RNase III enzymes are fundamental to the biogenesis of microRNAs (miRNAs) and small interfering RNAs (siRNAs) in all species studied. Although alternative miRNA pathways independent of Drosha or Dicer exist, each still requires one RNase III-type enzyme. Here, the authors describe two strategies that marry either RNase Z or the Integrator complex with the slicing activity of Argonaute2 to generate highly functional mature miRNAs. They provide stringent validation of their RNase III independence by demonstrating efficient miRNA biogenesis and activity in Drosha and Dicer knockout cells. These data provide proof-of-principle evidence for additional mechanistic possibilities for efficient generation of small regulatory RNAs, and represent novel silencing triggers that may be exploited for technical purposes.
Keywords: Argonaute, RNase III, Slicer, microRNA
Abstract
RNase III enzymes are fundamental to the biogenesis of microRNAs (miRNAs) and small interfering RNAs (siRNAs) in all species studied. Although alternative miRNA pathways independent of Drosha or Dicer exist, each still requires one RNase III-type enzyme. Here, we describe two strategies that marry either RNase Z or the Integrator complex with the slicing activity of Argonaute2 to generate highly functional mature miRNAs. We provide stringent validation of their RNase III independence by demonstrating efficient miRNA biogenesis and activity in Drosha and Dicer knockout cells. These data provide proof-of-principle evidence for additional mechanistic possibilities for efficient generation of small regulatory RNAs, and represent novel silencing triggers that may be exploited for technical purposes.
INTRODUCTION
RNA interference (RNAi) was coined to describe the phenomenon whereby injection of synthetic double-stranded RNA into Caenorhabditis elegans efficiently silences homologous transcripts (Fire et al. 1998). RNAi is now recognized as an ancient strategy for homology-based post-transcriptional silencing mediated by ∼20–32 nt RNAs, and it plays critical roles in gene regulation in diverse plant, fungal, and animal systems (Lai 2003; Axtell et al. 2011). At the heart of the conserved RNAi machinery lie two factors, Dicer and Argonaute (Czech and Hannon 2010; Yang and Lai 2011). Dicer is an enzyme of the RNase III family that cleaves longer substrates into short RNAs, which load into Argonaute (Ago) proteins and serve as guides to identify complementary targets for regulation. Ago proteins contain a signature Piwi domain, which adopts an RNase H-like fold that serves as a catalytic center for target cleavage (Parker et al. 2004; Song et al. 2004; Yuan et al. 2005). While some Ago proteins lack catalytic activity, at least one Ago protein in all species that have RNAi has the capacity to cleave targets, also known as “Slicer” activity (Hutvagner and Simard 2008).
One hypothesis concerning the origin of RNAi is that it evolved as a defense against invasive nucleic acids such as viruses and transposons, using small interfering RNAs (siRNAs) generated from such invaders to guide their destruction (Ding and Voinnet 2007). With a system for homology-based repression in place, RNAi may have been redirected to endogenous gene regulation. A wide variety of processes, ranging from post-transcriptional repression of gene expression during development and physiology, establishment and maintenance of silenced chromatin, to programmed genome rearrangements, have been shown to be directed by RNAi pathways and short RNAs in various organisms (Flynt and Lai 2008; Ketting 2011).
The RNAi pathway appears to have engendered the biogenesis of short RNAs from endogenous inverted repeat transcripts several times during evolution, and this capacity may have emerged independently in plants, fungi, and animals (Axtell et al. 2011). Depending on the specificity of processing and the enzymes involved, inverted repeat transcripts can yield microRNAs (miRNAs) or siRNAs. The specificity of small RNA sequences forms a general basis for hairpin categorization. It is generally accepted that miRNA hairpins generate a specific small miRNA/star duplex, whereas siRNA hairpins generate multiple small RNA duplexes, which can either be phased or exhibit heterogenous and/or overlapping termini. Nevertheless, the categories of inverted repeat transcripts exist along a continuum. For example, some plant hairpins are processed by the miRNA machinery into multiple small RNA species that associate with Ago proteins (Rajagopalan et al. 2006; Zhang et al. 2010), while others are concomitantly processed by both miRNA and siRNA machinery (Vazquez et al. 2008). On the other hand, while Drosophila hairpin RNAs are exclusively processed by the siRNA machinery (Czech et al. 2008; Kawamura et al. 2008; Okamura et al. 2008), some hairpin RNA-derived small RNAs load into the miRNA effector Ago1 (Miyoshi et al. 2010; Ameres et al. 2011; Okamura et al. 2011).
In any case, a fundamental commonality of siRNA and miRNA pathways is that RNase III-like enzymes are required for efficient biogenesis. mi/siRNA biogenesis in fungi and plants relies on Dicer family enzymes, and miRNA biogenesis in animals requires RNase III-like enzymes of both the Drosha and Dicer families (Fig. 1A). While canonical pathways generate most miRNAs, a cornucopia of alternative strategies have emerged recently, including animal pathways that are independent of Drosha or Dicer (Westholm and Lai 2011; Yang and Lai 2011). For example, mirtrons are short hairpin introns whose splicing defines their pre-miRNA hairpin ends (Okamura et al. 2007; Ruby et al. 2007), thus bypassing Drosha (Fig. 1B). The “3′ tailed” mirtrons combine splicing with action of the RNA exosome to generate the Dicer substrate (Flynt et al. 2010). Still other RNases can substitute for Drosha to cleave pre-miRNA hairpins from longer precursor transcripts, such as RNase Z (Bogerd et al. 2010) or the Integrator complex (Cazalla et al. 2011). On the other hand, Drosha cleavage of mammalian mir-451 generates a 42-nt-long hairpin that is too short to serve as a Dicer substrate (Fig. 1C). Instead, pre-mir-451 is loaded directly into Ago2 and relies on its “slicer” activity for maturation (Cheloufi et al. 2010; Cifuentes et al. 2010; Yang et al. 2010).
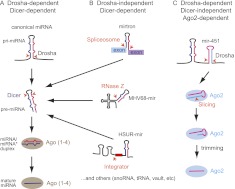
FIGURE 1.
Summary of major characterized miRNA biogenesis pathways in animal cells. (A) Canonical miRNAs are produced by sequential cleavage by the RNase III enzymes Drosha and Dicer. Resultant miRNA/star duplexes load into miRNA effector Argonautes (Ago1–4 in mammals), and the active regulatory complex contains a single-stranded miRNA. (B) Several alternative miRNA biogenesis pathways bypass Drosha cleavage by exploiting the spliceosome (which processes introns), RNase Z (which processes tRNAs), and Integrator (which processes snRNAs); other sources of Drosha-independent miRNAs have also been described. (C) Vertebrate mir-451 has a short hairpin that is processed by Drosha but then bypasses Dicer. Instead, the pre-miRNA is directly loaded into Ago proteins; those that enter the sole catalytic member Ago2 are sliced on the 3′ arm and subsequently trimmed to yield mature miR-451.
The existence of these sundry pathways highlights the flexibility of small RNA biogenesis during evolution (Westholm and Lai 2011; Yang and Lai 2011). Nevertheless, an RNase III-type enzyme remains essential in all alternative miRNA pathways. In this study, we successfully designed both RNA pol II- and pol III-dependent systems for effective miRNA biogenesis in mammalian cells that are completely independent of RNase III-family enzymes. Stringent evidence is provided by demonstrating efficient maturation and function of miRNAs in both Drosha and Dicer knockout cells. These constructs may be exploited as novel triggers of gene silencing, and they support the intriguing hypothesis that RNAi may have initially emerged as an Ago-dependent biogenesis pathway.
RESULTS
RNase III-independent miRNA biogenesis mediated by Integrator and Ago2
We wondered whether it was possible, by appropriate design, to generate substrates that bypass both RNase III-type enzymes that are considered obligate for miRNA biogenesis in mammalian cells. Our initial attempts to generate RNase III-independent miRNAs involved mirtrons of pre-mir-451 hairpin length. In Drosophila and C. elegans, many mirtrons can be expressed at sufficient levels to be detected using Northern analysis (Okamura et al. 2007; Ruby et al. 2007; Chung et al. 2011). However, the mammalian mirtrons that we tested (mir-877 and mir-1228), although conserved and/or reasonably represented in small RNA libraries (Berezikov et al. 2007; Westholm et al. 2012), were poorly active on cognate sensors when expressed in HeLa cells (Supplemental Fig. 1). Moreover, we did not observe robust accumulation of specific miRNA-sized species when total RNA from cells transfected with these mirtron constructs was assayed by Northern blot (Supplemental Fig. 1). This was consistent with other recent studies of mirtron biogenesis in mammalian cells, which did not utilize Northern analysis and relied on amplification by stem–loop–rtPCR to detect mature small RNAs (Havens et al. 2012; Sibley et al. 2012).
As the splicing-derived miRNA pathway did not appear promising for this effort, we investigated other strategies. We recently reported that the Integrator complex (Baillat et al. 2005), normally involved in biogenesis of Sm-class small nuclear RNAs (snRNAs), can substitute for Drosha/DGCR8 during miRNA biogenesis (Cazalla et al. 2011). Our studies of Herpesvirus saimiri snRNAs (HSURs) revealed that some are encoded upstream of a pre-miRNA hairpin. The Integrator complex cleaves the 3′ box downstream from the snRNA, releasing the pre-miRNA for nuclear export and cleavage by Dicer. A minigene containing the U1 promoter, 3′ cleavage box and pre-mir-HSUR4 hairpin but no snRNA (U1-mir-dd) (Fig. 2A; Supplemental Fig. 2) generates a Drosha-independent, Integrator/Dicer-dependent miRNA (Cazalla et al. 2011). Therefore, an snRNA-like moiety is not required for miRNA biogenesis.
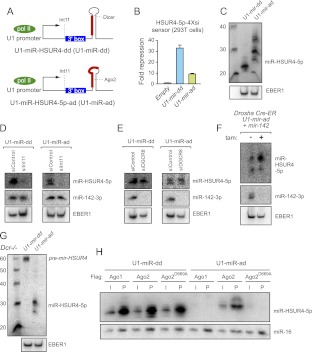
FIGURE 2.
Biogenesis and function of a Drosha/Dicer-independent miRNA generated by the Integrator pathway. (A) A minigene in which the normally Dicer-dependent (dd) pre-mir-HSUR4 hairpin (U1-mir-dd) (see also Supplemental Fig. 2) has been shortened to Ago2-dependent (ad) length (U1-mir-ad); mature miR-HSUR4-5p is indicated by the red box. (B) Functionality of U1-mir-dd and U1-mir-ad against a luciferase sensor bearing bulged miR-451 sites, assayed in 293T cells. Averages of three independent experiments with SD are shown. (C) Accumulation of mature miR-HSUR4-5p following transfection of U1-mir-dd and U1-mir-ad into 293T cells; EBER1 was cotransfected and detected as a control. (D–F) Integrator-dependent, microprocessor-independent processing of U1-mir constructs. 293T cells were transfected with control siRNA (siControl) or siRNA against Integrator11 (siInt11) for 48 h, followed by retransfection with siRNA and a mixture of expression plasmids for EBER1, mir-142, and either U1-mir-dd or U1-mir-ad. Expression of mature miR-HSUR4-5p from both U1-mir-dd and U1-mir-ad was lost following knockdown of Int11, while miR-142-3p and EBER1 were unaffected. (E) The same experimental setup as in D, except using siRNA against DGCR8. Mature miRNAs were effectively produced from both U1-mir constructs, but not from canonical mir-142. (F) Drosha-fl/fl Cre-ER mouse ES cells were mock-treated (−) or treated (+) with 4-OH-Tamoxifen (tam) for 72 h, followed by cotransfection of U1-mir-ad, mir-142, and EBER1. miR-HSUR4-5p but not miR-142-3p accumulated in Drosha knockout cells. (G) Dicer-independent processing of U1-mir-ad. Dicer knockout MEFs were transfected with U1-mir-dd and U1-mir-ad; biogenesis of the former was arrested at the pre-miRNA stage while the latter continued to generate mature miRNAs in the absence of Dicer. (H) Slicer-dependent processing of U1-mir-ad. Northern blot showing coimmunoprecipitation of miR-HSUR4-5p or endogenous miR-16 from 293T cells cotransfected with either U1-mir-dd or U1-mir-ad, and either FLAG-Ago1, FLAG-Ago2, or FLAG-Ago2D669A. (I) Input (5%); (P) Pellet (100%). Dicer-dependent mature miRNAs associate with all of these tagged Ago proteins, but Ago2-dependent mature miRNAs are present only in wt Ago2 immunoprecipitates.
We modified the HSUR4 pre-miRNA in the U1-mir-dd construct to mimic pre-mir-451 hairpin length and structure, yielding U1-mir-ad (Fig. 2A; Supplemental Fig. 2). We compared the functional properties of the two U1-miRNA constructs. When assayed in 293T cells, both constructs repressed a sensor bearing sites perfectly complementary to HSUR4-5p. Although less effective than the Dicer-dependent version, U1-mir-ad repressed its sensor nearly 10-fold (Fig. 2B). We confirmed that these constructs generate specific short RNAs using Northern assays. The Dicer-generated HSUR4-5p miRNAs were more homogenously sized and shorter than the ladder of RNAs produced by U1-mir-ad (Fig. 2C), consistent with the idea that the latter matures via a 3′ exoribonucleolytic activity.
We assayed the biogenesis of the U1-miRNA constructs in greater detail. We reported earlier that the generation of HSUR miRNAs is dependent on Integrator (Cazalla et al. 2011). Consistent with this, siRNA-mediated depletion of Int11 completely abrogated maturation of U1-mir-dd, while canonical mir-142 was unaffected (Fig. 2D). The maturation of U1-mir-ad similarly was fully dependent on Int11, as cells treated with siRNAs against Int11 failed to generate mature miR-HSUR4-5p (Fig. 2D). We next assayed cells depleted of the Drosha cofactor DGCR8 and observed that this treatment eliminated biogenesis of mir-142, but did not substantially affect either U1-mir-dd or U1-mir-ad (Fig. 2E). Thus, the biogenesis of U1-mir-ad appeared independent of the canonical nuclear miRNA pathway. We performed a strict test of this by assaying its properties in Drosha-knockout cells, generated by treating Drosha[fl/fl] Cre-ER mES cells with tamoxifen (Karginov et al. 2010); mock-treated cells served as a control. We waited 3 d to allow depletion of Drosha mRNA and protein, a timepoint at which mir-142 was no longer processed (Fig. 2F). On the other hand, U1-mir-ad generated mature miR-HSUR4-5p in Drosha knockout cells (Fig. 2F). Therefore, both Dicer-dependent and Ago-dependent U1-miRNA constructs are independent of the Microprocessor, but require Integrator.
We next examined the cytoplasmic steps in the biogenesis of U1-miRNAs. We transfected the U1-miRNA constructs into cells stably deleted for Dicer, which are unable to generate mature canonical miRNAs (Yi et al. 2006; Yang et al. 2010). Indeed, these cells were unable to process U1-mir-dd beyond the pre-miRNA stage (Fig. 2G). In contrast, mature miR-HSUR4-5p was generated upon introduction of U1-mir-ad into Dicer knockout cells (Fig. 2G). Consistent with these biogenesis data, U1-mir-ad was active for target regulation in Dicer knockout cells, whereas U1-mir-dd was not (Supplemental Fig. 3A). To our knowledge, this constitutes the first demonstration of efficient RNase III-independent miRNA biogenesis and function in metazoan cells.
Finally, comparison of anti-Ago1- and Ago2-immunoprecipitates (IP) confirmed the distinct properties of the “dd” and “ad” variants. Consistent with the association of canonical miRNAs (e.g., miR-16) with all mammalian Ago proteins, mature miR-HSUR4-5p from U1-mir-dd co-IPed with both Ago1 and Ago2 (Fig. 2H). Both of these mature miRNAs could also be co-IPed with catalytically inactive Ago2[D669A]. In contrast, U1-mir-ad matured only within Ago2-Slicer, as mature miR-HSUR4-5p from this construct was not detected in Ago1 or Ago2[D669A] (Fig. 2H). Consistent with these data, the regulatory activity of U1-mir-ad was abolished in Ago2-KO MEFs (Supplemental Fig. 3B). In summary, we have shown that a short hairpin substrate driven by the U1 promoter yields a Drosha/Dicer-independent, Integrator/Ago2-dependent, miRNA substrate that can function in target silencing.
RNase III-independent miRNA biogenesis mediated by RNase Z and Ago2
We explored a second RNase III-independent biogenesis scheme based on the capacity of RNase Z to cleave 3′ products from tRNA precursors. Murine gamma herpesvirus 68 encodes Drosha-independent miRNAs whose precursor hairpins lie downstream from tRNA transcripts (Bogerd et al. 2010), and the tRNA-hairpin system can also be used to express synthetic shRNAs (Scherer et al. 2007). We first generated a tRNAlys expression construct fused to a canonical Dicer-dependent (“dd”) pre-miRNA hairpin programmed with mature miR-451 (tRNA-mir-dd) (Fig. 3A; Supplemental Fig. 2). This generated functionally active miRNAs that could repress both perfect and bulged miR-451 sensors (Fig. 3B). Its ability to repress the bulged miR-451 sensor provided evidence for accurate 5′ end generation by tRNA-mir-dd. In fact, this construct was more active on both sensors than the endogenous human mir-144/451 construct that we characterized recently (Yang et al. 2010). In contrast to endogenous mir-451, however, tRNA-mir-dd was not able to repress its sensor in Dicer knockout cells (Fig. 3C). This indicated that this construct was truly dependent on Dicer for its regulatory activity, and could not be compensated for by potential loading of its pre-miRNA into Argonaute as has been observed (Diederichs and Haber 2007; Tan et al. 2009).
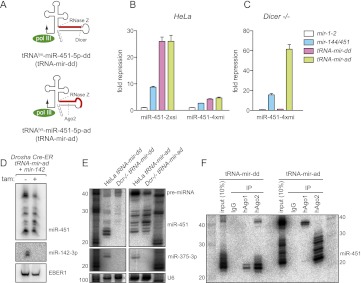
FIGURE 3.
Biogenesis and function of a Drosha/Dicer-independent miRNA generated by the tRNA pathway. (A) Design of tRNA–miRNA fusion constructs in which either a Dicer-dependent (dd) or Ago2-dependent (ad) pre-mir-451 hairpins reside downstream from the RNase Z cleavage site of a tRNAlys construct; mature miR-451 is represented by the red box. (B) Functionality of mir-451 expression constructs on perfect (si) and bulged (mi) miR-451 luciferase sensors. Compared with control mir-1-2 construct, the endogenous mir-144/451 and both “dd” and “ad” versions of tRNA-mir-451 fusions could repress these sensors. As expected, repression of the perfect sensor was more efficient with all constructs. (C) Functionality of miRNA expression constructs in Dicer−/− MEFs. Only Dicer-independent hsa-mir-144/451 and tRNA-451-ad constructs were functional in this setting. (D) Drosha-independent processing of tRNA-mir-ad. Drosha-fl/fl Cre-ER mouse ES cells were mock-treated (−) or treated (+) with 4-OH-Tamoxifen (tam) for 72 h, followed by cotransfection of tRNA-mir-ad, mir-142, and EBER1. Only miR-451 derived from tRNA-mir-ad, but not miR-142-3p, accumulated in Drosha knockout cells. (E) Dicer-independent processing of tRNA-mir-ad. Northern blot analysis of Hela cells and Dicer−/− MEFs transfected with tRNA-mir-dd or tRNA-mir-ad constructs; mir-375 was cotransfected as a control. (F) tRNA-mir-ad matures exclusively within Ago2. Northern blot showing coimmunoprecipitation of tRNA-mir-dd or tRNA-mir-ad with control (IgG), anti-human Ago1 (hAgo1), or anti-human Ago2 (hAgo2).
We then exchanged the Dicer-dependent hairpin for the endogenous Ago2-dependent (“ad”) pre-mir-451 hairpin (Fig. 3A; Supplemental Fig. 2). tRNA-mir-ad repressed both perfect and bulged target sensors (Fig. 3B) and was more active on both targets than endogenous mir-144/451, similar to tRNA-mir-dd. Moreover, tRNA-mir-ad was highly active in Dicer knockout cells, about threefold more than endogenous mir-144/451 (Fig. 3C).
We analyzed the biogenesis of tRNA-mir-dd and tRNA-mir-ad constructs in detail. When assayed in Drosha knockout cells that were completely unable to generate canonical mature miR-142-3p, tRNA-mir-ad were effectively matured (Fig. 3D). Analysis of Dicer knockout MEFs distinguished tRNA-mir-dd and tRNA-mir-ad since biogenesis of the former was dependent on Dicer, whereas the latter was not (Fig. 3E). The relative accumulation of pre-mir-451-ad versus its matured miRNAs was decreased in Dicer−/− cells, consistent with the possibility that Dicer-independent hairpins are processed more efficiently in the absence of a competing canonical miRNA pathway.
Finally, IP analysis confirmed that mature miR-451 generated by mir-451-dd accumulated in both Ago1 and Ago2 complexes, whereas mir-451-ad exhibited strictly Ago2-dependent biogenesis (Fig. 3F). Indeed, pre-mir-451-ad accumulated exclusively in Ago1 complexes, whereas mature miR-451 species of <30 nt accumulated exclusively in Ago2 complexes. Consistent with the notion of Ago2-dependence, we found that mir-451-ad, similar to mir-451-dd, were nonfunctional for target regulation in Ago2-KO MEFs, both on a perfect sensor (Supplemental Fig. 3C) and an imperfect sensor (Supplemental Fig. 3D).
In summary, diverse cellular ribonucleases can fully bypass RNase III-type enzymes for efficient miRNA biogenesis in mammalian cells, via RNA pol-II (snRNA pathway) or RNA pol-III (tRNA pathway) substrates.
DISCUSSION
In this study, we describe two designed miRNA pathways that fully bypass both of the RNase III enzymes that are generally considered obligate for miRNA biogenesis. These pathways, which marry either the Integrator complex or RNase Z with Ago2-slicer activity, are highly efficient at generating discrete small RNA species with miRNA functionality. While we have not identified endogenous versions of these substrates, our findings stoke the concept that additional hybrid biogenesis pathways may await discovery. It is relevant to appreciate that many technical innovations in RNA silencing preceded the discovery of naturally occurring counterparts. The discovery of RNAi in C. elegans preceded the discovery of endogenous RNAi pathways (Fire et al. 1998); transgenic RNAi in Drosophila using long inverted repeat transgenes (Kennerdell and Carthew 2000) preceded the recognition of endogenous hairpin RNAs that are processed by the RNAi machinery (Czech et al. 2008; Kawamura et al. 2008; Okamura et al. 2008); tRNA–shRNA fusion constructs were developed (Scherer et al. 2007) before endogenous tRNA–miRNAs were characterized (Bogerd et al. 2010), and synthetic shRNAs were used for gene silencing (Paddison et al. 2002) before endo-shRNAs were discovered (Babiarz et al. 2008). In a similar vein, intron-interrupted miRNAs were recently found to be functional in C. elegans (Zhang et al. 2011), although such endogenous loci remain to be found. We infer that additional exotic small RNA biogenesis mechanisms await characterization.
The RNase III-independent miRNA biogenesis strategies that we describe complement existing vectors for expressing silencing RNAs and may have potential technical advantages, in that they do not compete with the central enzymes in canonical miRNA biogenesis. In addition, they are endowed with other advantages of mir-451-type triggers, in that they do not produce star-strand species and do not populate nonslicing Ago proteins, both of which contribute to off-target effects. The recent description of optimal features of Slicer-mediated small RNA function (Yang et al. 2012) can be exploited to further increase the efficacy of these novel silencing triggers. Finally, our demonstration of flexible strategies for RNase III-independent miRNA biogenesis in present-day metazoan cells may support the notion that Argonaute-mediated gene silencing could have initially emerged prior to molecular coupling to RNase III family enzymes (Halic and Moazed 2010).
MATERIALS AND METHODS
Mirtron expression constructs and sensors
We cloned mirtron constructs by amplifying genomic regions, including flanking exonic sequences, from HEK 293 genomic DNA as template. We cloned the products into pcDNA6.2 TA vector (Invitrogen). The following primers were used for PCR.
clo877f2: AGGTGGTAGCAGATGAGACAC;
clo877r2: CTGGCCAGGGAGACACGCATG;
clo1228f1: GTGCCACCGACTGCAGCAGCTAC;
clo1228r1: TGCAGTGGGTCTCGTCCTCGC.
We used the following primers to generate perfect-matched or bulged targets of mirtron-derived miRNAs, cloned into psiCHECK2 vector.
877-2si_F: GGCCGCcTGTCCCCTGCGCCATCTCCTCTACtcaacaatcaccTGTCCCCTGCGCCATCTCCTCTACtG;
877-2si_R: TCGACaGTAGAGGAGATGGCGCAGGGGACAggtgattgttgaGTAGAGGAGATGGCGCAGGGGACAgGC;
1228-5p-2si_F: GGCCGCcCACACACCTGCCCCCGCCCACtcaacaatcaccCACACACCTGCCCCCGCCCACtG;
1228-5p-2si_R: TCGACaGTGGGCGGGGGCAGGTGTGTGggtgattgttgaGTGGGCGGGGGCAGGTGTGTGgGC;
1228-3p-2si_F: GGCCGCcTGGGGGGCGAGGCAGGTGTGAtcaacaatcaccTGGGGGGCGAGGCAGGTGTGAtG;
1228-3p-2si_R: TCGACaTCACACCTGCCTCGCCCCCCAggtgattgttgaTCACACCTGCCTCGCCCCCCAgGC.
U1-mir-HSUR constructs
The plasmid expressing Dicer-dependent hvsA-miR-HSUR4-5p (U1-mir-dd) (Fig. 2A, upper panel) has been described as U1-H4ΔsnRNA (Cazalla et al. 2011). This plasmid was further modified by site-directed mutagenesis to generate the plasmid U1-mir-ad (Fig. 2A, lower panel) that expresses hvsA-miR-HSUR4-5p in an Ago2-dependent manner.
tRNAlys3–miRNA constructs
To generate the tRNA–miRNA constructs (Fig. 2C,D), partially overlapping DNA oligonucleotides (IDT) were diluted to 10 μM and used in a standard PCR reaction (20 μL final volume) with Accuprime Taq HiFi DNA polymerase. Amplicons were gel purified and used in a ligation reaction with pCR2.1 TA vector (Invitrogen). Positive clones were verified by sequencing using the M13 forward and reverse primers. Cloning primers are as follows.
tRNAlys3_451ad_F: GCCCGGATAGCTCAGTCGGTAGAGCATCAGACTTTTAATCTGAGGGTCCAGGGTTCAAGTCCCTG;
tRNAlys3_451ad_R: AAAAACCATTACCATTACTAAACTCAGTAATGGTAACGGTTTCGCCCGAACAGGGACTTGAACCC;
tRNAlys3_451dd_5p_F: GCCCGGATAGCTCAGTCGGTAGAGCATCAGACTTTTAATCTGAGGGTCCAGGGTTCAAGTCCCTGTTCGGGC;
tRNAlys3_451dd_5p_R: AAAAACCGTTACCATTACTGAGTTTAACTAACTAACTCAGTAATGGTAACGGTTTCGCCCGAACAGGGACTT.
Other plasmids
We used the following published expression plasmids: pcDNA6-hsa-mir-144/451 and hsa-mir-375 (Yang et al. 2010, 2011), pcDNA3-pri-mir-142 (Cazalla et al. 2011) kindly provided by Jan Pawlicki, and pEBV RIJ expressing EBER 1 (Rosa et al. 1981). A plasmid expressing human FLAG-Ago1 (Meister et al. 2004) was obtained from Addgene (plasmid #10820). Plasmids expressing mouse FLAG-Ago2 and FLAG-Ago2D669A (O'Carroll et al. 2007) were kindly provided by Donal O'Carroll.
Cell culture and transfections
Mammalian cells were maintained in DMEM 10% FBS with glutamax supplemented with antibiotics (Invitrogen). Cells were transfected with Lipofectamine 2000 (Invitrogen) in DMEM 10% FBS without antibiotics. Transient RNAi knockdown assays were performed as described (Cazalla et al. 2011). Dicer−/− MEFs (Yang et al. 2010) and 293T cells were transfected with Lipofectamine 2000, and 48 h later total RNA was isolated and analyzed by Northern blot. To test dependence on Drosha, we took advantage of the Drosha[fl/fl] Cre-ER mES cell line (Karginov et al. 2010) kindly provided by Greg Hannon, in which Drosha can be deleted upon treatment with 4-Hydroxytamoxifen (“Tam,” Sigma). Cells were either mock-treated or treated with 100 nM 4-Hydroxytamoxifen for 3 d and transfected with 0.2 μg of EBER1-expressing plasmid, 0.8 μg of pCDNA3-pri-miR-142, and 1 μg of either U1-miR-HSUR4-ad or tRNA-451-ad using Lipofectamine 2000 (Invitrogen). After transfection, cells were further treated with 4-Hydroxytamoxifen. RNA was extracted 48 h later.
Northern blot
Total RNA were extracted with Trizol LS reagent (Invitrogen) following the manufacturer's instructions. RNA were separated on 20% urea polyacrylamide denaturing gels (National Diagnostics) and transfered to GeneScreen Plus nylon membranes (PerkinElmer) and UV cross-linked (stratalinker). Membranes were hybridized overnight with [γ-32P]labeled DNA oligonucleotides, washed (2× SSC, 0.1% SDS), and then exposed to PhosphorImager screens. For sequential detection, membranes were stripped in 0.1% boiling SDS. We used DNA probes complementary to miR-375 TCACGCGAGCCGAACGAACAAA, miR-451-5p AACTCAGTAATGGTAACGGTTT, miR-HSUR4 TTATAGCTGTAGCAACACGGT, hsa-1228-5p GGGGGGCGAGGCAGGTGTGA, hsa-1228-3p CACACACCTGCCCCCGCCCAC, hsa-mir-877-5p CCCTGCGCCATCTCCTCTAC, and U6 ATTTGCGTGTCATCCTTGCGCAG.
Sensor assays
Sensor assays in HeLa cells were performed as described using luciferase reporters for mir-451 (miR-451-2xsi and miR-451-4xmi) (Yang et al. 2010). For mir-877 and mir-1228 a modified psiCHECK2 vector was used for the cloning of miRNA perfect-matched targets. The oligonucleotide sequences are listed in Supplemental Table S1. Cells were seeded in 96-well plates and were transfected the following day with lipofectamine 2000 following the manufacturer's instructions with slight modification as follows: 60 ng of DNA were used per well (12 ng of sensor, 48 ng of microRNA expression vector) with 0.5 μL of lipofectamine 2000. After 48 h, medium was removed and cells were lysed in 1× passive lysis buffer (promega); renilla and firefly activities were measured with the Dual-glo luciferase assay (Promega).
Sensor assays in 293T cells were performed as described using luciferase reporters for miR-HSUR4-5p (4xsi) (Cazalla et al. 2011). 293T cells were seeded in 24-well plates and cotransfected the next day with 1 pg of Luc reporter with four perfectly complementary sites for hvsA-miR-HSUR4-5p and 800 ng of a plasmid expressing either Dicer-dependent hvsA-miR-HSUR4-5p (U1-miRHSUR4-dd, also called U1-H4ΔsnRNA in Cazalla et al. 2011) or Ago2-dependent hvsA-miR-HSUR4-5p (U1-miR-HSUR4-ad).
Immunoprecipitations
Immunoprecipitations of HeLa cells were performed by growing cells in 150-mm dishes to confluence, washed once, scraped off in ice-cold PBS, and collected by centrifugation (500g, 10 min, 4°C). Cells were lysed in NP40 buffer (50 mM Hepes-KOH at pH 7.5, 150 mM KCl, 2 mM EDTA, 0.5% NP40, 0.5 mM DTT, 1× Complete EDTA free protease inhibitor [Roche]) for 30 min on ice and the lysates were spun (10 min, 4°, 20,000g). Supernatants were incubated with Dynabeads protein-G coated with the appropriate antibodies for 3 h at 4°C. Anti hAgo2: 11A9 (Sigma-Aldrich), Anti hAgo1: 4B8 (Sigma-Aldrich), anti mAgo2: 2D4 (Wako). Beads were washed four times in NP40 lysis buffer and bound ribonucleoprotein complexes were eluted with 300 mM NaCl and an equal volume of phenol-chloroform-isoamylic acid solution (Sigma-Aldrich). RNA were then precipitated with ethanol overnight and pellets were directly resuspended in RNA loading buffer2 (Ambion).
Immunoprecipitations of 293T cells were performed using extracts prepared from 5 × 106 293T cells cotransfected 48 h earlier with 8 μg of a plasmid expressing either FLAG-Ago1, FLAG-Ago2, or FLAG-Ago2D669A, and 8 μg of either U1-miR-HSUR4-ad or U1-miR-HSUR4-dd. Anti-FLAG M2 affinity gel (Sigma) was used for immunoprecipitation (Cazalla et al. 2010). Coimmunoprecipitated RNA was extracted from beads with Trizol (Invitrogen) and analyzed by Northern blot.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Alexander Tarakhovsky, Donal O'Carroll, Greg Hannon, and Jidong Liu for knockout cell lines and plasmids used in this study. J.-S.Y. was supported by a Frank Lappin Horsfall Fellowship. T.M. received support from the Tourre Foundation. We thank Mohamed Benahmed (INSERM Nice) for hosting T.M. for part of this work. We thank Joan Steitz for critical reading, and the work of D.C. in her laboratory was supported by the NIH (P01-CA016038). Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund, the Starr Cancer Consortium (I3-A139), and the National Institute of General Medical Sciences of the National Institutes of Health (R01-GM083300). The content is solely the responsibility of the authors and does not necessarily represent the official views of these funding agencies.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.036194.112.
REFERENCES
- Ameres SL, Hung JH, Xu J, Weng Z, Zamore PD 2011. Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins. RNA 17: 54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Westholm JO, Lai EC 2011. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol 12: 221 doi: 10.1186/gb-2011-12-4-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123: 265–276 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung W-J, Willis J, Cuppen E, Lai EC 2007. Mammalian mirtron genes. Mol Cell 28: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR 2010. A mammalian herpesvirus uses non-canonical expression and processing mechanisms to generate viral microRNAs. Mol Cell 37: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328: 1563–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Xie M, Steitz JA 2011. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol Cell 43: 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Agius P, Westholm JO, Chen M, Okamura K, Robine N, Leslie CS, Lai EC 2011. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res 21: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson N, et al. 2010. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328: 1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ 2010. Small RNA sorting: Matchmaking for Argonautes. Nat Rev Genet 12: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. 2008. An endogenous siRNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA 2007. Dual role for Argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O 2007. Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC 2008. Biological principles of microRNA-mediated regulation: Shared themes amid diversity. Nat Rev Genet 9: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Chung WJ, Greimann JC, Lima CD, Lai EC 2010. microRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell 38: 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D 2010. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140: 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens MA, Reich AA, Duelli DM, Hastings ML 2012. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res 40: 4626–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ 2008. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ 2010. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell 38: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada T, Siomi MC, Siomi H 2008. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature 453: 793–797 [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW 2000. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol 18: 896–898 [DOI] [PubMed] [Google Scholar]
- Ketting RF 2011. The many faces of RNAi. Dev Cell 20: 148–161 [DOI] [PubMed] [Google Scholar]
- Lai EC 2003. microRNAs: Runts of the genome assert themselves. Curr Biol 13: R925–R936 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC 2010. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA 16: 506–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A 2007. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21: 1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W-J, Ruby JG, Guo H, Bartel DP, Lai EC 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Robine N, Liu Y, Liu Q, Lai EC 2011. R2D2 organizes small regulatory RNA pathways in Drosophila. Mol Cell Biol 31: 884–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 16: 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D 2004. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J 23: 4727–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa MD, Gottlieb E, Lerner MR, Steitz JA 1981. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol 1: 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer LJ, Frank R, Rossi JJ 2007. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res 35: 2620–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CR, Seow Y, Saayman S, Dijkstra KK, El Andaloussi S, Weinberg MS, Wood MJ 2012. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res 40: 438–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L 2004. Crystal structure of Argonaute and its implications for RISC Slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Tan GS, Garchow BG, Liu X, Yeung J, Morris JP 4th, Cuellar TL, McManus MT, Kiriakidou M 2009. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res 37: 7533–7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Blevins T, Ailhas J, Boller T, Meins F Jr 2008. Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res 36: 6429–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Lai EC 2011. Mirtrons: microRNA biogenesis via splicing. Biochimie 93: 1897–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Ladewig E, Okamura K, Robine N, Lai EC 2012. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA 18: 177–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC 2011. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 43: 892–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC 2010. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci 107: 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC 2011. Widespread regulatory activity of vertebrate microRNA* species. RNA 17: 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Lai EC 2012. Functional parameters of Dicer-independent microRNA biogenesis. RNA 18: 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, O’Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E 2006. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 38: 356–362 [DOI] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ 2005. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 19: 405–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Xia J, Chellappan P, Zhang X, Jin H 2010. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol 11: R81 doi: 10.1186/gb-2010-11-8-r81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Maniar JM, Fire AZ 2011. “Inc-miRs”: Functional intron-interrupted miRNA genes. Genes Dev 25: 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]