Pnkp is the end-healing and end-sealing component of an RNA repair system present in diverse bacteria from many phyla. Pnkp is composed of three catalytic modules: an N-terminal polynucleotide 5′-kinase, a central 2′,3′ phosphatase, and a C-terminal ligase. Here the authors report the crystal structure of the kinase domain of Clostridium thermocellum Pnkp bound to ATP•Mg2+ (substrate complex) and ADP•Mg2+ (product complex). The protein consists of a core P-loop phosphotransferase fold embellished by a distinctive homodimerization module composed of secondary structure elements derived from the N and C termini of the kinase domain. The studies suggest a catalytic mechanism whereby Asp38 (as general base) activates the polynucleotide 5′-OH for its nucleophilic attack on the γ phosphorus and Lys21 and Mg2+ stabilize the transition state.
Keywords: P-loop phosphotransferase, RNA end-healing
Abstract
Pnkp is the end-healing and end-sealing component of an RNA repair system present in diverse bacteria from many phyla. Pnkp is composed of three catalytic modules: an N-terminal polynucleotide 5′-kinase, a central 2′,3′ phosphatase, and a C-terminal ligase. Here we report the crystal structure of the kinase domain of Clostridium thermocellum Pnkp bound to ATP•Mg2+ (substrate complex) and ADP•Mg2+ (product complex). The protein consists of a core P-loop phosphotransferase fold embellished by a distinctive homodimerization module composed of secondary structure elements derived from the N and C termini of the kinase domain. ATP is bound within a crescent-shaped groove formed by the P-loop (15GSSGSGKST23) and an overlying helix-loop-helix “lid.” The α and β phosphates are engaged by a network of hydrogen bonds from Thr23 and the P-loop main-chain amides; the γ phosphate is anchored by the lid residues Arg120 and Arg123. The P-loop lysine (Lys21) and the catalytic Mg2+ bridge the ATP β and γ phosphates. The P-loop serine (Ser22) is the sole enzymic constituent of the octahedral metal coordination complex. Structure-guided mutational analysis underscored the essential contributions of Lys21 and Ser22 in the ATP donor site and Asp38 and Arg41 in the phosphoacceptor site. Our studies suggest a catalytic mechanism whereby Asp38 (as general base) activates the polynucleotide 5′-OH for its nucleophilic attack on the γ phosphorus and Lys21 and Mg2+ stabilize the transition state.
INTRODUCTION
RNA repair is a mechanism to rectify programmed RNA breaks incurred during RNA processing and under conditions of cellular stress (Amitsur et al. 1987; Schwer et al. 2004; Nandakumar et al. 2008). RNA repair enzymes capable of sealing 2′,3′-cyclic phosphate and 5′-OH ends are present in diverse taxa in all phylogenetic domains of life. Two general strategies of RNA repair have been described that differ with respect to the chemical mechanisms by which the 3′,5′ phosphodiester repair junction is synthesized. In repair pathways driven by “classic” ATP-dependent RNA ligases, phosphodiester formation entails three nucleotidyl transfer reactions: ligase reacts with ATP to form a covalent ligase-(lysyl-N)–AMP intermediate plus pyrophosphate; AMP is transferred from ligase-adenylate to the 5′-PO4 RNA end to form an RNA-adenylate intermediate (AppRNA); and ligase catalyzes attack by an RNA 3′-OH on the RNA-adenylate to seal the two ends via a phosphodiester bond, with release of AMP (Nandakumar et al. 2006). To satisfy the classic ligase specificity for 3′-OH/5′-PO4 ends, the original broken 2′,3′-cyclic phosphate and 5′-OH ends must be “healed” before they can be sealed. Healing entails hydrolysis of the 2′,3′-cyclic phosphate (by a phosphoesterase enzyme) to form a 3′-OH and phosphorylation of the 5′-OH (by a polynucleotide kinase enzyme) to form a 5′-PO4. A different repair strategy, which circumvents the need for 5′ end-healing, is exemplified by the RNA ligase RtcB, which splices 3′-phosphate and 5′-OH RNA ends via a pathway that requires GTP and involves covalently activated RtcB-(histidinyl-N)–GMP and RNA(3′)ppG intermediates (Tanaka et al. 2011; Chakravarty et al. 2012).
RNA repair systems that follow the “healing and sealing” paradigm differ with respect to (1) whether the 5′ end-healing, 3′ end-healing, and ATP-dependent ligase components are physically linked within the same polypeptide or encoded as separate polypeptides; (2) whether the outcome of the 3′ end-healing reaction is complete removal of the 2′,3′ cyclic phosphate (to yield a 3′-OH, 2′-OH) or hydrolysis of the cyclic phosphate to a 3′-OH, 2′-PO4; and (3) whether the repair junction formed by the ligase is a standard 3′,5′ phosphodiester or contains a covalent “mark” at the ribose O2′ atom, e.g., a 2′-PO4 or a 2′-OCH3 (Schwer et al. 2004; Keppetipola et al. 2007; Nandakumar et al. 2008; Chan et al. 2009a; Englert et al. 2010). The marking of the splice junction has biological implications. A residual 2′-PO4 is a functional impediment that is eventually removed by an RNA-2′-phosphotransferase enzyme (Spinelli et al. 1999). In contrast, a 2′-OCH3 at the repair junction is immutable and potentially advantageous, insofar as it can protect the marked junction from recurrent incision (Chan et al. 2009a).
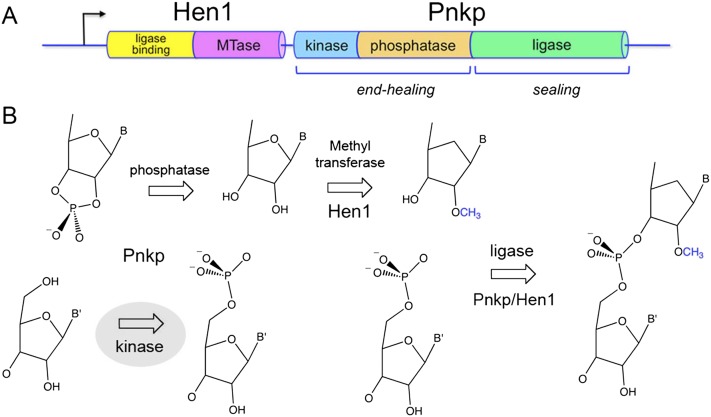
The ability to confer protective “immunity” during RNA repair is illustrated by the bacterial Pnkp-Hen1 system encoded in an operon-like gene cassette (Fig. 1A). Pnkp is composed of three catalytic domains: (1) an N-terminal kinase module that belongs to the P-loop phosphotransferase superfamily, (2) a central phosphatase domain that belongs to the binuclear metallophosphoesterase superfamily, and (3) C-terminal ATP-dependent ligase domain that belongs to the covalent nucleotidyltransferase superfamily (Martins and Shuman 2005). The kinase module catalyzes phosphoryl transfer from ATP to the 5′-OH RNA end. The phosphatase domain releases Pi from 2′-PO4, 3′-PO4, or 2′,3′-cyclic phosphate ribonucleotides (Fig. 1B; Keppetipola and Shuman 2006a,b, 2007). The ligase domain reacts with ATP to form a covalent LIG–AMP adduct, but it is unable per se to seal RNA strands (Martins and Shuman 2005; Smith et al. 2012).
FIGURE 1.
The Pnkp-Hen1 RNA repair pathway confers protective immunity to recurrent RNA damage. (A) The adjacent co-oriented ORFs encoding the Hen1 and Pnkp polypeptides comprise a bacterial RNA repair cassette conserved in taxa from 10 different bacterial phyla. Pnkp is a trifunctional RNA repair enzyme composed of 5′-OH polynucleotide kinase, 2′,3′ phosphatase, and ligase domains. Hen1 is composed of a C-terminal methyltransferase catalytic domain (MTase) fused to a unique N-terminal module that binds to the Pnkp ligase domain. (B) 2′,3′ cyclic phosphate and 5′-OH ends are substrates for healing and sealing by Pnkp and Hen1. The 5′ end is phosphorylated by the Pnkp kinase and the 2′,3′ cyclic phosphate is removed by the Pnkp phosphatase. The Hen1 methyltransferase installs a 2′-OCH3 mark at the terminal ribonucleoside prior to ligation of the ends. The ligation reaction is mediated by a heterodimer of the Pnkp ligase domain and the Hen1 N-terminal domain. The repair junction with the methyl mark is then resistant to scission by transesterifying endoribonucleases.
The sealing function of Pnkp is activated by its physical association with the Hen1 protein, which is encoded by the flanking genomic ORF (Chan et al. 2009a; Wang et al. 2012). This is the first instance in which RNA strand sealing by a covalently adenylylated ligase requires a separate protein partner. The C-terminal half of Hen1 is an autonomous manganese-dependent 3′-terminal ribose 2′-O-methyltransferase that installs the protective methyl mark (Fig. 1B; Jain and Shuman 2010, 2011; Huang 2012). The N-terminal half of Hen1 has no enzymatic activity per se but forms a heterodimer with the C-terminal ligase domain of Pnkp; the LIG/Hen1(N) heterodimer suffices for 3′-OH/5′-PO4 ligation (Wang et al. 2012).
The physical and functional properties of the Pnkp-Hen1 RNA repair system are distinctive vis à vis all others that have been characterized. Thus, it is of keen interest to elucidate the structure of the Pnkp-Hen1 holoenzyme and, en route, its constituent catalytic domains, so as to understand the principles of catalysis and substrate recognition, the relatedness of the CthPnkp-Hen1 domains to other enzymes that perform nucleic acid repair reactions, and, if possible, the logic of the domain organization and its implications for the order of the healing and sealing steps.
To date, crystal structures have been solved for the isolated Hen1 C-terminal methyltransferase domain (a monomer) bound to the reaction product AdoHcy (Chan et al. 2009b), the isolated Pnkp C-terminal ligase domain (a monomer) in a complex with the ATP substrate and as the covalent ligase–AMP intermediate (Smith et al. 2012), and the LIG–AMP•Mg2+-Hen1(N) heterodimer (Wang et al. 2012). The structure of the LIG domain is distinctive, being composed of a classic ligase adenylyltransferase fold that is embellished by a unique α-helical insert module and a unique C-terminal α-helical module. The insert module of the LIG domain mediates the interaction with the N-terminal domain of Hen1, which is itself a novel tertiary structure.
Here we delineate an N-terminal 170-amino acid segment of Clostridium thermocellum Pnkp as an autonomous polynucleotide kinase module. We determined the crystal structure of the kinase domain (a homodimer) bound to ATP•Mg2+ and ADP•Mg2+, reflective of substrate and product complexes, respectively, and used these structures to guide a mutational analysis of the kinase active site. Our findings highlight structural and functional similarities to another RNA repair enzyme, phage T4 polynucleotide kinase (Galburt et al. 2002; Wang et al. 2002), while revealing unique structural features of the bacterial polynucleotide kinase clade.
RESULTS AND DISCUSSION
An autonomous kinase domain of CthPnkp
It was shown previously that the N-terminal segment of CthPnkp, spanning residues 1 to 425, is active as a polynucleotide kinase and a 2′,3′ phosphoesterase (Martins and Shuman 2005). Further deletion analysis defined the central segment CthPnkp-(171-424) as an autonomous phosphoesterase domain (Keppetipola and Shuman 2006a). Here we asked whether the N-terminal segment of CthPnkp, which includes a 15GSSGSGKS22 P-loop motif, comprises an autonomous kinase domain. To this end, we produced CthPnkp-(1-425), CthPnkp-(1-180), CthPnkp-(1-175), CthPnkp-(1-170), and CthPnkp-(1-165) in Escherichia coli as His10 fusions and purified the recombinant proteins from soluble bacterial extracts by Ni-Agarose chromatography (Supplemental Fig. S1A). The kinase activity of the Pnkp proteins was assayed by the transfer of 32Pi from [γ-32P]ATP to a 36-mer 5′-OH DNA oligonucleotide acceptor. Kinase specific activities were determined from the slope of the respective protein titration curves and are plotted in the bar graph in Supplemental Figure S1B. The CthPnkp-(1-180), CthPnkp-(1-175), and CthPnkp-(1-170) preparations had similar specific activities; CthPnkp-(1-165) was about half as active as CthPnkp-(1-170). Shorter derivatives were not tested. We concluded that the N-terminal segment comprises a stand-alone kinase domain, and proceeded to conduct crystallization trials with a tag-free version of the CthPnkp-(1-170) protein.
Crystallization of the kinase domain
Crystals of the native kinase domain were grown by hanging drop vapor diffusion after mixing a sample of the protein solution containing 0.6 mM kinase, 2 mM ATP, and 10 mM MgCl2 with an equal volume of precipitant solution containing 100 mM HEPES (pH 7.5), 200 mM MgCl2, and 30% PEG-400. The crystals diffracted to 2.1 Å resolution and belonged to space group P212121. Our attempts to solve the structure by molecular replacement using the kinase domain of bacteriophage T4 Pnkp or mammalian Pnkp as search models were fruitless. Implementation of SeMet-SAD methods was complicated by the fact that the CthPnkp kinase domain contains only a single methionine at the translation start site. Therefore, we modified the protein by introducing three new internal methionines in lieu of Val44, Ile93, and Leu137 before producing and purifying a SeMet-substituted kinase domain. Crystals of the SeMet-kinase were grown by hanging drop vapor diffusion after mixing a protein solution containing 0.3 mM SeMet-kinase, 2 mM ATP, and 10 mM MgCl2 with an equal volume of precipitant solution containing 100 mM sodium citrate (pH 5.6), 5 mM dithiothreitol, 100 mM MgCl2, and 16% PEG-3350. These crystals diffracted to 2.0 Å resolution and belonged to space group P212121. SAD phasing located eight selenium atoms in the asymmetric unit, which contained two kinase protomers; these Se sites corresponded to Met1 and the three methionines installed by mutation. The refined model of the SeMet-kinase at 2.0 Å resolution (Rwork/Rfree 0.175/0.230) (Supplemental Table S1) comprised a kinase homodimer, each subunit of which had ATP and Mg2+ bound in the active site. The structure of the native kinase domain was then determined by molecular replacement with the SeMet-kinase model, from which the nucleotide and metal ligands had been deleted. The refined model of the native kinase at 2.1 Å resolution (Rwork/Rfree 0.177/0.228) (Supplemental Table S1) comprised a homodimeric protein virtually identical to the SeMet-kinase, except for the presence of the native Val44, Ile93, and Leu137 side-chains instead of SeMet. The native kinase protomers each had ADP and Mg2+ bound in the active site. These structures exemplify the substrate and product complexes with respect to the nucleotide phosphate donor.
Overview of the kinase structure
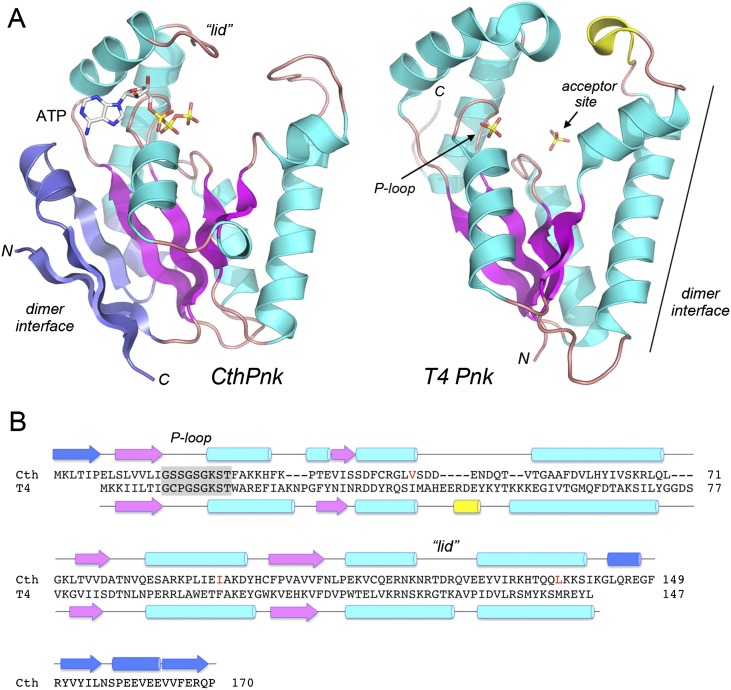
The tertiary structure consists of a central five-stranded parallel β-sheet with topology β6↑β5↑β2↑β4↑β3↑, proceeding from left to right in the view in Figure 2A. Strands β6 and β5 form a β sandwich with a β1↓β7↓ sheet. The central sheet is flanked on both sides by α helices (Fig. 2A). The P-loop motif (15GxxGxGKS22) is located between the β2 strand and the α1 helix. ATP is bound within a crescent-shaped groove formed by the P-loop and an overlying “lid” domain composed of helices α6 and α7 and the connecting 120RTDRQVE126 peptide (Fig. 2A,B). A surface model of the kinase protomer with ATP•Mg2+ in the NTP donor site (Supplemental Fig. S2) highlights the deep nucleotide binding groove and the positive electrostatic potential surrounding the ATP.
FIGURE 2.
Tertiary structure of the CthPnkp kinase domain and comparison to phage T4 polynucleotide kinase. (A) The tertiary structures of the SeMet-kinase protomer A (CthPnk) and the kinase domain of bacteriophage T4 Pnkp (T4Pnk; from pdb 1LY1) were superimposed and then offset horizontally. Common secondary structural elements in CthPnk and T4Pnk are colored magenta (for β strands) and cyan (for α helices). Unique secondary structure elements in CthPnk that form the dimer interface are colored blue. N and C denote the N and C termini of the kinase polypeptides. ATP in the CthPnk active site is shown as a stick model. T4Pnk has two sulfate anions (shown as stick models) in the active site. The sulfate at left is in the NTP phosphate donor site and is coordinated by the P-loop, as indicated by the arrow. The 5′-OH acceptor site in the T4Pnk structure is demarcated by a second sulfate at right that mimics the 5′-terminal phosphodiester of the polynucleotide, HONpN–. (B) The secondary structure elements of CthPnk and T4Pnk (colored as in panel A) are shown above and below their aligned amino acid sequences, with β strands rendered as arrows and α helices as cylinders. Gaps in the alignment are indicated by dashes. The P-loop motifs are highlighted in gray boxes. Three native amino acid residues that were replaced by methionine in the SeMet-kinase are indicated in red.
A DALI search (Holm et al. 2008) with the CthPnkp kinase structure identified numerous members of the P-loop phosphotransferase superfamily as homologs. Top DALI hits were archaeal seryl-tRNA(Sec) kinase (Protein Data Bank [pdb] 3A4L), the C-terminal kinase domain of mammalian polynucleotide kinase-phosphatase (pdb 1YJ5), and the N-terminal kinase domain of phage T4 polynucleotide kinase-phosphatase (pdb 1RRC). The superimposed tertiary structures of the RNA repair kinases CthPnk and T4Pnk (Z score 16.1; RMSD of 1.9 Å over 135 Cα positions) are shown side by side in Figure 2A, with the conserved strands colored magenta and the conserved helices colored cyan. The CthPnk and T4Pnk primary and secondary structures are aligned in Figure 2B. The bacterial and phage enzymes have a common core structure, comprising a central four-stranded parallel β-sheet, plus the P-loop and two-helix lid that form the binding site for the ATP phosphate donor. The donor site is demarcated by a sulfate in the T4Pnk structure in Figure 2A, which occupies the position of the ATP β-phosphate in the CthPnk structure.
The binding site for the 5′-OH phosphate acceptor in T4Pnk is within a tunnel formed by approximating the tip of the lid and the tip of an inter-helix loop (colored yellow in Fig. 2A). This site accommodates a single-stranded oligonucleotide, but not a duplex (Eastberg et al. 2004). A surface view of the analogous 5′-OH acceptor site in the CthPnk structure is shown in Supplemental Figure S2, which highlights a tunnel that leads directly from the protein surface to the ATP γ phosphate in the donor site. The acceptor tunnel is lined by positive surface potential and is wide enough to fit a 5′-OH single-stranded DNA or RNA, albeit not a duplex 5′-OH end.
The most striking difference between the bacterial and phage kinases is the presence in CthPnk of an N-terminal β-strand and a C-terminal αβαβ module (colored blue in Fig. 2A) that have no counterparts in T4Pnk. These distinctive N- and C-terminal elements in CthPnk come together in the tertiary structure to create a homodimer interface (on the left side of the kinase protomer in the view in Fig. 2A). In contrast, the homodimerization surface of T4Pnk is located of the opposite side of the protomer and is composed of secondary structure elements of the core phosphotransferase fold (Fig. 2A).
Kinase homodimer
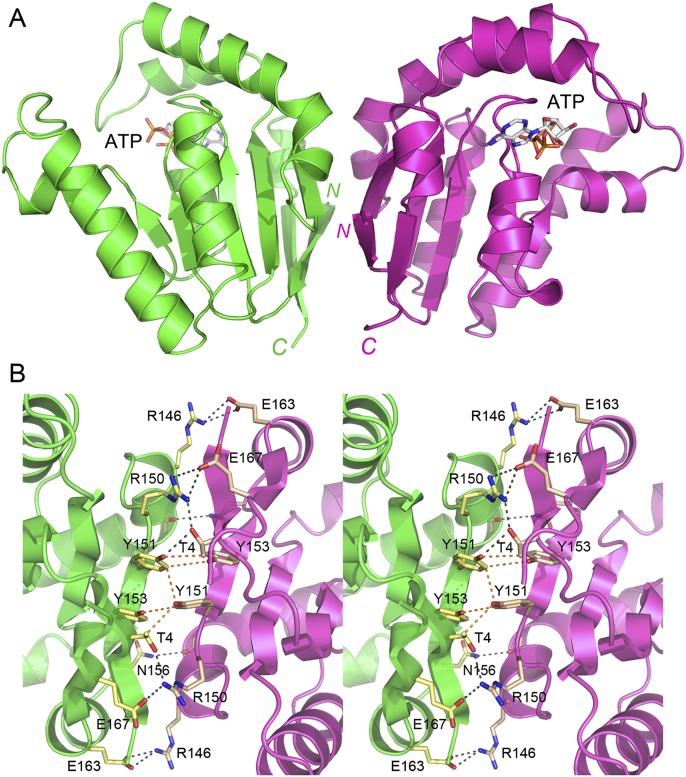
The two protomers in the asymmetric unit of the SeMet kinase crystal form a homodimer, depicted in Figure 3A with the protomer A colored green and protomer B colored magenta. The protomers are related by pseudo twofold symmetry. The subunit interface buries 1882 Å2 of surface area per protomer (calculated in PISA) (Krissinel and Henrick 2007). The two protomers in the asymmetric unit of the native kinase crystals also comprised a homodimer, with an interface of 1662 Å2 of buried surface area per protomer. The SeMet-kinase and native kinase dimers, which crystallized under different conditions and with different unit cell dimensions, superimposed with an RMSD of 0.88 Å at 339 Cα positions. The subunits of the kinase dimer are in a parallel orientation, wherein both C termini project downward in the view shown in Figure 3A. Thus, the phosphatase domains that follow the kinase module in the linear structure of CthPnkp will likely be located near each other on the inferior side of the kinase dimer. A close-up stereoview of the kinase dimer interface is shown in Figure 3B. The cross-protomer interactions entail (1) pseudo-symmetric pairs of van der Waals contacts between Tyr151, Tyr153, and Thr4; (2) pairs of salt bridges from Arg150 to Glu167 and Arg146 to Glu163; and (3) hydrogen bonds from Arg150 to Thr4, and Asn156 to the main-chain carbonyl of Lys142.
FIGURE 3.
The CthPnkp kinase domain crystallized as a homodimer. (A) The two CthPnk-(1-170) protomers in the asymmetric unit of the SeMet-kinase crystal are shown with protomers A and B colored green and magenta, respectively. The ATP in the active site is depicted as a stick model. N and C denote the N and C termini. (B) Stereoview of the dimer interface highlighting cross-protomer atomic contacts. Amino acid side-chains are shown as stick models. Ionic and hydrogen-bonding interactions are indicated by black dashed lines; van der Waals contacts are denoted by orange dashed lines.
The kinase active site
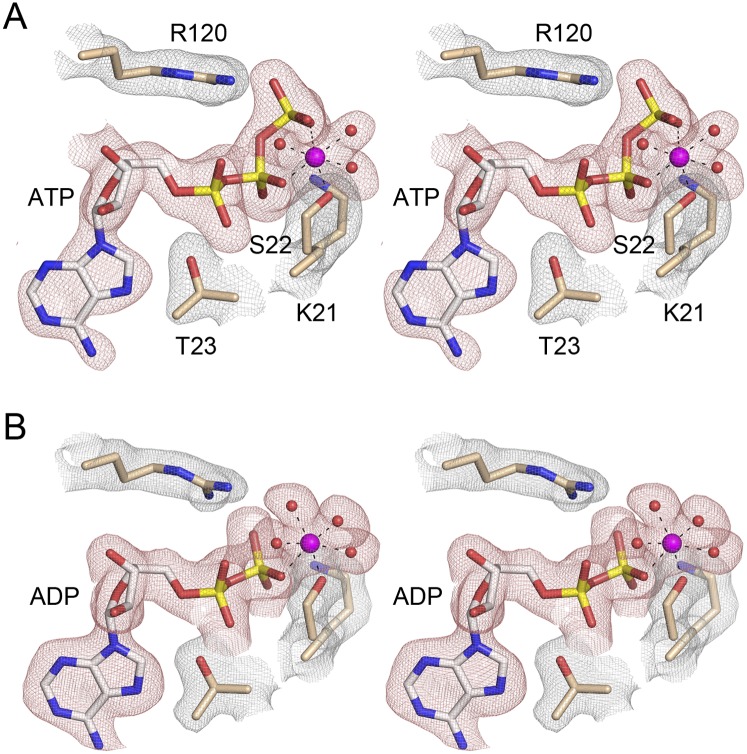
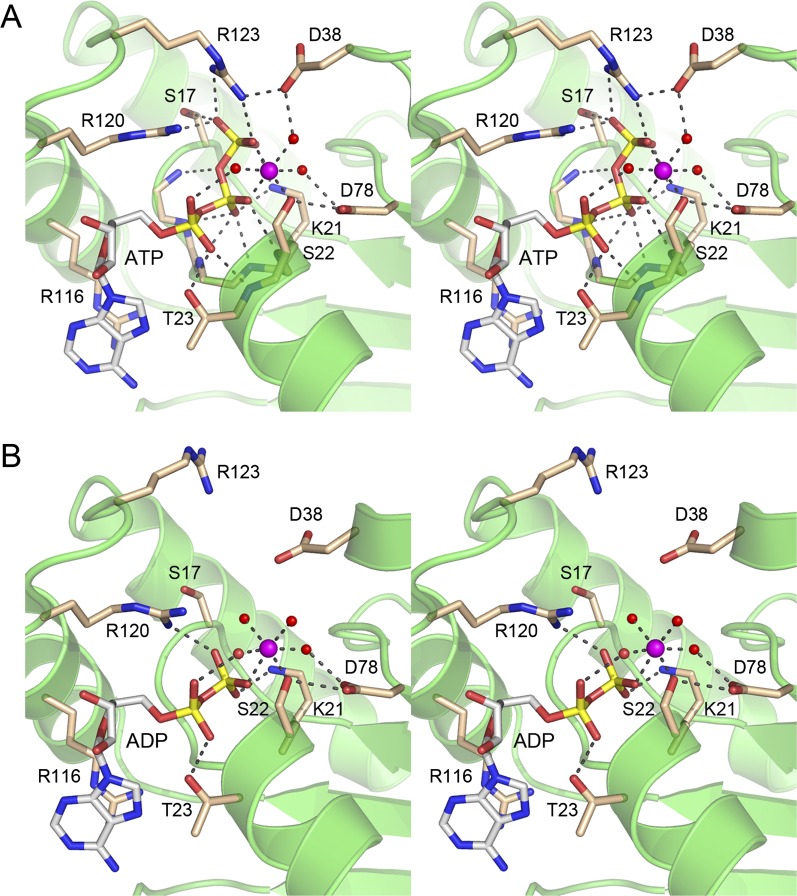
The Fo − Fc maps revealed electron density for ATP and an octahedral metal coordination complex in the active site of SeMet-kinase (Fig. 4A), consistent with the addition of 2 mM ATP and 10 mM MgCl2 to the protein solution prior to crystallization. A detailed view of the active site architecture and the atomic contacts to ATP and Mg2+ in the SeMet-kinase structure is shown in Figure 5A. Six consecutive main-chain amide nitrogens of the 18GSGKST22 P-loop donate hydrogen bonds to the α and β phosphates of ATP. The Thr23 hydroxyl donates a hydrogen bond to the ATP α phosphate. The Lys21 side-chain makes a bifurcated ionic interaction with the β and γ phosphates. In addition, the ATP γ phosphate is anchored by electrostatic interactions with the Arg120 and Arg123 side-chains emanating from the lid and by a hydrogen bond from the P-loop Ser17 side-chain hydroxyl. Two of the sites in the octahedral coordination complex of the Mg2+ ion are occupied by nonbridging β and γ phosphate oxygens of ATP. The other sites are filled by the Ser22 Oγ atom and three waters. Thus, the kinase protein makes only a single direct atomic contact to the metal cofactor. A secondary shell of atomic contacts to the metal-bound waters in the metal complex includes the Asp38 and Asp78 side-chains (Fig. 5A). There are no protein contacts to the ATP ribose sugar. The adenine nucleobase makes a π-cation stack on the Arg116 side-chain of the lid. These features of the ATP•Mg2+ substrate complex in the phosphate donor site suggest a mechanism whereby the protein and the metal orient the γ phosphate for nucleophilic attack by the polynucleotide 5′-OH and stabilize the extra negative charge developed in the pentacoordinate transition state during an associative in-line phosphoryl transfer.
FIGURE 4.
Active site density in the kinase•ATP•Mg2+ and kinase•ADP•Mg2+ complexes. Stereoviews of the finely sampled electron density maps of the active sites of the A protomers of the SeMet-kinase (panel A; 2.0 Å) and native kinase (panel B; 2.1 Å) crystals. The red mesh depicts Fo − Fc difference density (contoured at 3.0 σ; grid spacing 0.13 Å) calculated prior to the inclusion of nucleotide or a metal complex in the model. The difference density was modeled as ATP•Mg2+ for the SeMet-kinase structure and ADP•Mg2+ for the native kinase structure. The nucleotides are depicted as stick models with gray carbons; Mg2+ is shown as a magenta sphere. The contacts to the ligands of the octahedral Mg2+ coordination complex are denoted by dashed lines. Waters in the metal complex are red spheres. The gray mesh depicts the refined 2Fo − Fc density maps (at 1.5 σ; grid spacing 0.13 Å) of P-loop residues Lys21, Ser22, and Thr23 and lid residue Arg120 that are situated close to the ATP/ADP nucleotides.
FIGURE 5.
Atomic contacts in the active site. Stereoviews of the phosphate donor site of the A protomers of the SeMet-kinase•ATP•Mg2+ substrate complex (panel A) and the native kinase•ADP•Mg2+ product complex (panel B). Amino acids and ATP/ADP are shown as stick models with beige and gray carbons, respectively. Mg2+ is depicted a magenta sphere in the center of an octahedral coordination complex. Waters in the metal coordination complex are denoted by red spheres. Atomic contacts are indicated by dashed lines.
The Fo − Fc maps for the native kinase domain revealed electron density for ADP and an octahedral Mg2+ complex in the active site (Fig. 4B), suggesting that the γ phosphate was hydrolyzed during crystal growth. The transition from kinase•ATP•Mg2+ substrate complex to kinase•ADP•Mg2+ product complex elicited changes in the active site (Fig. 5B). For example, Arg123, having lost its bidentate interaction with the γ phosphate, also severed its salt bridge to Asp38, the upshot of which is that the Arg123 side-chain shifts orientation and now points out of the active site in the ADP complex. Also, the Mg2+ ligand site occupied by the γ phosphate oxygen in the ATP substrate complex is filled by a water molecule in the kinase•ADP•Mg2+ structure (Fig. 5B). Other contacts in the active site are unchanged.
Although the polynucleotide 5′-OH phosphate acceptor site is vacant in the CthPnk structures (Supplemental Fig. S2), comparison to the structures of T4Pnk with 5′-OH oligonucleotides bound (Eastberg et al. 2004) highlights conservation of two key constituents of the acceptor site. Asp38 in CthPnk corresponds to T4Pnk residue Asp35, which coordinates the 5′-OH and is thought to serve as a general base catalyst of phosphoryl transfer (Wang and Shuman 2010). Arg41 in CthPnk corresponds to T4Pnk residue Arg38, which coordinates the 5′-terminal phosphodiester of the polynucleotide acceptor strand or its sulfate mimetic (Wang et al. 2002; Eastberg et al. 2004).
Mutational analysis
We tested the effects of 10 single-alanine mutations on the polynucleotide kinase activity of CthPnkp-(1-425). The residues selected for alanine scanning were Lys21 and Ser22 in the P-loop; Asp38 and Arg41 in the predicted polynucleotide phosphoacceptor site; Arg116 and Arg120 in the lid; Asp78, which coordinates two of the metal ligands (Ser22-Oγ and a water); Arg87, a “structural” residue that tethers vicinal secondary structure elements by making a bidentate salt bridge to Glu147 and hydrogen bonds to Ser140-Oγ and the Asn81 and Val82 main-chain carbonyls; Arg150, a constituent of the kinase dimer interface; and Lys95, a surface residue, conserved in T4Pnk, but far from the active site and making no atomic interactions, chosen as a “negative control” for the effects of a presumptively benign change.
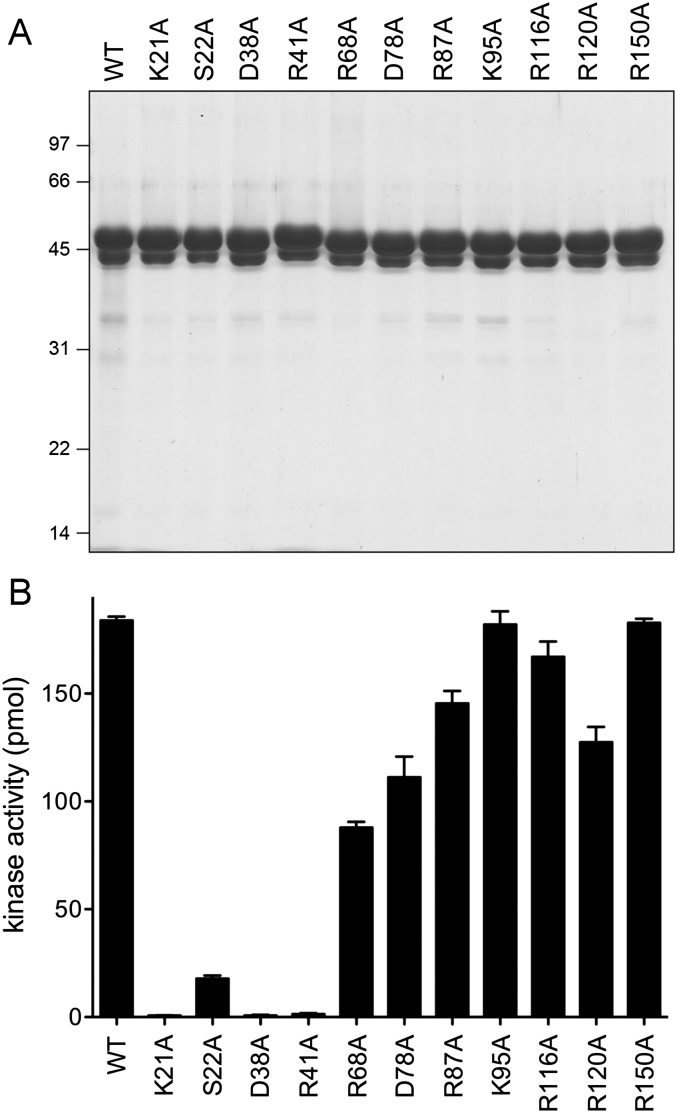
The wild-type and mutated proteins were produced in E. coli as His10 fusions and purified from soluble lysates by Ni-affinity chromatography. SDS-PAGE analysis revealed a predominant ∼48-kDa polypeptide corresponding to the kinase-phosphatase domain CthPnkp-(1-425) (Fig. 6A). As noted previously (Keppetipola and Shuman 2006a), the preparations also contained a ∼44-kDa polypeptide arising from proteolytic cleavage of CthPnkp at a discrete site located near the C terminus of the phosphatase domain (Fig. 6A). The preparations were assayed for kinase activity in reaction mixtures containing 100 ng of input enzyme; this amount of wild-type protein sufficed for near-quantitative radiolabeling of the input 5′-OH DNA oligonucleotide acceptor. Conducting the screening assays in this fashion highlighted the most severe mutational effects on catalysis. The extents of product formation are plotted in the bar graph in Figure 6B. The negative control K95A change was apparently benign, as predicted. Kinase activity was ablated by subtraction of Lys21, whereas elimination of the Ser22 hydroxyl group reduced activity by an order of magnitude. In contrast, replacing lid residue Arg116 with alanine had little impact on the extent of DNA labeling; a similar benign effect was reported for the corresponding R122A mutation of T4Pnk (Wang and Shuman 2002). It would appear that the π-cation stacking of this arginine on the nucleobase is not critical for kinase activity at the ATP concentrations employed in routine biochemical assays, i.e., 100 μM ATP and 25 μM ATP for the bacterial and phage kinases, respectively (the bacterial kinase has an apparent Km of 16 μM ATP) (Martins and Shuman 2005). Mutating lid residue Arg120 to alanine also had little impact. Kinase activity was abolished by alanine substitutions for Asp38 and Arg41, consistent with their postulated roles in orienting and activating the polynucleotide 5′-OH nucleophile. Alanine mutations of Asp78, Arg87, and Arg150 had either no effect or only a modest effect on the extent of product formation (Fig. 6B).
FIGURE 6.
Structure-guided mutagenesis. (A) Aliquots (8 μg) of the Ni-Agarose preparations of the indicated His10CthPnkp-(1-425) proteins were analyzed by SDS-PAGE. The polypeptides were visualized by staining with Coomassie blue dye. The positions and sizes (in kilodaltons) of marker polypeptides are indicated. (B) Kinase reaction mixtures contained 180 pmol of 36-mer 5′-OH DNA oligonucleotide and 100 ng of the indicated His10CthPnkp-(1-425) proteins. The extents of label transfer from [γ32P]ATP to the 36-mer DNA are shown. Each datum in the bar graph is the average of three experiments ±SEM.
Having implicated Lys21, Ser22, Asp38, and Arg41 in catalysis, we gleaned structure–activity relations by testing the effects of conservative substitutions. Lys21 was replaced by arginine and glutamine, Ser22 by threonine, Asp38 by asparagine and glutamate, and Arg41 by lysine and glutamine. The wild-type and conservatively mutated CthPnkp-(1-425) proteins were produced in E. coli as His10 fusions and purified from soluble lysates by Ni-affinity chromatography (Supplemental Fig. S3A). They were then assayed for kinase activity by enzyme titration. Specific activities are plotted in the bar graph in Supplemental Figure S3B. Lys21 proved to be essential, insofar as the K21Q and K21R changes resulted in a greater than 100-fold and 15-fold decrements in activity, respectively. It is likely that an arginine is too large to be accommodated in the CthPnk active site and still make the dual contacts to the β and γ phosphates that are achieved by the P-loop lysine. Threonine could functionally substitute for the P-loop Ser22 (at ∼80% of wild-type activity), consistent with the essential role of the side-chain Oγ atom as a metal ligand. Replacing Asp38 with the isosteric asparagine abolished kinase activity, consistent with its putative role as a general base. Installing a glutamate at position 38 restored kinase activity to ∼6% of wild type, signifying a steric constraint on the longer main-chain to carboxylate linker. Whereas changing Arg41 to glutamine caused a greater than 100-fold activity decrement, introducing a lysine at this position restored activity to one-third of the wild-type level, highlighting the key role of positive charge on the side-chain, consistent with the imputed function of Arg41 in coordinating a phosphodiester in the acceptor strand. Because similar conservative mutational effects were reported for T4Pnk at the P-loop lysine and serine positions, the aspartate general base, and the polynucleotide-binding arginine (Wang and Shuman 2001, 2002), we surmise that the bacterial and phage kinases rely on much the same catalytic strategy.
Concluding remarks
The present study provides new insights to the structure and mechanism of the kinase domain of bacterial Pnkp by capturing the enzyme in complexes with the ATP•Mg2+ substrate and the ADP•Mg2+ product. The core fold of the bacterial kinase and the amino acid constituents of the phosphate donor site are similar to those of T4 polynucleotide kinase. The several published T4 Pnkp structures have ADP (or a sulfate anion) in the donor site (Galburt et al. 2002; Wang et al. 2002; Eastberg et al. 2004; Zhu et al. 2007). There is no metal ion present in the kinase active site of any of the available T4 Pnkp structures, and there is no structure of T4 Pnkp bound to ATP. The CthPnkp kinase•ATP•Mg2+ structure highlights how the metal ion is coordinated between the β and γ phosphates and how the P-loop lysine coordinates a different pair of nonbridging β and γ phosphate oxygens. The bacterial kinase structures reveal that the P-loop serine is the sole enzymic constituent of the octahedral metal complex. (In the available T4 Pnkp structures lacking a metal ion in the kinase active site, the P-loop serine donates a hydrogen bond to the ADP β-phosphate or its sulfate mimetic [Galburt et al. 2002; Wang et al. 2002].)
The core phosphotransferase fold of the bacterial kinase is embellished by a distinctive homodimerization module composed of secondary structure elements derived from the N and C termini of the kinase domain. This dimer interface presumably contributes to the homodimeric quaternary structure of the isolated full-length CthPnkp protein (Martins and Shuman 2005) and the formation of a complete (Pnkp)2(Hen1)2 tetramer (Chan et al. 2009a). The parallel orientation of the kinase domain subunits in the crystal structure, combined with knowledge of the structures of component modules (Chan et al. 2009b; Wang et al. 2012), suggests a model for the (Pnkp)2(Hen1)2 tetramer (Supplemental Fig. S4) in which the central phosphatase domains reside on the same face of the kinase dimer and serve as linkers to pairs of LIG-Hen1 heterodimers. Because the structure of the phosphoesterase domain is not known, we do not rule out the possibility that it too contributes to the quaternary structure of the Pnkp–Hen1 complex.
MATERIALS AND METHODS
Purification of tag-free native CthPnkp-(1-170)
The CthPnkp-(1-170) ORF was amplified by PCR with primers that introduced a BamHI site at the translation start codon and an XhoI site flanking the stop codon. The PCR product was digested with BamHI and XhoI and then inserted into pET28-His10Smt3. The resulting pET28-His10Smt3-Pnkp-(1-170) plasmid was transformed into E. coli BL21(DE3). A 4-L culture derived from a single kanamycin-resistant transformant was grown at 37°C in Luria-Bertani medium containing 60 μg/mL kanamycin until the A600 reached 0.6. The culture was then adjusted to 0.3 mM isopropyl-β-D-thiogalactoside and 2% (v/v) ethanol and incubated for 15 h at 17°C with continuous shaking. Cells were harvested by centrifugation, and the pellet was stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 120 mL of lysis buffer (50 mM Tris-HCl at pH 7.5, 1.2 M NaCl, 25 mM imidazole, 10% glycerol). Lysozyme, PMSF, benzamidine, and Triton X-100 were added to final concentrations of 1 mg/mL, 1 mM, 1 mM, and 0.2%, respectively. After mixing for ∼40 min, the resulting lysate was sonicated to reduce viscosity and insoluble material was removed by centrifugation for 45 min at 17,000 rpm in a Sorvall SS34 rotor. The soluble extract (120 mL; 1.4 g protein) was applied to an 11-mL column of Ni-nitrilotriacetic acid–Agarose (Qiagen) that had been equilibrated with lysis buffer. The column was washed sequentially with 40 mL of buffer A (50 mM Tris-HCl at pH 7.5, 200 mM NaCl, 10% glycerol), 25 mL of buffer A with 100 mM imidazole, and 25 mL of buffer A with 200 mM imidazole. The bound protein was serially step-eluted with buffer A containing 300 mM and 500 mM imidazole. The elution profile was monitored by SDS-PAGE. The peak His10-CthPnkp-(1-170)–containing fractions were pooled (90 mL; 156 mg protein) and supplemented with 0.31 mg of His-tagged Smt3-specific protease Ulp1. The mixture was then dialyzed overnight against 4 L of 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol. SDS-PAGE analysis of the dialysates indicated that the His10Smt3 tag had been cleaved from the kinase domain. The dialysate was then passed over a second Ni-Agarose column (3 mL). The tag-free CthPnkp-(1-170) protein was recovered in the flow-through fraction and adjusted to 5 mM DTT. The kinase domain preparation was concentrated to 12 mg/mL (0.6 mM) by centrifugal ultrafiltration.
Purification of tag-free SeMet-substituted Pnkp-(1-170)
To facilitate structure determination by SAD methods using crystals of SeMet-substituted kinase, we replaced the native Val44, Ile93, and Leu137 side-chains with methionine. Methionine codons were introduced into the CthPnkp-(1-170) ORF by PCR amplification with mutagenic primers. The resulting pET28-His10Smt3-SeMet-kinase expression plasmid was transformed into E. coli B834. A single transformant was inoculated into 5 mL of LB medium containing 60 μg/mL kanamycin and incubated for 7 h at 37°C. The bacteria were harvested by centrifugation and then resuspended in 200 mL of complete LeMaster medium containing 60 μg/mL kanamycin and 50 μg/mL SeMet; after overnight incubation, the culture volume was increased to 4 L with fresh LeMaster medium (+kanamycin +SeMet), and growth was continued at 37°C with constant shaking until the A600 reached 0.6. The culture was adjusted to 0.3 mM isopropyl-β-D-thiogalactoside and 2% (v/v) ethanol and then incubated for 4.5 h at 30°C with continuous shaking. Cells were harvested by centrifugation, and the pellet was stored at −80°C. A soluble extract (125 mL; 1.3 g protein) was prepared as described above. The recombinant His10Smt3-SeMet-kinase was purified by Ni-Agarose chromatography (70 mL, 160 mg protein) and digested with Ulp1 during dialysis as described above. A light precipitate that formed during dialysis was removed by centrifugation. A second round of Ni-Agarose chromatography yielded tag-free SeMet-kinase in the flow-through fraction. This material was adjusted to 5 mM DTT and 80% saturation with ammonium sulfate. The precipitate was collected by centrifugation and resuspended in 2 mL of 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM DTT, 10% glycerol. The protein solution was dialyzed against 200 mL of the same buffer to remove residual ammonium sulfate. The dialysate was clarified by centrifugation to yield a solution of 6.6 mg/mL SeMet-kinase.
Crystallization
The native kinase preparation was adjusted to 2 mM ATP and 10 mM MgCl2 and incubated for 10 min before aliquots of the protein solution (2 μL) were mixed on a cover slip with an equal volume of precipitant solution containing 100 mM HEPES (pH 7.5), 200 mM MgCl2, 30% (v/v) PEG-400. Crystals were grown at 22°C by hanging drop vapor diffusion against a reservoir of the same precipitant solution. Rod-shaped crystals appeared after 2 d; single crystals were frozen directly in liquid nitrogen.
The SeMet-kinase preparation was adjusted to 2 mM ATP and 10 mM MgCl2 and incubated for 10 min before aliquots of the protein solution (2 μL) were mixed on a cover slip with an equal volume of precipitant solution containing 100 mM sodium citrate (pH 5.6), 5 mM DTT, 100 mM MgCl2, 16% (v/v) PEG-3350. Crystals were grown at 22°C by hanging drop vapor diffusion against a reservoir of the same precipitant solution. Needle-shaped crystals appeared the next day; single crystals were transferred to a solution containing 100 mM sodium citrate (pH 5.6), 5 mM DTT, 2 mM ATP, 100 mM MgCl2, 16% PEG-3350, and 15% glycerol prior to freezing the crystals in liquid nitrogen.
Diffraction data collection and structure determination
The structure of the SeMet kinase was solved using single-wavelength anomalous dispersion (SAD) data from a single crystal, as follows. Diffraction data at 2.0 Å resolution were collected at the selenium anomalous peak wavelength at NSLS beamline X25 equipped with a Pilatus 6M detector. Three sweeps of consecutive 0.5° rotations were performed, each with different crystal orientations, exposure times and crystal-to-detector distances. Data from 1860 images were indexed and integrated in HKL2000 and scaled in SCALEPACK. The SeMet-kinase crystals belonged to orthorhombic space group P212121. Data collection statistics are compiled in Supplemental Table S1. Substructure solution and initial phase calculations were performed in PHENIX.AUTOSOL (Adams et al. 2002). Eight selenium sites were located. Density modification was performed in PHENIX.AUTOSOL assuming two protomers per asymmetric unit and 50% solvent content, after which ∼90% of the amino acids in protomers A and B were placed by the auto-build function. The model was iteratively adjusted in COOT (Emsley and Cowtan 2004) and refined in PHENIX without using noncrystallographic symmetry (NCS) restraints. The maps revealed Fo − Fc density for ATP and an octahedral metal coordination complex in the active sites of both protomers, consistent with exposure of the protein to ATP and MgCl2 prior to crystallization. The final 2.0 Å SeMet-kinase structure (Rwork/Rfree of 0.175/0.230) had excellent geometry, no Ramachandran outliers and no large Fo − Fc difference Fourier peaks (Supplemental Table S1).
Diffraction data to 2.1 Å resolution for the native kinase crystal were collected in 370 consecutive 1° rotations with 1-sec exposure times at NSLS beamline X25. Data were indexed and integrated in MOSFLM and scaled with SCALA. The native kinase crystals belonged to orthorhombic space group P212121, but were not isomorphous with the SeMet-kinase crystals. Phases for the native kinase were obtained via molecular replacement in PHASER using a search model that was constructed by removing all nonprotein atoms from the SeMet-kinase structure. Following the placement of the dimer and rigid body refinement in PHASER, the native kinase model was iteratively adjusted in COOT, refined in PHENIX, and subjected as a single group to TLS B-factor refinements in PHENIX without using NCS restraints. The maps revealed Fo − Fc density for ADP and an octahedral metal coordination complex in the active sites of both native kinase protomers. The refinement and model statistics for the final 2.1 Å native kinase structure (Rwork/Rfree of 0.177/0.228) are compiled in Supplemental Table S1.
Pnkp truncations and kinase domain mutations
The pET16-His10CthPnkp-(1-425) expression plasmid encoding a bifunctional kinase-phosphoesterase “end-healing” domain was described previously (Martins and Shuman 2005). Further C-terminal truncations of the ORF at residues 180, 175, 170, and 165 were generated by PCR amplification with antisense primers that introduced new stop codons and a 3′-flanking BamHI site. Missense mutations were introduced into the pET16-His10CthPnkp-(1-425) vector by the two-stage PCR-based overlap extension method. The Pnkp inserts of all plasmids were sequenced to confirm the presence of the desired mutations and the absence of any unwanted coding changes. The pET16-His10Pnkp plasmids were transformed into E. coli BL21(DE3). The recombinant proteins were produced by IPTG-induction for 15 h at 17°C of 100-mL bacterial cultures derived from single ampicillin-resistant transformants. The recombinant His10Pnkp proteins were purified from soluble bacterial lysates by Ni-Agarose chromatography. Protein concentrations were determined by using the BioRad dye binding reagent with bovine serum albumin as the standard.
Polynucleotide kinase assay
Reaction mixtures (10 μL) containing 50 mM Tris-acetate (pH 7.0), 10 mM MgCl2, 5 mM dithiothreitol, 100 μM [γ-32P]ATP, 180 pmol of a synthetic 36-mer 5′-OH DNA oligonucleotide d(CCTGTTCTTATTGGCCTCCTGGCATACCTTTTCCGG), and CthPnkp as specified were incubated for 30 min at 45°C. The reactions were quenched by adding 6 μL of 95% formamide, 20 mM EDTA, 0.05% bromphenol blue, 0.05% xylene cyanol. The mixtures were analyzed by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 M urea in 90 mM Tris-borate, 2.5 mM EDTA. The radiolabeled 36-mer oligonucleotide products were quantified by scanning the gel with a Fujifilm FLA-7000 imaging device.
DATA DEPOSITION
The coordinates for the refined models of kinase•ATP•Mg2+ and kinase•ADP•Mg2+ have been deposited in the RCSB protein structure database (pdb codes 4GP7 and 4GP6).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank NSLS staff members Annie Heroux and Neil Whalen for their assistance with data collection. This research was supported by NIH grant GM42498. S.S. is an American Cancer Society Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.036061.112.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Amitsur M, Levitz R, Kaufman G 1987. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J 6: 2499–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Subbotin R, Chait BT, Shuman S 2012. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci 109: 6072–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Zhou C, Huang R 2009a. Reconstituting bacterial RNA repair and modification in vitro. Science 326: 247 doi: 10.1126/science.1179480 [DOI] [PubMed] [Google Scholar]
- Chan CM, Zhou C, Brunzelle JS, Huang RH 2009b. Structural and biochemical insights into 2′-O-methylation at the 3′-terminal nucleotide of RNA by Hen1. Proc Natl Acad Sci 106: 17699–17704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastberg JH, Pelletier J, Stoddard BL 2004. Recognition of DNA substrates by bacteriophage T4 polynucleotide kinase. Nucleic Acids Res 32: 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Gundllapalli S, Beier H, Söll D 2010. Branchiostoma floridae has separate healing and sealing enzymes for 5′-phosphate RNA ligation. Proc Natl Acad Sci 107: 16834–16839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galburt EA, Pelletier J, Wilson G, Stoddard BL 2002. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure 10: 1249–1260 [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A 2008. Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RH 2012. Unique 2′-O-methylation by Hen1 in eukaryotic RNA interference and bacterial RNA repair. Biochemistry 51: 4087–4095 [DOI] [PubMed] [Google Scholar]
- Jain R, Shuman S 2010. Bacterial Hen1 is a 3′ terminal RNA ribose 2′-O-methyltransferase component of a bacterial RNA repair cassette. RNA 16: 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Shuman S 2011. Active site mapping and substrate specificity of bacterial Hen1, a manganese-dependent 3′ terminal RNA ribose 2′O-methyltransferase. RNA 17: 429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Shuman S 2006a. Mechanism of the phosphatase component of Clostridium thermocellum polynucleotide kinase-phosphatase. RNA 12: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Shuman S 2006b. Distinct enzymic functional groups are required for the phosphomonoesterase and phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase. J Biol Chem 281: 19251–19259 [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Shuman S 2007. Characterization of the 2′,3′ cyclic phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase and bacteriophage lambda phosphatase. Nucleic Acids Res 35: 7721–7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Nandakumar J, Shuman S 2007. Reprogramming the tRNA splicing activity of a bacterial RNA repair enzyme. Nucleic Acids Res 35: 3624–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick L 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Martins A, Shuman S 2005. An end-healing enzyme from Clostridium thermocellum with 5′ kinase, 2′,3′ phosphatase, and adenylyltransferase activities. RNA 11: 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Shuman S, Lima CD 2006. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell 127: 71–84 [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S 2008. RNA repair: An antidote to cytotoxic eukaryal RNA damage. Mol Cell 31: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Sawaya R, Ho CK, Shuman S 2004. Portability and fidelity of RNA-repair systems. Proc Natl Acad Sci 101: 2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Wang LK, Nair PA, Shuman S 2012. The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family. Proc Natl Acad Sci 109: 2296–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SL, Kierzek R, Turner DH, Phizicky EM 1999. Transient ADP-ribosylation of a 2′-phosphate implicated in its removal from ligated tRNA during splicing in yeast. J Biol Chem 274: 2637–2644 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Chakravarty AK, Maughan B, Shuman S 2011. A novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J Biol Chem 286: 43134–43143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Shuman S 2001. Domain structure and mutational analysis of T4 polynucleotide kinase. J Biol Chem 276: 26868–26874 [DOI] [PubMed] [Google Scholar]
- Wang LK, Shuman S 2002. Mutational analysis defines the 5′-kinase and 3′-phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids Res 30: 1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Shuman S 2010. Mutational analysis of the 5′-OH oligonucleotide phosphate acceptor site of T4 polynucleotide kinase. Nucleic Acids Res 38: 1304–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Lima CD, Shuman S 2002. Structure and mechanism of T4 polynucleotide kinase: An RNA repair enzyme. EMBO J 21: 3873–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Chan CM, Christensen D, Zhang C, Selvadurai K, Huang RH 2012. Molecular basis of bacterial protein Hen1 activating the ligase activity of bacterial protein Pnkp for RNA repair. Proc Natl Acad Sci 109: 13248–13253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Smith P, Wang LK, Shuman S 2007. Structure-function analysis of the 3′-phosphatase component of T4 polynucleotide kinase/phosphatase. Virology 366: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]