Analysis of available RNA crystal structures has allowed the authors to identify a new family of RNA arrangements that they call double twist-joints, or DTJs. Each DTJ arrangement is composed of a double helix that contains two bulges incorporated into different strands and separated from each other by 2 or 3 bp. At each bulge, the double helix is over-twisted, while the unpaired nucleotides of both bulges form a complex network of stacking and hydrogen-bonding with nucleotides of helical regions. In total, the authors identified 14 DTJ cases, which can be combined in three groups based on common structural characteristics. Two DTJs form parts of functional centers in the ribosome and in the RNase P.
Keywords: RNA structure, RNA motif, nucleotide bulge, helical over-twist
Abstract
Analysis of available RNA crystal structures has allowed us to identify a new family of RNA arrangements that we call double twist-joints, or DTJs. Each DTJ is composed of a double helix that contains two bulges incorporated into different strands and separated from each other by 2 or 3 bp. At each bulge, the double helix is over-twisted, while the unpaired nucleotides of both bulges form a complex network of stacking and hydrogen-bonding with nucleotides of helical regions. In total, we identified 14 DTJ cases, which can be combined in three groups based on common structural characteristics. One DTJ is found in a functional center of the ribosome, another DTJ mediates binding of the pre-tRNA to the RNase P, and two more DTJs form the sensing domains in the glycine riboswitch.
INTRODUCTION
The twist-joint structures
In the available RNA structures, there are many situations when one double helix stacks on top of another helix in such a way that the two contacting base pairs become over-twisted, compared to the juxtaposition of two neighboring base pairs in the standard A-RNA conformation (Fig. 1A). Henceforth, we name such arrangements twist-joints (TJs). Usually in a TJ, two strands in both helices are directly connected, while the other two strands are separated by an unpaired region or belong to different parts of the polynucleotide chain. In the TJs of this kind, the angle of the over-twist varies between 20° and 70°, and in most cases, it is between 30° and 50°. In this article, we analyze a particular set of RNA structures that are based on a combination of two or more closely positioned TJs.
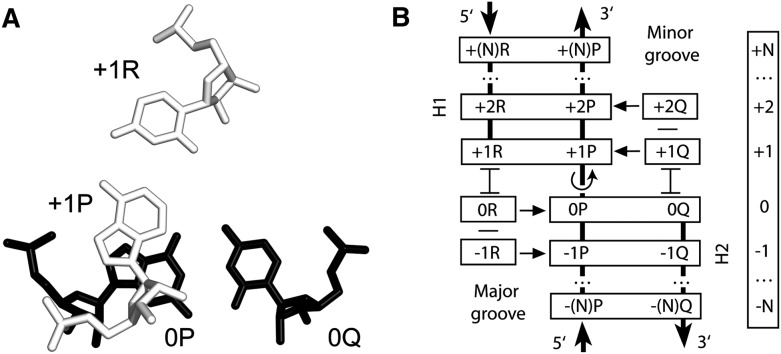
FIGURE 1.
Definition of a TJ arrangement. (A) A typical juxtaposition of 2 bp, [0P;0Q] (black) and [+1P;+1R] (white), forming a TJ. Nucleotide +1P is next to 0P in the polynucleotide chain, while nucleotides 0Q and +1R are not directly connected. Compared to the standard A-RNA conformation, there is an over-twist between 0P and +1P (in the figure, it is ∼35°), which reduces the area of stacking between the 2 nt. (B) Schematic representation of a TJ arrangement. All individual nucleotides (short horizontally oriented rectangles) and base pairs (long rectangles) are positioned in layers. Each layer has its own number, as indicated in the vertical bar on the right. Base pairs [+1R;+1P] and [+2R;+2P] form helix 1 (H1), while base pairs [0P;0Q] and [−1P;−1Q] form helix 2 (H2). The over-twist occurs between base pairs [+1R;+1P] and [0P;0Q], as indicated by the rotating arrow. Unpaired nucleotides +1Q and +2Q stack on 0Q and interact with the minor groove of H1 (indicated by horizontal arrows). Nucleotides 0R and −1R stack on +1R and interact with the major groove of H2. Nucleotides 0R and +1Q make the direct stabilization of the over-twist. Outside double helical regions, the interbase stacking is indicated either by letter “H” rotated for 90° (symbol  ) or by a short horizontal line.
) or by a short horizontal line.
For the description of TJ structures, we will use the nomenclature shown in Figure 1B. The two helices composing a TJ are named H1 and H2. Strand P is common for both helices: Its 5′-half belongs to H2, while its 3′-half is a part of H1. In helix H1, strand P forms base pairs with strand R, while in H2, it does it with strand Q. The inter-helix contact is formed by base pairs [0P;0Q] and [+1R;+1P]. Due to the over-twist between these base pairs, only 1 nucleotide (nt) in each base pair forms a contact with a nucleotide of the other helix (Fig. 1, nucleotides 0P and +1P), while the other 2 nt, +1R and 0Q, are clearly separated from each other. On most occasions, nucleotides +1R and 0Q are connected by a relatively short loop containing no more than 3 nt (data not shown).
Compared to the standard A-RNA conformation, the over-twist between helices H1 and H2 corresponds to the mutual displacement of the contacting base pairs [+1P;+1R] and [0P;0Q] in the direction of the major and minor grooves, respectively (Fig. 1A). Such displacement weakens the interaction between the 2 bp, which thus may require additional stabilization. Indeed, inspection of the TJs existing in the available RNA structures shows that in practically all of them, the juxtaposition of the two helices is stabilized through interaction with other elements of the same RNA molecule. One can identify two different types of stabilization, which we call here direct and indirect.
Direct stabilization of TJ
The direct stabilization is a more common type of stabilization of TJ arrangements. It is associated with presence of an additional nucleotide at the junction between the two helices that stacks on the top of one helix and forms H-bonds with a nucleotide of the other helix. The general scheme of the direct stabilization is shown in Figure 1B. Two nucleotides, 0R and +1Q, are involved in the direct stabilization of the over-twist between base pairs [0P;0Q] and [+1R;+1P]. Nucleotide 0R stacks to helix H1 and forms H-bonds with the major groove of helix H2, while nucleotide +1Q stacks to H2 and forms H-bonds with the minor groove of H1. The additional nucleotides −1R and +2Q can stack on 0R and +1Q, respectively, and form hydrogen bonds with the corresponding base pairs in helices H2 and H1. The presence of −1R and +2Q thus provides additional direct stabilization of the TJ.
As an example of a directly stabilized TJ, we can name the previously described C-loop motif (Lescoute et al. 2005), a typical case of which (arrangement 2AW4-389) is shown in Figure 2, A and B (henceforth, each discussed arrangement will be named by the Protein Data Bank [PDB] code of the structure from which it has been extracted [here, 2AW4] followed by the number of the nucleotide in position 0P [here, 389]). Another case of a directly stabilized TJ is present in the G-ribo motif (Steinberg and Boutorine 2007b), where it favors the formation of G-ribo pseudoknots (Steinberg and Boutorine 2007a). In different TJs, the particular patterns of direct stabilization can differ by the presence or absence of each of the 4 nt 1R, 0R, +1Q, and +2Q; by the position of these nucleotides in the RNA chain; and by the mode of their interaction with helices H1 and H2.
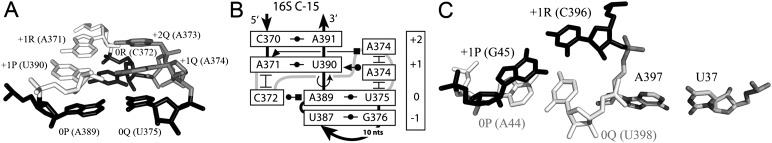
FIGURE 2.
Examples of the direct and indirect stabilization of a TJ arrangement. (A,B) The C-loop motif as an example of a directly stabilized TJ. The tertiary structure (A) and schematic representation (B) of the C-loop motif 2AW4-389, originally defined as 16S C-15 (Lescoute et al. 2005). The motif is extracted from the structure of the E. coli 16S rRNA (PDB 2AW4). For depicting the scheme of internucleotide H-bonding, we use the standard annotation of noncanonical base pairs (Leontis and Westhof 2001). The curved lines show backbone connectivity. (C) An example of indirect TJ stabilization observed in arrangement 2AVY-44. The over-twist between base pairs [+1P;+1R] (black) and [0P; 0Q] (white) is stabilized due to the immediate connection of nucleotides 0R and 0Q to adenosine A397 (gray). The position of A397 is fixed through base-pairing with U37, which comes from another part of the structure.
Indirect stabilization of TJ
Indirect TJ stabilization takes place when the positions of nucleotides 0Q and +1R are constrained through immediate covalent connection to nucleotides that are firmly integrated with other parts of the same RNA structure. An example of indirect TJ stabilization represents arrangement 2AVY-44 found in the 30S subunit of the Escherichia coli ribosome (Fig. 2C). In this case, adenosine A397, which intervenes between nucleotides 0Q (U398) and +1R (C396), interacts with uridine U37 coming from another part of the 16S rRNA structure. The fixation of the position of A397 through base-pairing with U37 indirectly stabilizes the positions of both flanking nucleotides 0Q and +1R, thus stabilizing the over-twisting between base pairs [0P;0Q] (base pair [A44;U398]), and [+1P;+1R] ([G45;C396]). Although in the available RNA structures there are many indirectly stabilized TJs, this type of stabilization is less common than the direct one.
Double TJs
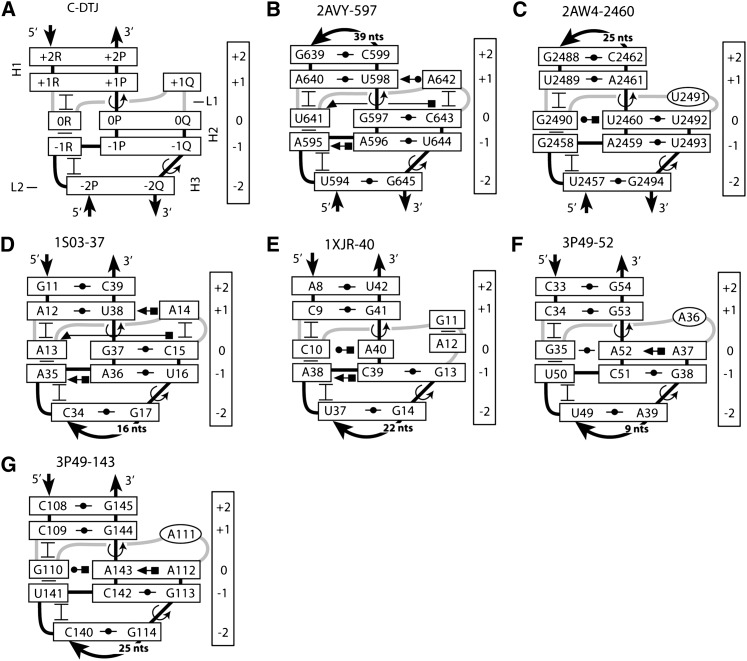
Among TJs occurring in the known RNA structures, one can distinguish a special class of arrangements in which two TJs are present in the same duplex, being separated from each other by only a few base pairs. We call such arrangements double TJs (DTJs). We found more than a dozen such cases in the available RNA structures. As demonstrated below, the essential feature of DTJs that distinguishes them from the merger of two individual TJs is the presence of additional stabilizing interactions between the two TJs. More specifically, due to the close position of the two TJs, each DTJ contains nucleotides that simultaneously stabilize both of them. We thus can tell about a common strategy for stabilization of both TJs within the same DTJ.
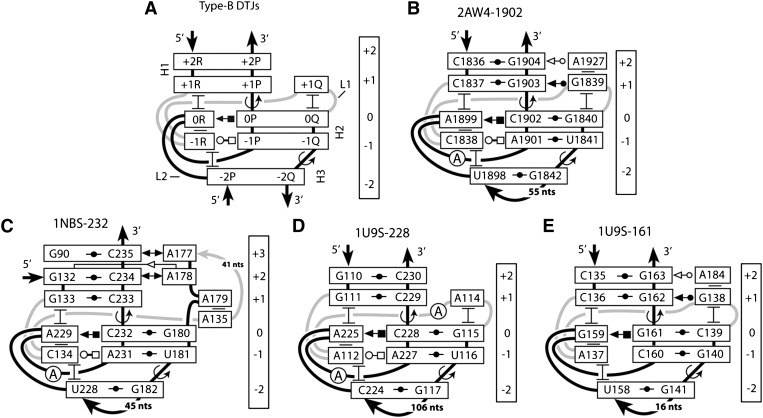
In the identified DTJs we found three different types of over-twist stabilization, which we name A, B, and C. All three types conform to the same pattern of secondary structure (Fig. 3), which will be used for their description. The pattern consists of three consecutive double helices H1, H2, and H3, forming two TJs—TJ1 and TJ2—and having two loops—L1 and L2. In spite of the apparent symmetry of this pattern, the actual DTJs are essentially asymmetric. First, one TJ always has a higher amplitude of over-twist; we call it TJ1 and note that it is always stabilized directly in the major groove and in some instances in the minor groove. The other TJ, designated TJ2, may lack direct stabilization entirely or be stabilized directly only in the major groove; it is never stabilized in the minor groove. For identification of layers in the whole DTJ structure, we use the numbering system shown in Figure 1B, by linking it to TJ1. In the following sections, the description of the identified motifs precedes the discussion of the strategy that we used for their identification.
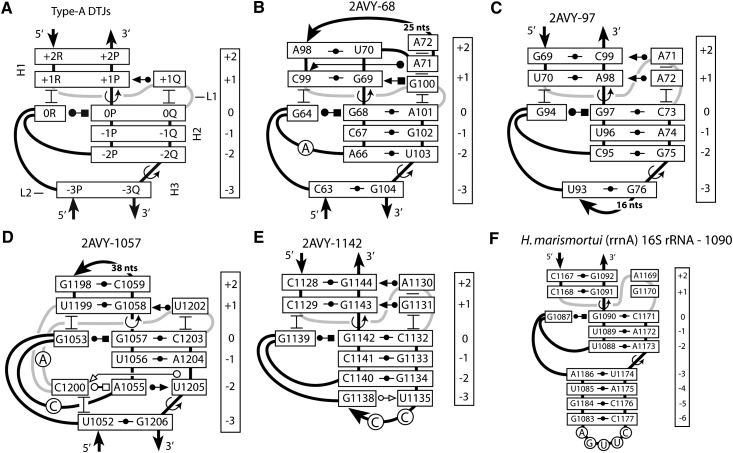
FIGURE 3.
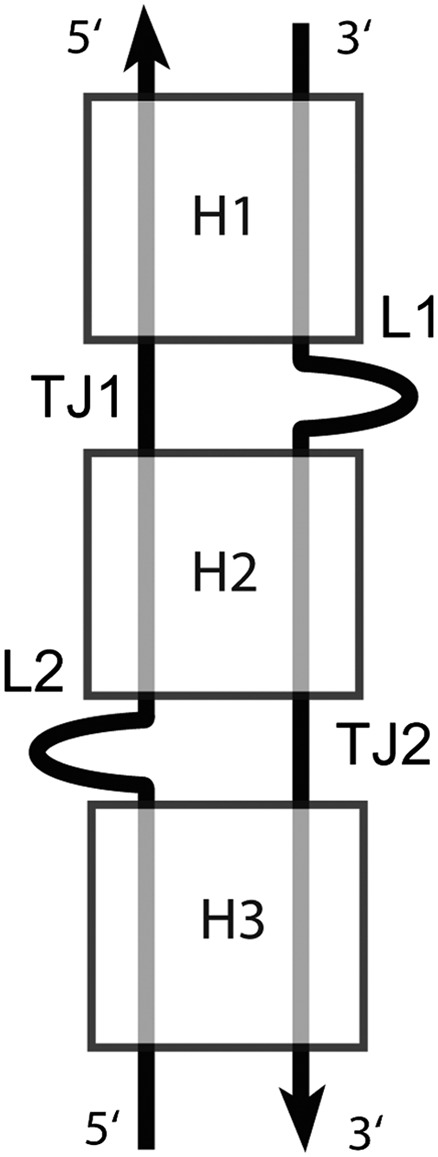
The secondary structure of a DTJ arrangement. A DTJ is composed of three consecutive double helices, H1, H2, and H3. In all known DTJs the central helix H2 contains either 2 or 3 bp. At both ends, H2 is flanked by helices H1 and H3, which, to be qualified as such, should contain at least one WC or GU base pair. At the junction points between the helices, there are two over-twists, which occur in the opposite strands. One over-twist (TJ1) has a higher amplitude than the other one (TJ2); this difference is used here for alignment of different DTJs.
RESULTS
A-double twist joints
A-double twist joints (A-DTJs) are defined as those DTJs in which helix H2 consists of 3 bp. In the available RNA structures, we found three A-DTJ arrangements, all in the small ribosomal subunit. The A-DTJs occurring in the E. coli ribosome are shown in Figure 4, B through D. All these arrangements contain nucleotides 0R and +1Q, which are, respectively, the first nucleotide of loop L2 and the last nucleotide of loop L1. Both nucleotides play important roles in stabilization of the TJ1.
FIGURE 4.
Structure of A-DTJ motifs. (A) The general scheme of the A-DTJ motif. TJ1 and TJ2 occur between base pairs [0P;0Q] and [+1R;+1P] and between base pairs [−2Q;−2P] and [−3Q;−3P], respectively. (B–D) The three A-DTJ motifs found in the E. coli 16S rRNA (Schuwirth et al. 2005). (E) The quasi-A-DTJ arrangement 2AVY-1142, which has the same structure as all other A-DTJs except that it does not have helix H3. (F) Reconstruction of motif 2AVY-1142 in the Haloarcula marismortui 16S rRNA (Wuyts et al. 2004). Circled nucleotides do not stack to other nucleotides of the motif.
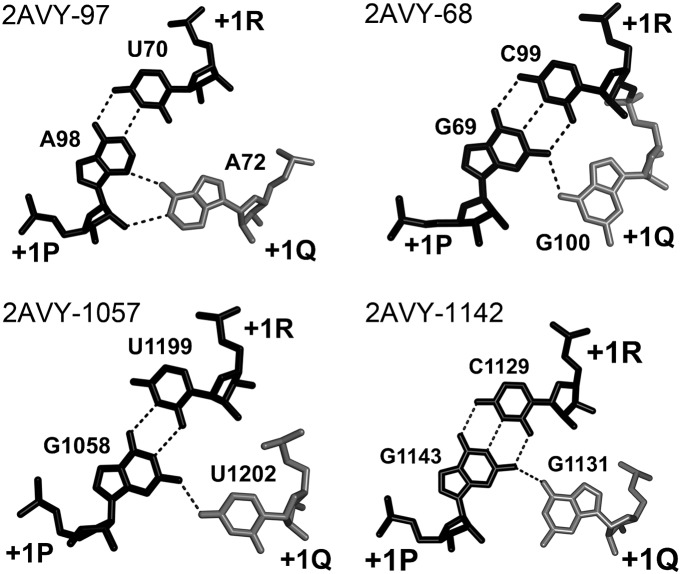
Nucleotide +1Q forms a hydrogen bond with purine +1P. Although in the A-DTJs shown in Figure 4, +1Q has different identities G, A, and U, its interaction with nucleotide +1P seems to be conserved. The identities of +1Q and +1P covary in the way that G and U in +1Q correspond to G in +1P, while A in +1Q corresponds to A in +1P. Such covariation always allows the formation of close-to-isosteric dinucleotide arrangements cis-WC-SE with a hydrogen bond between the two bases (Fig. 5).
FIGURE 5.
A-DTJ: structure of layer +1. In all A-DTJs, nucleotide +1Q (white) interacts with the minor groove of base pair [+1P;+1R] (black), making either the cis WC/SE (2AVY-97, 2AVY-1142) or cis HG/SE (2AVY-68 and 2AVY-1057) juxtaposition with +1P. Although in all motifs, the base of nucleotide +1Q is positioned in vicinity of the ribose +1P, only in motif 2AVY-97 are the two entities close enough to form an H-bond.
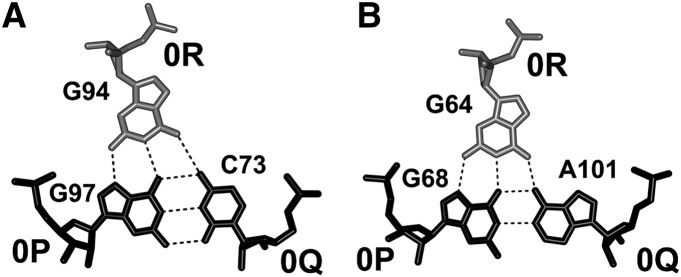
In all three A-DTJs existing in the E. coli ribosome, guanosine 0R stacks on pyrimidine +1R and forms two hydrogen bonds with guanosine 0P (Fig. 6). It also forms a hydrogen bond with nucleotide 0Q, which is either cytidine or adenosine. This interaction firmly attaches 0R to the whole helix H2. The particular conformation of the polynucleotide chain between nucleotides 0R and 0P in two A-DTJs, 2AVY-68 and 2AVY-1057, corresponds to the previously analyzed lonepair triloop motif (Lee et al. 2003). In the third A-DTJ, 2AVY-97, the conformation of this region is different due to the absence of the bulged nucleotide between 0R and −2P. The rigidity of the arrangement of 0R with the whole helix H2 virtually freezes the juxtaposition of nucleotides 0R and −2Q. The latter nucleotides are directly connected to −3P and −3Q, which form the first base pair of helix H3. As a result, the position of base pair [−3P;−3Q] becomes constrained with respect to the whole helix H2. We can thus say that in the A-DTJ structure, nucleotide 0R plays a dual role: Not only does it directly stabilize the TJ1, but also it provides for indirect stabilization of the TJ2.
FIGURE 6.
A-DTJ: structure of nucleotide triple [0R;0P;0Q] existing in the bacterial ribosome. In all three A-DTJs and in the quasi-A-DTJ 2AVY-1142, guanosine 0R (white) interacts with the major groove of base pair [0P;0Q] (black). In motifs 2AVY-97, 2AVY-1057, and 2AVY-1142, base pair [0P;0Q] is GC (A), while in motif 2AVY-68, it is GA (B). The interbase hydrogen bonds are shown by dotted lines.
Finally, our analysis showed that the interaction of 0R with base pair [0P;0Q] as it is formed in A-DTJs (Fig. 6) requires that helix H2 contain strictly 3 bp and is not possible if helix H2 is either longer or shorter than that. Although in the situation when helix H2 contains only 2 bp, nucleotide 0R is able to reach the major groove of base pair [0P;0Q] (see Supplemental Fig. S1); the mutual orientation of 0R and [0P;0Q] in this case would be inappropriate for formation of more than one H-bond. In another extreme case, when helix H2 contains 4 bp, base pair [0P;0Q] becomes too far from nucleotide 0R for any reasonable interaction (data not shown). These aspects make the pattern of DTJ stabilization observed in A-DTJs specific to this type of DTJ arrangements.
A quasi-A-DTJ arrangement
In addition to the three A-DTJ motifs discussed above, our screening of the available RNA structures revealed another arrangement (2AVY-1142) (Fig. 4E) whose structure in many aspects is similar to that of the A-DTJs but does not contain helix H3, which is replaced by a tetraloop. The absence of helix H3 leaves no place for a TJ2 and does not allow us to qualify 2AVY-1142 as a double-twisted arrangement. In a following section we, however, argue that even though in the available structures of the 30S ribosomal subunit arrangement 2AVY-1142 does not form an A-DTJ motif, it may do so in other organisms.
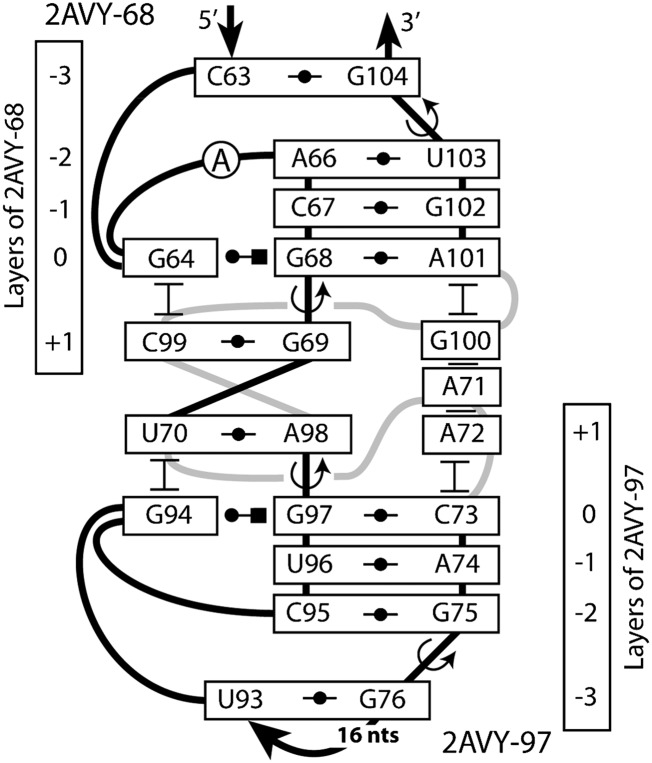
The conglomerate of 2AVY-68 and 2AVY-97
In the tertiary structure of 16S rRNA, the two A-DTJs 2AVY-68 and 2AVY-97 are located so closely to each other that they form a common arrangement (Fig. 7). In this arrangement, the two motifs are positioned head-to-head, so that helix H1 becomes common. More specifically, base pair [+2R;+2P] of 2AVY-97 serves as [+1P;+1R] in 2AVY-68 and vice versa. Two nucleotides of 2AVY-97, +1Q (A72) and +2Q (A71), as well as nucleotide +1Q (G100) of 2AVY-68 form a stack, which interacts with the minor groove of the common helix H1. This stack is flanked by nucleotides 0Q of both motifs, which reinforces the direct stabilization of the two adjacent TJ1 over-twists.
FIGURE 7.
A-DTJ: the common head-to-head arrangement of 2AVY-68 and 2AVY-97. The double helix composed of base pairs G69-C99 and U70-A98 plays the role of helix H1 for each of the two motifs. Nucleotides A72, A71, and G100 form a continuous stack, which interacts with the minor groove of helix H2, thus stabilizing the TJ1 of both A-DTJs.
Evolutionary conservation of A-DTJs
As we describe in detail in Supplemental Text S1, the four identified A-DTJs demonstrate all four possible patterns of conservation, in which each of the two TJs can be conserved or nonconserved independently of the other TJ. The most conserved A-DTJ motif is 2AVY-1057, which exists in virtually all living organisms, while three other motifs demonstrate a notably lower degree of conservation. First, they are observed only in prokaryotes, and second, none of them is formed in all prokaryotes. In motifs 2AVY-1142 and 2AVY-68, only one of the two TJs, respectively, TJ1 and TJ2, is present in all prokaryotes, while in motif 2AVY-97, none of the TJs is conserved. The absence of conservation is always associated with location of the corresponding TJ on the surface of the 30S subunit. The possibility for conservation of only one of the two TJs demonstrates that although within an A-DTJ arrangement the two TJs cooperate with each other, the level of interdependence is not critical, so that, in principle, each of the two TJs can appear individually, without the other TJ.
B-DTJs
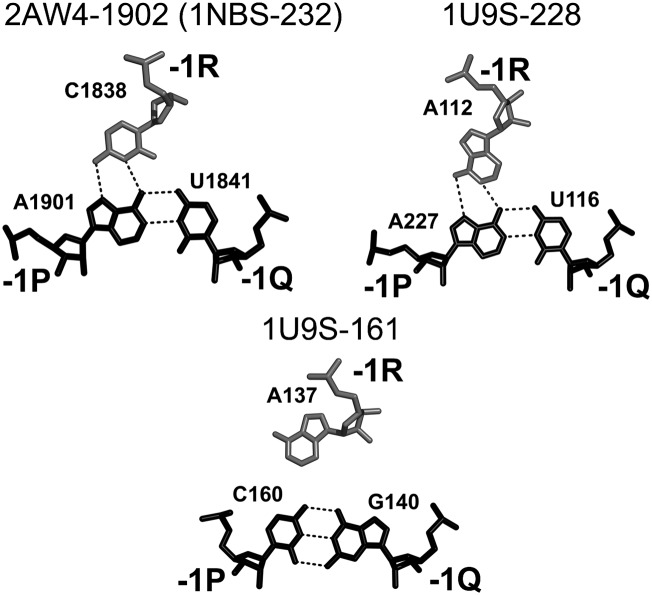
In the available RNA structure, we also found DTJ arrangements in which the number of base pairs in helix H2 is different from three. In all such cases, however, helix H2 contains only 2 bp. The shortening of helix H2 makes the type of stabilization observed in A-DTJs impossible. Correspondingly, the new DTJs use alternative strategies for stabilization of both TJs. Based on the particular scheme of stabilization, we divide all these arrangements in two groups B and C. The B-group is composed of those DTJs in which helix H2 contains 2 bp while 0R belongs to loop L2 (Fig. 8A). In the available RNA structures, we identified four B-DTJs, which are shown in Figure 8, B through E. Motif 2AW4-1902 is present in 23S rRNA, while the other three motifs are found in the RNase P.
FIGURE 8.
Structure of B-DTJ motifs. (A) The general scheme of the B-DTJ motif. The TJ1 and TJ2 occur between base pairs [0P;0Q] and [+1R;+1P] and between base pairs [−1Q;−1P] and [−2Q;−2P], respectively. (B–E) Motifs 1U9S-161 and 1U9S-228 exist in the T. thermophilus RNase P (Krasilnikov et al. 2004), motif 1NBS-232 exists in the Bacillus subtilis RNase P (Krasilnikov et al. 2003), while motif 2AW4-1902 exists in the E. coli 23S rRNA (Schuwirth et al. 2005). In the presented structures, base pairs [+2P;+2R], [+1P;+1R], [0P;0Q], and [−1P;−1Q] are always WC, while base pair [−2P;−2Q] is either UG or CG. Nucleotides 0R and 0P form a cis-SE-HG base pair. Nucleotide +1Q, if present, forms either a cis-HG-SE or cis-WC-SE base pair with nucleotide +1P. Nucleotide −1R in structures B through D forms a trans-HG-WC base pair with −1P.
Although the strategies of stabilization of B- and A-DTJs have some similarities, in many aspects they are essentially different. The most similar detail pertains to the stabilization of TJ1 in the minor groove. In the B-DTJs 2AW4-1902 and 1U9S-161, the first nucleotide of loop L1 occupies position +1Q and forms an H-bond with the minor groove of base pair [+1R;+1P] in about the same way as in A-DTJs (cf. Fig. 5 and Supplemental Fig. S3A,C). However, in the other two B-DTJs 1U9S-228 (Supplemental Fig. S3B) and 1NBS-232 (Fig. 8C), nucleotides +1Q and +1P do not form any contact at all, which suggests that for B-DTJs, the presence of this interaction is not critical.
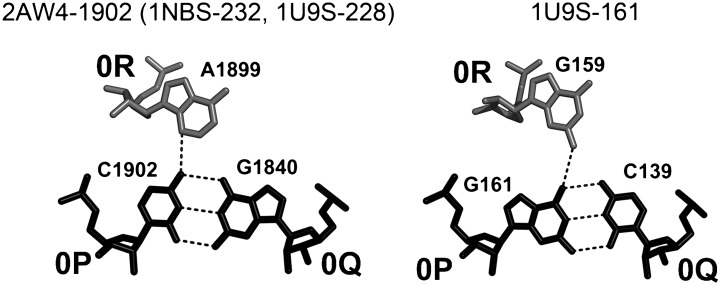
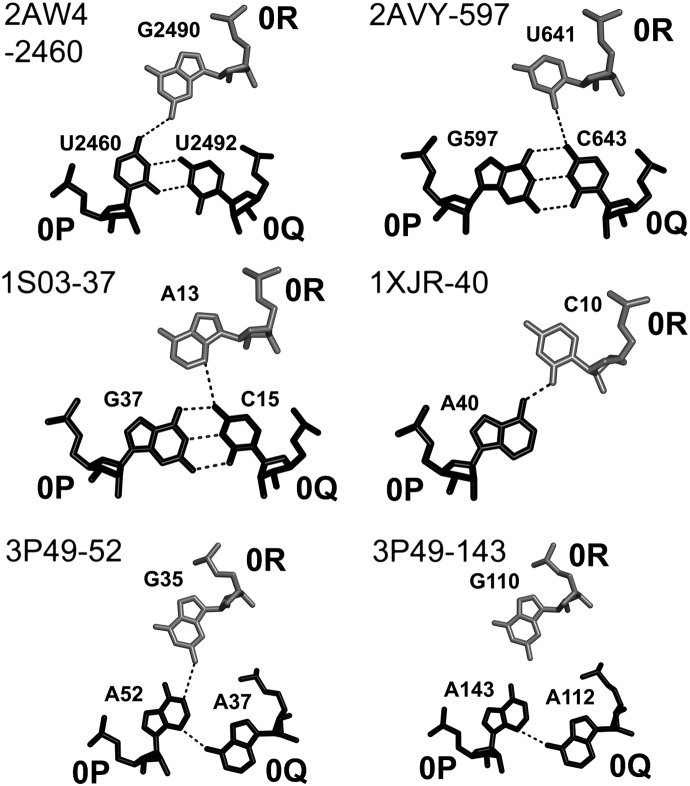
Also, like in A-DTJs, the first nucleotide of loop L2 occupies position 0R and interacts with the major groove of base pair [0P;0Q]. However, due to the shorter helix H2, this interaction in B-DTJs is essentially different from that in A-DTJs. The juxtapositions of 0R and [0P;0Q] found in B-DTJs are shown in Figure 9. In all four motifs, 0R forms only one H-bond with [0P;0Q], in contrast to three H-bonds observed in A-DTJs (Fig. 6). This only H-bond is formed between 0R and 0P and is maintained in all B-DTJs due to the fact that in all of them combination [0R;0P] is either AC or GG. The presence of this H-bond ensures the universal position of 0R with respect to base pair [0P;0Q].
FIGURE 9.
B-DTJ: interaction of nucleotide 0R (white) with the major groove of base pair [0P;0Q] (black). The juxtaposition of 0R and 0P can be described as base pair cis-SE-HG. In all cases, there is one hydrogen bond between 0R and 0P. Compared to the equivalent interaction in A-DTJs, the position of base pair [0P;0Q] with respect to nucleotide 0R is under-twisted, which is due to a shorter helix H2 (see also Supplemental Fig. S1).
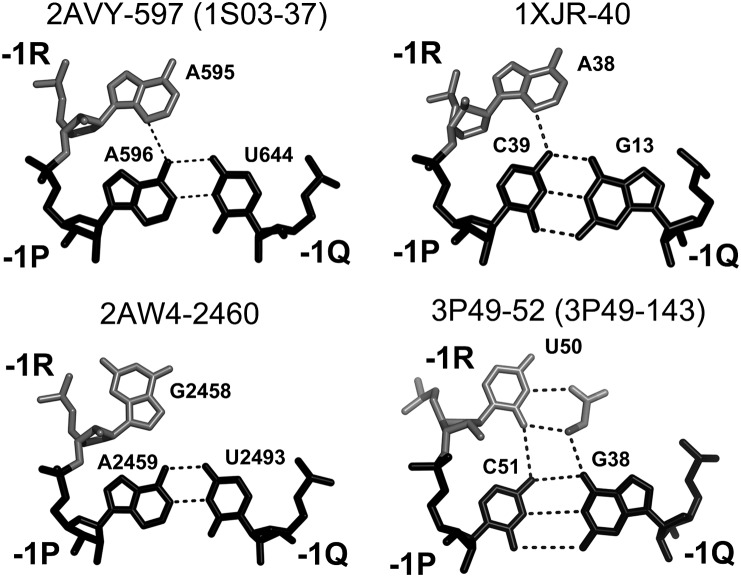
Given that the level of stabilization of TJ1 provided by nucleotide 0R is lower in B-DTJs than in the A-DTJs, it is not surprising that in B-DTJ one can always find an additional mode of stabilization not observed in A-DTJs. All B-DTJs contain a nucleotide in position −1R, which is the first nucleotide of loop L1 (Fig. 8). At the tertiary structure level, this nucleotide is stacked between 0R and −2P and can potentially form H-bonds with base pair [−1P;−1Q]. Indeed, in three B-DTJ motifs, 2AW4-1902, 1NBS-232, and 1U9S-228, −1R forms a trans-WC-HG base pair with −1P (Fig. 10). Due to the presence of adenosine in position −1P and either cytidine or adenosine in position −1R of all three B-DTJs, nucleotides −1R and −1P always form two H-bonds.
FIGURE 10.
B-DTJ: interaction of nucleotide −1R with base pair [−1P;−1Q]. In all B-DTJs, nucleotide −1R is either adenosine or cytosine. The arrangement of nucleotides at level −1 in motif 1NBS-232 is practically identical to that of 2AW4-1902. In motif 1U9S-161, nucleotide −1R (A137) is located too far from −1P (C160) to make any interaction.
Interestingly, we found that cytidine and adenosine in position −1R are not freely interchangeable. Our modeling experiments showed that the trans-WC-HG base pair between cytidine −1R and adenosine −1P (Fig. 10) brings the ribose of −1R close enough to +1Q, so that the 2 nt can be directly connected (motifs 2AW4-1902 and 1NBS-232). However, when cytidine −1R is replaced by adenosine, its ribose becomes positioned too far from +1Q, so that the connection between −1R and +1Q will require an additional intervening nucleotide (nucleotide A113 in motif 1U9S-228) (Fig. 8D).
In the fourth B-DTJ motif 1U9S-161, the identity of nucleotide −1P (C160) does not allow the formation of a stable [−1R;−1P] interaction (Fig. 10). Correspondingly, this B-DTJ contains no additional nucleotide between −1R and +1Q, even though −1R is adenosine. The example of motif 1U9S-161 demonstrates that the properly formed tertiary base pair [−1R;−1P], like the discussed above base pair [+1P;+1Q], is not critically important for the integrity of B-DTJs. We noticed, however, that although none of the triples at levels +1 and −1 exists in all B-DTJs, at least one of them is present in each B-DTJ. This observation allows us to suggest that even though +1 or −1 triple per se is not critical for the integrity of the motif, the presence of at least one of them is essential for the B-DTJ stability.
Each of the 2 nt 0R and −1R directly stabilizes one of the two TJs (for −1R, it is true in all cases except 1U9S-161) and, indirectly, the other TJ. In addition, the intercalation of nucleotide −1R between 0R and −2P provides B-DTJs with a new mode of stabilization inexistent in A-DTJs. In all B-DTJs, 4 nt—+1R, 0R, −1R, and −2P—form a stack, within which the positions of all 4 nt become stabilized through the contacts with its neighbors. The existence of such a 4-nt stack would directly stabilize the juxtaposition of base pairs [+1R;+1P] and [−2P;−2Q] located at the opposite ends of the DTJ arrangement. In view of such stabilization, the involvement of +1Q and −1R in interaction with, respectively, base pairs [+1R;+1P] and [−1P;−1Q] becomes of a secondary importance, which explains the absence of one of these interactions in some B-DTJs.
The 4-nt stack [+1R;0R;−1R;−2P] is additionally stabilized by two specific hydrogen bonds between the backbone of 0R and the base of −1R and between the backbone of −1R and the base of 0R (see Supplemental Fig. S4). The acceptor of each hydrogen bond is always one of three atoms O4′, O5′, or OP2 of the corresponding nucleotide, while the donor is the amino group of either cytidine or adenosine. The latter aspect explains the fact that in the identified B-DTJs, nucleotides 0R and −1R are almost exclusively adenosines and cytidines (Fig. 8). The only exception pertains to motif 1U9S-161, in which nucleotide 0R is guanosine. Consequently, this nucleotide is the only one out of all 0R and −1R nucleotides that does not form a hydrogen bond with the backbone of the opposite nucleotide (Supplemental Fig. S4).
Analysis of the available nucleotide sequences of motifs 2AW4-1902, 1U9S-228, and 1NBS-232 showed that all three of them are highly conserved in evolution (Supplemental Text S1). For motif 1U9S-161, information concerning its conservation is currently unavailable.
A B-DTJ-like element in the A-DTJ 2AVY-1057
One of the discussed above A-DTJs, motif 2AVY-1057, contains an interesting detail that is normally seen in B-DTJs. In this motif, unlike in all other A-DTJs, the first nucleotide of loop L1 (cytidine 1200) (Fig. 4D) is positioned between nucleotide 0R (G1053 in 16S rRNA) and base pair [−3P;−3Q] (U1052-G1206) and forms the trans-WC-HG base pair with adenosine −2P. Although due to such position, C1200 looks like nucleotide −1R in B-DTJs, the two situations are different. While in B-DTJs, nucleotide −1R stacks on both 0R and −2P, in the A-DTJ 2AVY-1057, due to the presence of the additional base pair in helix H2, the niche between 0R and [−3P;−3Q] becomes too big to allow nucleotide C1200 to simultaneously stack on 0R and −3P. In reality, the base of C1200 occupies position −2R and stacks only on −3P (U1052), leaving space between itself and nucleotide 0R (G1053). This example shows that for a stable stacked position of nucleotides 0R and −1R in a DTJ, helix H2 should contain strictly 2 bp (Supplemental Fig. S1). The longer distance between nucleotides +1Q and −2R also explains the existence of the bulged nucleotide A1201, which is here required for connection between nucleotides +1Q and −2R.
C-DTJs
The last group of DTJs, which we call C-DTJs, harbors six motifs (Fig. 11). The secondary structures of these motifs are virtually identical to those of the B-DTJs. Like in B-DTJs, helix H2 in C-DTJs consists of 2 bp, while the last nucleotide of loop L1 occupies position +1Q. The major difference between the two groups is that while in B-DTJs, −1R and 0R belong, respectively, to loops L1 and L2, in C-DTJs the situation is opposite: 0R is the last nucleotide of loop L1, while −1R is the first nucleotide of loop L2. A C-DTJ arrangement can thus be presented as a B-DTJ in which nucleotides 0R and −1R exchange in their positions (cf. Figs. 8A and 11A).
FIGURE 11.
Structure of C-DTJ motifs. (A) The general scheme of the C-DTJ motif. Like in B-DTJs, TJ1 and TJ2 are formed between base pairs [0P;0Q] and [+1R;+1P] and between base pairs [−1Q;−1P] and [−2Q;−2P], respectively. (B–G) The motifs 2AVY-597 and 2AW4-2460 are taken from the E. coli 16S and 23S rRNA, respectively (Schuwirth et al. 2005). Motif 1S03-37 is present in the mRNA of the spc operon of E. coli (Merianos et al. 2004). Motif 1XJR-40 exists in the RNA element of the stem–loop II motif (s2m) of the genome of the SARS virus (Robertson et al. 2005). Motifs 3P49-52 and 3P49-143 are found in the structure of the glycine riboswitch (Butler et al. 2011). In the presented structures, base pairs [+2P;+2R], [+1P;+1R], and [−1P;−1Q] are WC, while [−2P;−2Q] can be either UG or CG. Nucleotide 0R makes a triple with base pair [0P;0Q]. Nucleotide +1Q, if present, forms a cis-WC-SE base pair with nucleotide +1P. In structures B, D, and E, nucleotide −1R forms a trans-HG-WC base pair with −1P.
Two C-DTJ arrangements exist in the ribosome, one in 16S rRNA and the other one in 23S rRNA (motifs 2AVY-597 and 2AW4-2460) (Fig. 11B,C; Schuwirth et al. 2005). The third C-DTJ, 1S03-37, is present in the mRNA of operon spc complexed with the ribosomal protein S8 (Fig. 11D; Merianos et al. 2004), while the fourth one, 1XJR-40, is found in the stem–loop II motif (abbreviated as s2m) of the RNA element in the genome of the SARS virus (Fig. 11E; Robertson et al. 2005). The last two motifs, 3P49-52 and 3P49-143, are identified in the crystal structure of the glycine riboswitch (Butler et al. 2011).
Inspection of the C-DTJs presented in Figure 11, B through G, shows that some interactions in these structures are even less pronounced than in B-DTJs. In particular, nucleotide +1Q forms a triple with base pair [+1R;+1P] only in the two motifs 2AVY-597 and 1S03-37, where this triple has the same structure as in the A- and B-DTJs (Supplemental Fig. S3E). However, in the other four motifs, this interaction does not exist at all.
The differences between B- and C-DTJs exist at other levels as well. Thus, in the two motifs 2AVY-597 and 1S03-37 base pair [0P;0Q] is WC, while in the other C-DTJs, one observes a situation never seen in either A- or B-DTJs. In 2AW4-2460, base pair [0P;0Q] is U-U; in the riboswitch-derived motifs 3P49-52 and 3P49-143, it is a cis-HG-SE (sheared) base pair A-A, while in 1XJR-40, nucleotides 0P and 0Q do not interact at all. Despite such abnormalities, nucleotide 0R interacts with the major groove of base pair [0P;0Q] or with an unpaired nucleotide 0P (in 1XJR-40) in all C-DTJs except 3P49-143, where existence of this interaction is questionable (see Fig. 12). Also, despite the visible variation among C-DTJs in the structure of the zero-level, in all cases nucleotide 0R occupies about the same position. Interestingly, because in C-DTJs, nucleotide 0R comes from loop L2 and not from loop L1, as in B-DTJs, its position is flip-flopped compared with that in B-DTJs. Using structural language, one can say that in C-DTJs, nucleotide 0R is locally parallel to 0Q and not to 0P, as in B-DTJs. As a result, triple [0R;0P;0Q] in C-DTJs (Fig. 12) has an essentially different structure compared with B-DTJs (Fig. 9) and looks more like the B-DTJ triple [−1R;−1P;−1Q] (Fig. 10).
FIGURE 12.
C-DTJ: interaction of nucleotide 0R with the major groove of base pair [0P;0Q]. In the motifs 2AW4-2460, 1XJR-40, and 3P49-52, nucleotide 0R makes the cis-SE-WC base pair with nucleotide 0P, while in the motifs 2AVY-597 and 1S03-37, nucleotide 0R makes the cis-SE-HG base pair with nucleotide 0Q. Unlike in B-DTJs, nucleotide 0R in C-DTJs is locally parallel to 0Q and is anti-parallel to 0P.
In C-DTJs, the nucleotide arrangement at level −1 seems to be more conserved than at levels +1 and 0. Thus, nucleotide −1R is always directly connected to −1P, which, in its turn, always forms a WC base pair with −1Q. In addition to the direct covalent connection, nucleotides −1R and −1P can form an H-bond, which exists in most C-DTJs (Fig. 13).
FIGURE 13.
C-DTJ: interaction of nucleotide −1R with major groove of base pair [−1P;−1Q]. In all C-DTJ motifs except 2AW4-2460, nucleotide −1P forms a cis-SE-HG base pair with nucleotide −1P. In the motif 3P49-52, the molecule of glycine is coordinated by interactions with nucleotides of triple [−1R;−1P;−1Q] (shown in the figure) as well as with nucleotides 0Q, −2P, and −2Q (data not shown).
Contrary to B-DTJs, in which each of the two nucleotides 0R and −1R stabilizes one TJ directly and the other one indirectly, in C-DTJs each of the 2 nt stabilizes only one TJ: 0R stabilizes only TJ1, both directly and indirectly, and the same is true for −1R with respect to TJ2. Due to this difference, the common strategy for stabilization of both TJs is reduced in C-DTJs to the interaction between 0R and −1R. The stacking of these nucleotides leads to the formation of a 4-nt stack [+1R;0R;−1R;−2P] similar to that observed in B-DTJs. This 4-nt stack thus serves as a major stabilizing aspect for the whole C-DTJ structure; this stack coordinates the positions of nucleotides +1R and −2P, thus directly stabilizing both TJs.
Additional interactions between 0R and −1R include hydrogen bonds between the base of 1 nt and the backbone of the other nucleotide and vice versa (see examples in Supplemental Fig. S5). Although these hydrogen bonds look similar to the corresponding interactions in B-DTJs (Supplemental Fig. S4), the difference is essential. While in B-DTJs, the bases of 0R and of −1R interact with atoms O4′, O5′, or OP2 of the opposite nucleotide, all of which are acceptors of hydrogen bond, in C-DTJs, these bases interact exclusively with atom O2′. Given that O2′ can function both as a donor and as an acceptor of a hydrogen bond, it is able to associate with virtually any hydrophilic group. Therefore in C-DTJs, unlike in B-DTJs, the hydrogen bonds between 0R and −1R do not require particular nucleotide identities and thus are nonspecific.
Analysis of the available nucleotide sequences of motifs 2AVY-597, 2AW4-2460, and 1XJR-40 showed that all of them are highly conserved in evolution (Supplemental Text S1). For motif 1S03-37, information concerning its conservation is currently unavailable.
Base pair U-G in helix H3: an additional stabilizing element
Despite the differences between the structures of A-, B-, and C-DTJs, there is one element that can stabilize all of them. As one can see in Figures 4, 8, and 11, in most identified DTJs, the first base pair of helix H3 (base pair [−3P;−3Q] in A-DTJs and base pair [−2P;−2Q] in B- and C-DTJs) has the U-G identity. The particular geometry of the U-G base pair in which U and G are displaced, respectively, toward the major and minor grooves, compared with their position in a WC base pair, allows the guanosine of this base pair to optimize its stacking with the nucleotide of the P-strand of the last base pair in helix H2, thus additionally stabilizing the whole arrangement (Supplemental Fig. S6). Interestingly, out of the three DTJ types, the C-DTJ is the only one in which the motifs having the U-G identity of the first base pair of helix H3 do not constitute a majority (cf. Fig. 11 and Figs. 4, 8). However, out of the three C-DTJs having a WC base pair in this position, two motifs, 3P49-52 and 3P49-143, form riboswitches, whose function requires dissolution of the structure when the cellular concentration of glycine is not sufficiently high. We thus suggest that the presence or absence of the U-G base pair in the first position of helix H3 can regulate the stability of the whole DTJ arrangement. For some DTJs, like 2AW4-2460, the presence of the U-G base pair at this place seems to be a necessary element (in the latter motif, the U-G base pair at this place occurs in >99% of the sequences). In others, like 2AVY-597, the existence of the U-G base pair is not so critical (here, only two-thirds of the sequences contain it). Finally, for some DTJs, like the discussed above riboswitch-derived motifs 3P49-52 and 3P49-143, a U-G base pair at this place could even be harmful.
Identification of the motifs
The DTJ arrangements discussed in this article were identified by visual inspection of available RNA structures and with use of automated search. Those DTJs that exist in the bacterial ribosome were found as a result of visual inspection of the structures of the E. coli and Thermus thermophilus ribosomes. Given that together, these ribosome structures contain all three types of DTJ, the identified arrangements constituted the primary observation that unleashed the whole study. The found arrangements were classified, which in turn allowed us to suggest queries for automated search of similar structures.
Thus, for A-DTJs, the cis-WC-HG base pair between 2 nt separated in the polynucleotide chain by no more than four intervening nucleotides was found to be an important stabilizing element. Following this observation, we ran the automated search and looked for all cis-WC-HG base pairs between 3 nt separated by 0–4 nt in the nonredundant list of structures having a resolution of at least 4 Å available as of February 2012. The run of the FR3D software (Sarver et al. 2008) in the symbolic search regime gave 36 hits, only some of which could be qualified as DTJs. All these arrangements fitted to the A-DTJ pattern in which the identity of base pair [0P;0R] was G-G. All of them existed in the ribosome and repeated the arrangements that had been identified at the outset of the analysis. No A-DTJ was found outside the ribosome.
For collection of the B- and C-DTJ motifs, we used the geometric search option available in the FR3D algorithm. Thus, in arrangement 2AW4-1902 (B-DTJ) the formation of the cis-SE-HG base pair [0R;0P] accompanied by a dinucleotide stack [0R;−1R] was considered a specific characteristic of the motif and was used as a query in the automated search. Subsequently, 6 nt positions [−2P,−1P,0P,+1P,−1R,0R] of this motif were used as query motifs for geometric search of similar arrangements (the value of discrepancy was chosen to be 0.45). The obtained hits were analyzed visually, and most of them could be classified as DTJs. One of false-positives represented an interesting case that deserved a special attention. Supplemental Figure S7 shows arrangement 3UXR-B68, in which the positions and identities of individual nucleotides would have fitted it to the B-DTJ group. The only aspect that did not allow us to qualify this arrangement as a B-DTJ motif consisted in a different secondary structure, which did not fit to the general pattern shown in Figure 3. The completeness of the set of the found B-DTJ motifs was verified through cross-search of the arrangement of the same 6 nt [−2P,−1P,0P,+1P,−1R,0R] observed in each identified motif. The same procedure was used for the search of C-DTJ motifs. In this case, arrangement 2AVY-597, occurring in 16S rRNA, was taken as a prototype.
DISCUSSION
In this article, we consider a new family of RNA arrangements in which two covalently connected quasi-coaxial double helices are over-twisted with respect to each other. We call such arrangements TJs. In the available RNA structures, there is a variety of different kinds of TJs, some of which, like the C-loop motif (Lescoute et al. 2005), have already been discussed, while others await classification and analysis.
Here we focus on more complex arrangements consisting of two closely positioned TJs, which we call DTJs. In the known RNA structures we found 14 DTJs, which fell in three distinct groups named here as A-, B-, and C-DTJs. On the level of secondary structure, all DTJs can be represented as a double helix containing two bulges belonging to the opposite strands and separated from each other by either 2 bp (B- and C-DTJ) or 3 bp (A-DTJ). Although in a WC double helix, bulges are generally considered as a source of local instability, in the DTJs they are prerequisites for formation of both over-twists. In addition, the specific interactions formed by the bulged nucleotides with the double helical regions are essential for stabilization of the particular conformations of the motifs. We distinguish between the direct and indirect stabilization, the first of which is mostly made by nitrogen bases, while the second is mediated by the backbone of RNA chain. Interestingly, in A- and B-DTJs, the same bulged nucleotide can provide a direct stabilization of one over-twist and an indirect stabilization of the other over-twist.
On a more detailed level, we identified two major strategies of DTJ stabilization. The first strategy, exemplified by the A-DTJs, consists in the direct stabilization of TJ1 and indirect stabilization of TJ2. The direct stabilization is achieved through specific interactions of the bulged nucleotides 0R and +1Q with base pairs [0P;0Q] and [+1R;+1P], while the indirect stabilization is mediated by the covalent attachment of base pair [−3P;−3Q] to nucleotide 0R. Because the specific interaction between nucleotide 0R and base pair [0P;0Q] requires that helix H2 contain strictly 3 bp, this strategy is observed only in the A-DTJs.
The second strategy of stabilization, which is observed in B- and C-DTJs, is based on formation of the continuous 4-nt stack [+1R;0R;−1R;−2P]. The existence of such a stack will tune the positions of nucleotides +1R and −2P, thus simultaneously solidifying the conformations of both over-twists. In this situation, additional interactions of bulged nucleotides with the major and minor grooves of helices H1 and H2, which are present in all A-DTJs, become relatively less important and could be missing in individual B- and C-DTJs. Also, given that a nucleotide stack can be reasonably rigid only if it is sufficiently short, the length of helix H2 containing only 2 bp seems to be optimal. In this case, however, the specific interaction between 0R and [0P;0Q] observed in A-DTJs becomes sterically impossible. This aspect explains why the strategies of stabilization for DTJs containing 2 and 3 bp are so different.
Different levels of cooperation between TJs in DTJ motifs
Because in the analyzed DTJs, the strategies of the TJ1 and TJ2 stabilization cooperate with each other, it is possible to tell about a common strategy of stabilization for both TJs. Nevertheless, in different motifs the level of cooperation between the two strategies is different. In particular, compared to the B- and C-DTJs, the interdependence between the TJs in the A-DTJ is much less pronounced. Indeed, in A-DTJ, each of the two TJs can exist regardless of the existence of the other TJ, which was demonstrated by the specific evolutionary conservation patterns of the motifs 2AVY-1142 and 2AVY-68 (Fig. 4E,F; Supplemental Fig. S2; Supplemental Text S1). In the B- and C-DTJs, however, the situation is essentially different. Here, the particular mode of stabilization of a DTJ, which is based on formation of the 2-nt stack [+1R;0R;−1R;−2P], makes TJ1 and TJ2 inseparable from each other. Correspondingly, in those B- and C-DTJs for which the evolutionary conservation could be analyzed, both TJs were always present.
Role of DTJs in RNA structure
Analysis of the interactions formed by DTJs with their surroundings in the known RNA structures has allowed us to make general suggestions concerning possible roles of these motifs in RNA architecture and function. First, the presence of two consecutive TJs in a double helix provides a total over-twist of ∼70°. Such rearrangement could allow helices H1 and H3 to fit to their optimal structural contexts without breakage of the long stacked domain made of H1, H2, and H3.
A large cumulative over-twist is not, however, the only consequence of the introduction of two TJs. Because in the regular A-RNA conformation, base pairs are inclined to the axis of the helix by ∼20°, the axis of the over-twist rotation will not be colinear to the axis of the helix. As a result, introduction of an over-twist provides a bend in the double helix (Supplemental Fig. S8). In DTJs, the presence of two closely located over-twists makes the situation even more complex. The global effect of the two over-twists on the helix geometry will strongly depend on the distance between the two over-twists or, more specifically, on the length of helix H2. In particular, in A-DTJs, where helix H2 contains 3 bp, helix H1 inclines to helix H3 for ∼15° (Supplemental Fig. S8A). In B- and C-DTJs, where helix H2 has 2 bp, the angle between H1 and H3 becomes twice as big, i.e., ∼30° (Supplemental Fig. S8B,C). The possibility for introduction of different bends into the helical domain composed of H1, H2, and H3 thus provides additional opportunities for its fitting to the given structural context.
Another potential role of the identified motifs is based on the fact that formation of an over-twist between 2 bp opens their surfaces for harboring other nucleotides, like 0R and +1Q. The exposure of such surfaces can initiate the formation of longer stacks of nucleotides coming from different parts of the polynucleotide chain. In particular, in A-DTJs 2AVY-68, 2AVY-97, and 2AVY-1142, as well as in B-DTJs 2AW4-1902 and 1U9S-161, nucleotide +1Q initiates a long stack of nucleotides, which also interacts with the minor groove of H1 (Figs. 4B,C,E, 8B,E). In situations of this kind, the over-twist would play a role of nucleus, thus promoting folding of a domain and/or formation of interdomain interactions.
One more potential role of a DTJ relates to the presence of a bulge nucleotide at the end of loop L2. While the existence of a bulge nucleotide in loop L1 of the motifs 1U9S-228 and 2AVY-1057 is required for formation of the particular tertiary interaction with helix H2 (at level −1 and −2, respectively), a bulged nucleotide in loop L2, when it exists, always interacts with an RNA element outside the DTJ. In particular, in two ribosome-based motifs, 2AW4-1902 and 2AVY-68, this nucleotide (nucleotide A1900 and A65, respectively) mediates interaction of the DTJ with another part of the ribosomal RNA. Even more interesting could be the cases when the element interacting with the bulged nucleotide of L2 belongs to a different molecule. Thus, in motif 2AVY-1057, found in the small ribosomal subunit, the second nucleotide of L2 (C1054) coordinates the codon–anticodon mini-duplex in the ribosomal A-site (Jenner et al. 2010). An almost identical situation happens in the motifs 1U9S-228 and 1NBS-232, where the second nucleotide of L2 (nucleotide A226 in 1U9S-228 and A230 in 1NBS-232; structures 1U9S and 1NBS represent RNases P of classes A and B, respectively) forms an A-minor interaction with the T-stem of the tRNA. The latter interaction thus contributes to the correct recognition and positioning of the pre-tRNA substrate in the active site of the RNase P (Kazantsev et al. 2005; Reiter et al. 2010).
Finally, each of the two C-DTJs 3P49-52 and 3P49-143 forms a specific pocket for harboring a molecule of glycine, which is essential for the function of the glycine riboswitch. Both pockets are formed by nucleotides −1R, −1P, and −1Q of the −1 layer (Fig. 13), as well as by nucleotides 0Q and [−2P;−2Q] of levels 0 and −2 (data not shown). The binding of glycine to this pocket is essential for formation of the whole motif. To make the formation of a C-DTJ dependable on association with glycine, the structure of the motif should be relatively unstable. As mentioned above, such instability is partly achieved through avoiding of the U-G identity for base pair [−2P;−2Q] and also through the absence of stable interactions at the zero-level (Fig. 12).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by operating grant from Canadian Institutes of Health Research to S.V.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030940.111.
REFERENCES
- Butler EB, Xiong Y, Wang J, Strobel SA 2011. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol 18: 293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M 2010. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 17: 555–560 [DOI] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR 2005. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci 102: 13392–13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Yang X, Pan T, Mondragon A 2003. Crystal structure of the specificity domain of ribonuclease P. Nature 421: 760–764 [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Xiao Y, Pan T, Mondragon A 2004. Basis for structural diversity in homologous RNAs. Science 306: 104–107 [DOI] [PubMed] [Google Scholar]
- Lee JC, Cannone JJ, Gutell RR 2003. The lonepair triloop: A new motif in RNA structure. J Mol Biol 325: 65–83 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Leontis NB, Massire C, Westhof E 2005. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res 33: 2395–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianos HJ, Wang J, Moore PB 2004. The structure of a ribosomal protein S8/spc operon mRNA complex. RNA 10: 954–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A 2010. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468: 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MP, Igel H, Baertsch R, Haussler D, Ares M Jr, Scott WG 2005. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol 3: e5 doi: 10.1371/journal.pbio.0030005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB 2008. FR3D: Finding local and composite recurrent structural motifs in RNA 3D structures. J Math Biol 56: 215–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Steinberg SV, Boutorine YI 2007a. G-ribo motif favors the formation of pseudoknots in ribosomal RNA. RNA 13: 1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg SV, Boutorine YI 2007b. G-ribo: A new structural motif in ribosomal RNA. RNA 13: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, Perriere G, Van De Peer Y 2004. The European ribosomal RNA database. Nucleic Acids Res 32: D101–D103 [DOI] [PMC free article] [PubMed] [Google Scholar]