may play a conserved role in snRNA biogenesis (Malovannaya et al. 2010, 2011). On the basis of these observations, particularly the high efficiency of the co-IP of other Integrator subunits with both Asu and CG4585, we suggest that Asu be known as “Asun/IntS13” and CG4785 as “IntS14.” While we have not determined if Asun/IntS13 and IntS14 interact with each other, they have also been identified in other RNAi screens for factors required for the biosynthesis of Drosophila small RNAs (Zhou et al. 2008). Those screens distinctly did not identify any other Integrator subunits, suggesting that Asun/IntS13 and IntS14 may function as a module used by distinct RNA processing machinery analogous to how the CPFS73/100 heterodimer is used by the cleavage and polyadenylation and histone pre-mRNA processing factors (Sullivan et al. 2009).
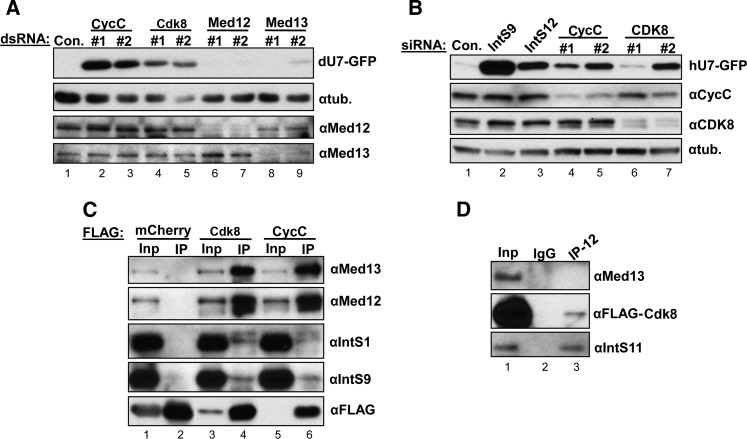
Requirement for cyclin C and Cdk8 in snRNA 3′-end formation is conserved and independent of its function in Mediator
There has been extensive previous characterization of CycC and CDK8 as members of the Mediator CDK8 module in conjunction with Mediator 12 and 13 (for review, see Galbraith et al. 2010; Conaway and Conaway 2011). Knockdown of these two Mediator subunits did not produce GFP expression in our screen, suggesting that either they are not required for snRNA processing or were not depleted effectively. To address the latter possibility, we created independent dsRNAs specific to Med12 and Med13 and observed that they elicited significant reduction in the levels of endogenous Med12 and Med13, yet failed to trigger misprocessing in the U7-GFP reporter (Fig. 5A). These results imply that CycC and Cdk8 are required for snRNA 3′-end formation independent of Med12 and Med13. To determine if CycC or CDK8 plays a conserved role in the 3′-end formation of human snRNA, we treated HeLa cells with two siRNAs targeting both transcripts and then transfected the human version of the U7-GFP reporter that we showed previously to express GFP in response to Integrator activity disruption (Albrecht and Wagner 2012). We observed that both siRNAs resulted in efficient reduction of the levels of endogenous CycC, and siRNA #2 resulted in significant depletion of CDK8 and gave rise to GFP expression comparable to those after depletion of IntS12 (Fig. 5B). We noted that CDK8 siRNA #1 did not generate as much GFP expression as siRNA #2, but the knockdown was not as effective and also not as effective as codepleting CycC. These results demonstrate the requirement for CycC and CDK8 in snRNA 3′-end formation in metazoans.
FIGURE 5.
CycC/Cdk8 function in snRNA 3′-end formation independent of Mediators 12/13 and are associated with Integrator subunits. (A) Western blot analysis of lysates from S2 cells treated with dsRNAs targeting members of the Drosophila Mediator Cdk8 module and transfected with the U7-GFP reporter. (B) Western blot analysis of lysates from HeLa cells treated with siRNA to CycC or CDK8 followed by transfection with a human version of the U7-GFP reporter. (C) Immunoprecipitations using αFlag-agarose to detect interactions between CycC/Cdk8 and Integrators 1 and 9. Input lanes represent 2% of input, and IP lanes represent 50% of immunoprecipitate. (D) Western blot analysis of immunoprecipitations using anti-IntS12 antibodies.
These results are consistent with what has been detected genetically for the Drosophila Mediator Cdk8 module, where sex comb phenotypes for CycC− and Cdk8− clones are indistinguishable from each other but clearly distinct from the phenotypes of Med12− and Med13− clones (Loncle et al. 2007). Taken together, these observations support a model predicting that Drosophila CycC and Cdk8 function in an additional pathway—snRNA 3′-end formation, not related to their association with Mediator.
The functional requirement for CycC/Cdk8 in fly snRNA 3′-end formation generates the question of whether these two proteins are associated with Integrator subunits. To test this possibility, we generated stable S2 cell lines expressing Flag-mcherry (negative control), Flag-Cdk8, or Flag-CycC and performed immunoprecipitations from nuclear extracts prepared from these cell lines. We observed substantial amounts of endogenous Med12 and Med13 associating with both Flag-CycC and Flag-Cdk8 as predicted, yet we also observed a reproducible interaction with Integrator subunits (Fig. 5C). The reduced levels of Integrator proteins in the co-IP suggest that the interaction is relatively weak or transient and that only a small pool of CycC/Cdk8 not associated with Med12 and Med13 binds to the Integrator subunits. To test these predictions, we used IntS12 antibodies to immunoprecipitate the Integrator complex from nuclear extract derived from S2 cells stably expressing Flag-Cdk8. We observed significant amount of IntS11 and a portion of Flag-Cdk8 associating with IntS12 (Fig. 5D). Importantly, we did not detect Med13 interacting with IntS12. This reciprocal pull-down confirms the presence of a small amount of Cdk8 associating with the fly Integrator complex in the absence of Mediator.
Our data suggest two potential functions for cyclin C/Cdk8 in snRNA 3′-end formation: phosphorylation of the RNAPII CTD, or as an Integrator subunit kinase. There have been no prior reports implicating Cdk8 as a Ser7 kinase, but it has been shown to phosphorylate the CTD of Rpb1 in vitro (Rickert et al. 1999). Alternatively, there could be an “Integrator Cdk8 module” regulating an Integrator subunit through direct phosphorylation in an analogous role to that in the Mediator complex, where CycC/CDK8 phosphorylates Mediator 13 (Knuesel et al. 2009). Indeed, phosphorylation of Integrator subunits may be an obligatory event similar to that observed for factors involved in mRNA 3′-end formation (Ryan 2007).
CONCLUSIONS
The genome-wide RNAi screen presented has redefined the Drosophila Integrator complex to comprise 14 core subunits. We argue that this core number is likely to be conserved in humans because the orthologs of Asun/IntS13 and IntS14 were present in the human Integrator immunoprecipitations at levels comparable to the known 12 members (Malovannaya et al. 2010). The conserved requirement of CycC/Cdk8 in snRNA 3′-end formation indicates that not only is the core complex conserved across metazoan species, but also the regulation of its activity is likely retained. The purpose of this regulation is not yet clear at present but may derive from a need to modulate Integrator activity to coincide with differential snRNA requirements.
An important consideration of the U7-GFP reporter is that in order for a knockdown to be scored as a positive “hit,” it must compromise snRNA processing but retain snRNA transcription. In this case, any factor that is important for both transcription and 3′-end processing will not be scored. This likely explains why depletion of members of the snRNA activating protein complex (SNAPc) do not trigger GFP expression, nor did the members of the recently defined Little Elongation Complex (Hung and Stumph 2011; Smith et al. 2011). Depletion of any of these factors would be predicted to extinguish U7-GFP promoter activity and would not result in GFP expression. Interestingly, we also did not score the fly ortholog of RPAP2 (CG34183), which has been suggested to play an important role in mediating Integrator's association to the RNAPII CTD (Egloff et al. 2012). Depleting fly RPAP2 did not give rise to snRNA misprocessing in our hands, suggesting that RPAP2 may fall into the group of factors we mentioned above or may not be critical in Drosophila. This latter would be consistent with observations that the fly CTD has a markedly distinct structure with very few serines at the seventh position within its heptad repeat, and the fly RPAP2 is significantly smaller than its human counterpart, possessing only the small zinc-finger domain recently found in the Kluyveromyces lactis RPAP2 crystal structure (Xiang et al. 2012).
A remarkable property of the Integrator complex is a network of essential interactions that make it intolerant to perturbation. Depletion of even a single subunit, in most cases results in snRNA misprocessing. Understanding the nature of these interactions and the biochemical functions of the individual subunits represents the most compelling questions in the field.
MATERIALS AND METHODS
Genome-wide RNAi screen
The dsRNA library was purchased from Open Biosystems (Thermo Scientific) as 5 μg per well resuspended in 100 μL of water. The dsRNA was realiquoted into 167 96-well plates (Greiner Bio-One) at 1 μg per well and allowed to dry down. Then 5 × 104 cells were plated into each well and allowed to incubate for 3 d. On the fourth day, the 30 ng of U7-GFP reporter was transfected into each well using Effectene according to the manufacturer's instructions (QIAGEN). Two days following reporter transfection, cells were imaged live using a Cellomics Array Scan V high-throughput automated microscope (Thermo Scientific). Exposure times were calculated on a plate-to-plate basis, and fluorescence levels were calculated using pre-written algorithms incorporated into the manufacturer's software. The two algorithms used in the data presentation are the standard deviation in fluorescence intensity and the mean differential intensity because these two methods were most effective in blindly identifying positive controls in pilot assays. Raw data for the screen are accessible at the Wagner laboratory homepage (http://www-bmb.med.uth.tmc.edu/faculty/Wagner/documents/Chen%20et%20al%20RNA-raw%20data%20from%20RNAi%20screen.pdf).
S2 cell culture
Drosophila S2 cells were cultured as described previously. RNAi/U7-GFP reporter experiments were conducted by plating 5 × 104 cells per 96-well plate in 100 μL of Sf-900 II media (Invitrogen). To each well, 1 μg of dsRNA was added each day for 2 d followed by transfection of 30 ng of U7-GFP reporter. The following day, an additional 1 μg of dsRNA was added, and then cells were visualized by fluorescence microscopy in the sixth day and then harvested for Western blot analysis. For stable cell line generation, Flag-tagged expressing cDNA cloned into the pUB plasmid was cotransfected with a Blastocidin-expressing plasmid, and selection was performed in Drosophila media (Invitrogen) containing 10% FBS and 25 μg/mL blastocidin. Stable cell lines were amplified and nuclear extract was prepared using previously described methods (Dignam et al. 1983).
Immunoprecipitation and Western blot analysis
For immunoprecipitation of Flag-tagged protein, 1 mg of nuclear extract was incubated with 10 μL of anti-Flag antibody-conjugated agarose beads (anti-Flag M2 affinity gel; Sigma-Aldrich) with constant rotation for 2 h at 4°C in buffer D (20 mM HEPES at pH 7.9, 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM PMSF) plus 0.1% Triton X-100. Beads were washed twice with 1× TBS (50 mM Tris-HCl at pH 7.4, 150 mM NaCl) and then twice with buffer D plus 0.1% Triton X-100. During the last wash, switch to a new tube. For immunoprecipitation of endogenous protein, 2 μg of antigen-purified antibody was pre-incubated with 20 μL of Protein A/G beads (Santa Cruz Biotechnology, Inc.) with constant rotation in buffer D for 1 h at 4°C, and then 400 μg of nuclear extracts was added and incubated with rotation for another 2 h at 4°C. The wash condition is the same as mentioned above. Finally, 50 μL of 1× SDS loading buffer was added to the beads, boiled for 3 min at 95°C, and resolved in 12.5% SDS-PAGE gel. Western blot analysis was performed using standard procedure, and the antibodies for IntS1, IntS9, IntS11, IntS12, and SLBP were described previously (Ezzeddine et al. 2011). Antibodies against Med12/kto and Med13/skd were a gift from Jessica Treisman (New York University). The human CycC and CDK8 antibodies were purchased from Bethyl Laboratories, Inc.
Yeast two-hybrid analysis
Full-length coding regions of the 12 Drosophila Integrator subunits were obtained from the Drosophila Genomics Resource Center and cloned in-frame into the pGBKT7 vector (BD). Asu/Ints13 and CG4785/Ints14 were cloned into pGADT7 (AD). Pairwise cotransformations of AD and BD constructs into yeast strain AH109 were according to the manufacturer's instructions (Matchmaker 3 System; Clontech). Empty vectors were cotransformed to control for construct autoactivation. Positive interactions were analyzed using nutritional selection by spotting four serial 10-fold dilutions on SD-plates lacking leucine/tryptophan (vector control), or additionally histidine (medium stringency) and histidine/adenine (high stringency). Images were taken after 3 d of incubation at 30°C.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Phillip Carpenter for discussions and reviewing the manuscript; Steve and Greg Rogers for their help with the Cellomics Array Scan; and members of the Marzluff laboratory, in particular, Steve Lyons, Mike Slevin, and Patrick Lackey, for help in the RNAi screen. This work was supported by grants to E.J.W. (K99GM080477 and 5R00GM080447) and W.F.M. (GM58912). Investigations in the Warren laboratory were conducted with the generous support of the Queensland Cancer Fund and the NHMRC through the Australian Drosophila Biomedical Research Support Facility. The Wagner laboratory also acknowledges support from a T.C. Hsu Faculty Development Award.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.035725.112.
REFERENCES
- Ach RA, Weiner AM 1987. The highly conserved U small nuclear RNA 3′-end formation signal is quite tolerant to mutation. Mol Cell Biol 7: 2070–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoulitchev S, Chuikov S, Reinberg D 2000. TFIIH is negatively regulated by cdk8-containing Mediator complexes. Nature 407: 102–106 [DOI] [PubMed] [Google Scholar]
- Albrecht TR, Wagner EJ 2012. snRNA 3′ end formation requires heterodimeric association of integrator subunits. Mol Cell Biol 32: 1112–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MA, Jodoin JN, Lee E, Hales KG, Hays TS, Lee LA 2009. Asunder is a critical regulator of dynein-dynactin localization during Drosophila spermatogenesis. Mol Biol Cell 20: 2709–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123: 265–276 [DOI] [PubMed] [Google Scholar]
- Cazalla D, Xie M, Steitz JA 2011. A primate herpesvirus uses the Integrator complex to generate viral microRNAs. Mol Cell 43: 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW 2011. Function and regulation of the Mediator complex. Curr Opin Genet Dev 21: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vegvar HE, Lund E, Dahlberg JE 1986. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell 47: 259–266 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S 2007. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S 2008. Expression of human snRNA genes from beginning to end. Biochem Soc Trans 36: 590–594 [DOI] [PubMed] [Google Scholar]
- Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S 2010. The Integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem 285: 20564–20569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S 2012. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell 45: 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chen J, Waltenspiel B, Burch B, Albrecht T, Zhuo M, Warren WD, Marzluff WF, Wagner EJ 2011. A subset of Drosophila Integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3′-end formation. Mol Cell Biol 31: 328–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Donner AJ, Espinosa JM 2010. CDK8: A positive regulator of transcription. Transcription 1: 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N 1985. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J 4: 1827–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N, Weiner AM 1986. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell 47: 249–258 [DOI] [PubMed] [Google Scholar]
- Hung KH, Stumph WE 2011. Regulation of snRNA gene expression by the Drosophila melanogaster small nuclear RNA activating protein complex (DmSNAPc). Crit Rev Biochem Mol Biol 46: 11–26 [DOI] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ 2009. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol 29: 650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc V, Tassan JP, O'Farrell PH, Nigg EA, Leopold P 1996. Drosophila Cdk8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II. Mol Biol Cell 7: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, Bourbon HM 2007. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J 26: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, O'Malley BW, Qin J 2010. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci 107: 2431–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. 2011. Analysis of the human endogenous coregulator complexome. Cell 145: 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP 2007. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220 [DOI] [PubMed] [Google Scholar]
- Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S 2003. The C-terminal domain of Pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J 22: 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert P, Corden JL, Lees E 1999. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene 18: 1093–1102 [DOI] [PubMed] [Google Scholar]
- Ryan K 2007. Pre-mRNA 3′ cleavage is reversibly inhibited in vitro by cleavage factor dephosphorylation. RNA Biol 4: 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Lin C, Garrett AS, Thornton J, Mohaghegh N, Hu D, Jackson J, Saraf A, Swanson SK, Seidel C, et al. 2011. The little elongation complex regulates small nuclear RNA transcription. Mol Cell 44: 954–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbings L, Grimes BR, Bownes M 1998. A testis-specifically expressed gene is embedded within a cluster of maternally expressed genes at 89B in Drosophila melanogaster. Dev Genes Evol 208: 523–530 [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34: 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell 28: 692–699 [DOI] [PubMed] [Google Scholar]
- Xiang K, Manley JL, Tong L 2012. The yeast regulator of transcription protein Rtr1 lacks an active site and phosphatase activity. Nat Commun 3: 946 doi: 10.1038/ncomms1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell 36: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ 2008. Comparative analysis of Argonaute-dependent small RNA pathways in Drosophila. Mol Cell 32: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]